Individual variations in ‘brain age’ relate to early-life factors more than to longitudinal brain change
Figures
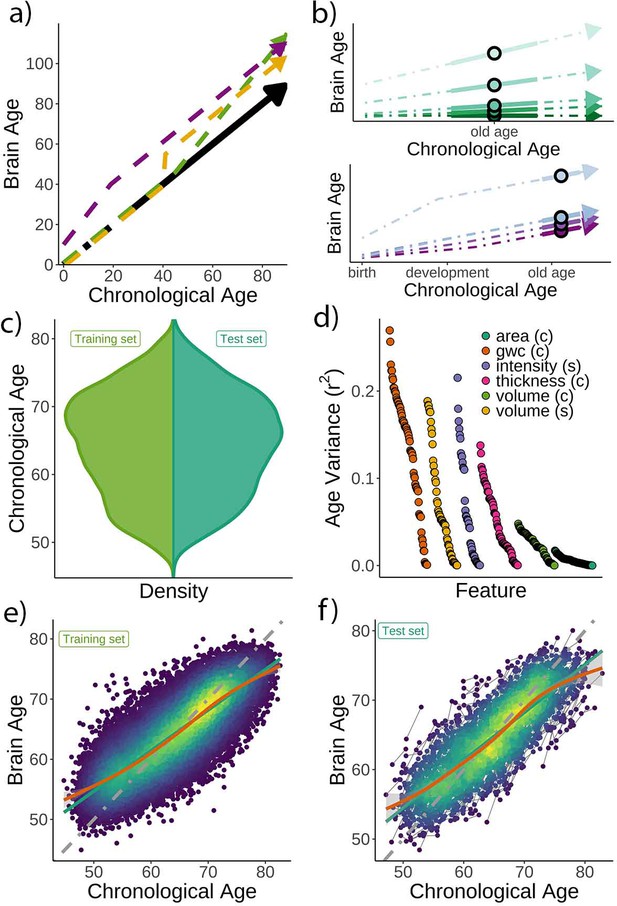
Theoretical expectations and study characteristics.
(a) Three hypothetical trajectories leading to higher brain age delta. Higher brain age delta can be explained by a steeper rate of neurobiological aging (green), distinct events that led to the accumulation of brain damage in the past (yellow), or early-life genetic and developmental factors (purple). The black arrow represents normative values of brain age through the lifespan. (b) Brain aging (green) vs. early-life (blue-purple) accounts of brain age in older age. For the brain aging notion, cross-sectional brain age (points) relates to the slope of brain age as assessed by two or more observations across time (continuous line), reflecting ongoing differences in the rate of aging (dashed line, green scale). For the early-life notion, cross-sectional brain age (points) relates to early environmental, genetic, and/or developmental differences such as birth weight (blue-purple scale). (c) Relative age distribution for the UK Biobank test and training datasets. (d) Age variance explained (r2) for each MRI feature in the training dataset. Features are grouped by modality and ordered by the variance explained. (e) Brain age model as estimated on the training (n = 38,682), and (f) test datasets (participants = 1372; two observations each). In (e) and (f), lines represent the identity (gray; i.e., f(x) = x or diagonal fit), the linear (green), and the generalized additive models (GAM; orange) fits of chronological age to brain age. Confidence intervals (CIs) around the GAM fit represent 99.9% CIs for the mean. In (d), gwc = gray-white matter contrast, (c) = cortical, and (s) = subcortical.
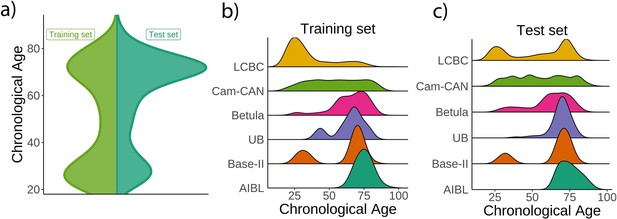
Age distribution for the Lifebrain replication dataset.
(a) Relative age distribution for the Lifebrain training and test datasets. Relative age distribution for the different cohorts of the Lifebrain (b) training and (c) test datasets.
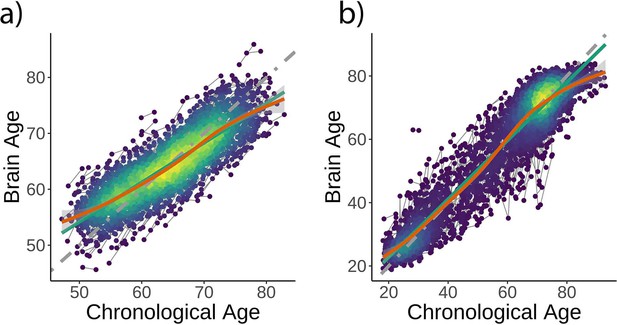
Brain age model predictions.
Brain age model prediction (i.e., on test data) as estimated (a) using LASSO in the UK Biobank dataset and (b) extreme boosting gradient in the Lifebrain sample. Gray, green, and orange lines represent the identity, the linear, and the generalized additive models (GAM) functions fitting brain on chronological age.
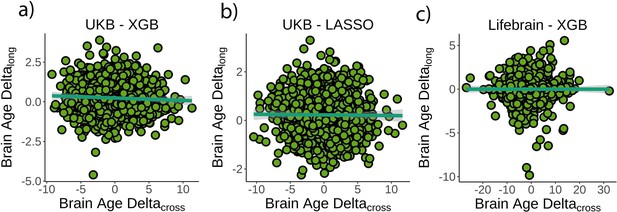
Relationship between cross-sectional and longitudinal brain age delta.
(a) Main analysis using the UK Biobank dataset and boosting gradient (n = 1372, p=0.04, r2 = 0.002). (b) Replication analyses using a different training algorithm (LASSO; n = 1372, p=0.65, r2 = 0.001) and (c) an independent dataset (Lifebrain; n = 1500, p=0.53, r2 = 0.001). XGB = boosting gradient as implemented in XGBoost. Confidence intervals (CIs) represent 99.9% CI for the fit. Longitudinal brain age delta (brain age deltalong) refers to the rate of change in delta between baseline and follow-up MRI measurements. Cross-sectional brain age delta (brain age deltacross) refers to centercept brain age delta; that is, at mean age.
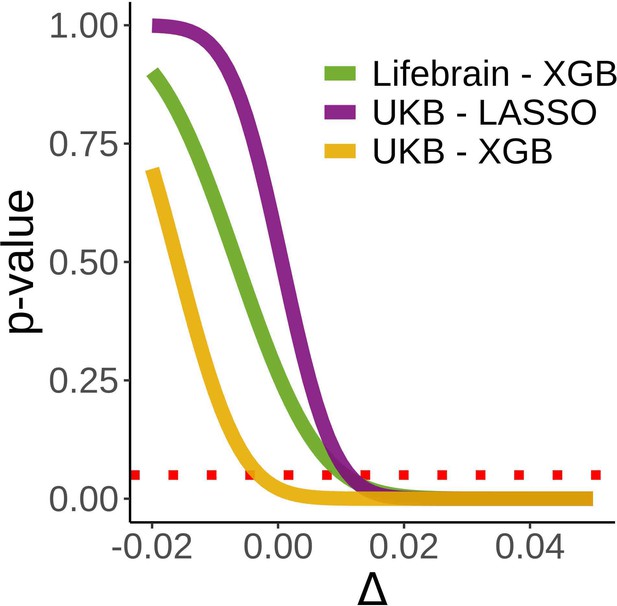
Equivalence tests.
Inferiority tests for the three main models used to assess the relationship between cross-sectional and brain age deltalong. Inferiority tests test whether a null hypothesis of an effect as large as Δ can be rejected. In the x-axis, Δ reflects the null hypothesis as βetas (years/delta). A null hypothesis of an effect at least as large as 0.11 years/delta can be rejected (p<0.05) in all three tests. Δ has been evaluated at [–0.02, 0.05, 0.001]. The dashed red line indicates a p=0.05 criterion for the null hypothesis rejection.
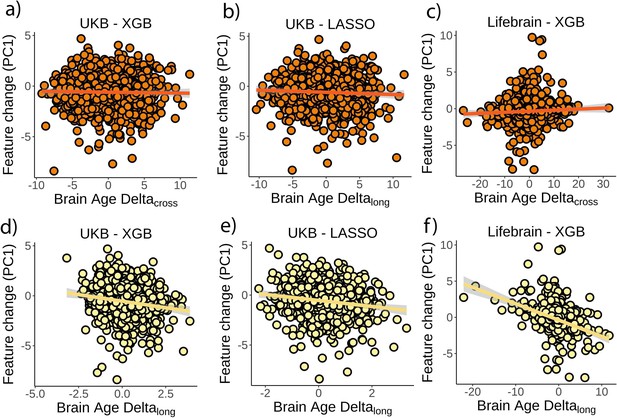
Relationship between brain age delta and composite measures of change.
Relationship between a composite measure of change as captured by the first principal component on feature change and cross-sectional brain age delta in (a) the UK Biobank and boosting gradient, (b) the UK Biobank and the LASSO algorithm, and (c) the Lifebrain dataset. Relationship between the composite measure of change and (longitudinal) brain age deltalong in (d) the UK Biobank and boosting gradient, (e) the UK Biobank and the LASSO algorithm, and (f) the Lifebrain dataset. Negative values in the principal component reflect brain decline (e.g., steeper cortical thinning, higher ventricle volume, etc.). n = 1369 and 1497 for the UK Biobank and the Lifebrain datasets.
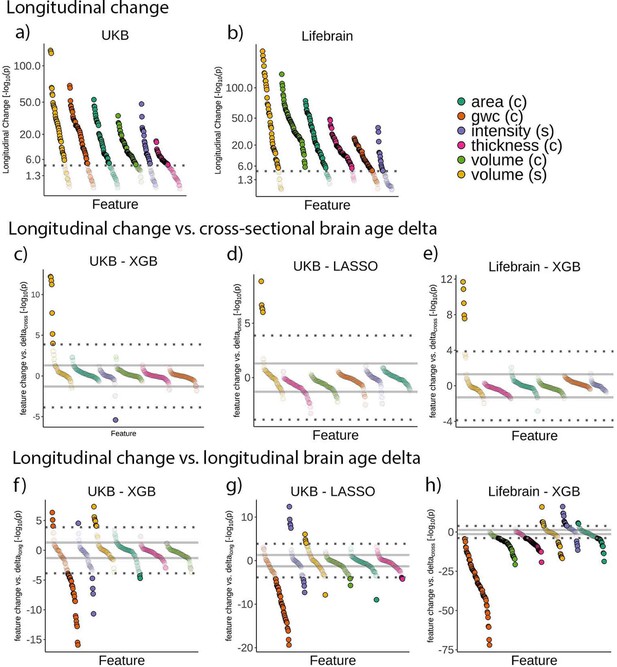
Relationship between brain age delta and change in raw features.
Feature change over time in the (a) UK Biobank and (b) Lifebrain datasets. Signed relationship between cross-sectional brain age delta and longitudinal change in the raw features in (c) the UK Biobank using a boosting gradient algorithm, (d) the UK Biobank using a LASSO algorithm, and (e) the Lifebrain dataset using the boosting gradient algorithm. Signed relationship between change in brain age delta (brain age deltalong) and longitudinal change in the raw features in (f) the UK Biobank using a boosting gradient algorithm, (g) the UK Biobank using a LASSO algorithm, and (h) the Lifebrain dataset using the boosting gradient algorithm. Dashed lines represent a Bonferroni-corrected significance threshold (|n| = 365 and 372 features for UK Biobank and Lifebrain datasets, respectively). The solid line represents an uncorrected p=0.05 significance threshold. n = 1372 and 1500 for the UK Biobank and the Lifebrain datasets.
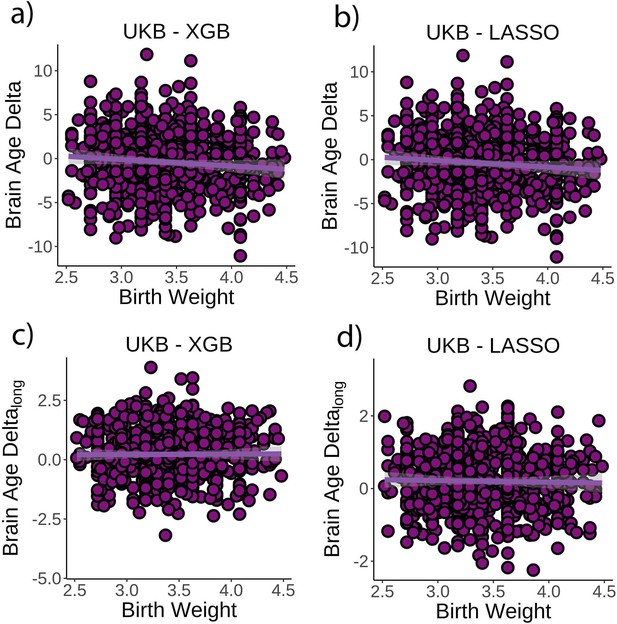
Relationship between cross-sectional brain age delta and birth weight.
(a) Main effect of birth weight on brain age delta using the UK Biobank dataset and boosting gradient (n = 770, p=0.02, r2 = 0.009). (b) This effect was replicated using a different training algorithm (LASSO) (n = 770, p=0.005, r2 = 0.009). Relationship between longitudinal change in brain age delta and birth weight was not significant either (c) in the main test or (d) in the LASSO replication analysis (p>0.5). Note that we used delta at time point 1 to illustrate the main effect of birth weight at time 0 and brain age deltalong to represent the birth weight × time interaction of the linear mixed models. Confidence intervals (CIs) represent 99.9% CI for the fit. XGB = boosting gradient as implemented in XGBoost.
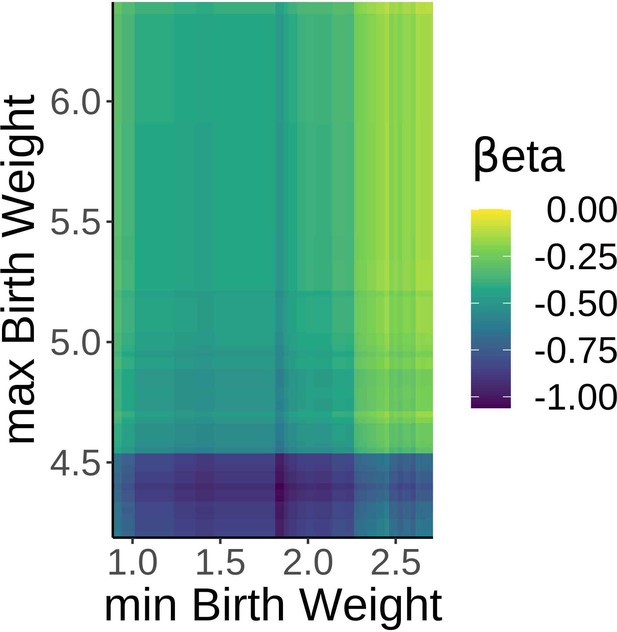
Robust effects of birth weight on brain age delta.
βeta estimates showing the relationship between brain age delta and birth weight with variable minimum and maximum birth weight exclusion thresholds. Note negative βetas irrespective of the minimum and maximum self-reported birthweight thresholds.
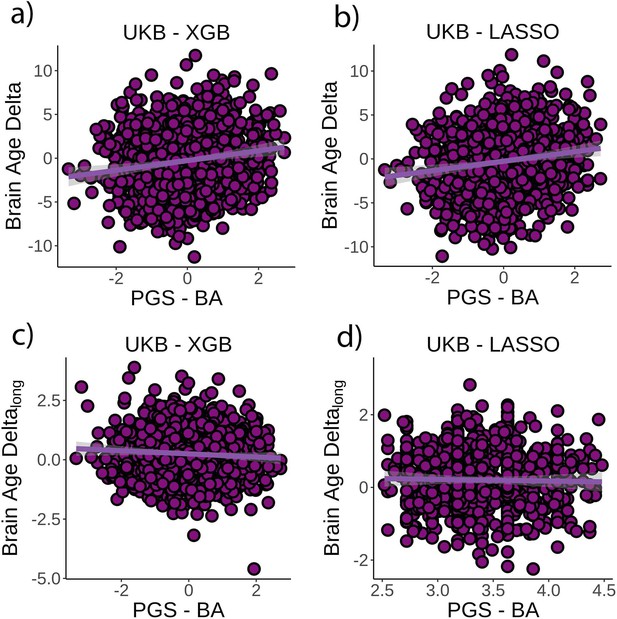
Relationship between cross-sectional brain age delta and polygenic scores of brain age delta (PGS-BA).
(a) Main effect of PGS-BA on brain age delta using the UK Biobank dataset and boosting gradient (n = 1339, p<0.001, r2 = 0.02). (b) This effect was replicated using a different training algorithm (LASSO) (n = 1339, p<0.001, r2 = 0.02). (c) We found a negative association between longitudinal change in brain age delta and PGS-BA (=0.02; higher genetic liability to brain age related to negative change in brain age delta), which was not found (d) in the LASSO replication analysis (p=1.0). Note that we used delta at time point 1 to illustrate the main effect of PGS-BA at time 0 and brain age deltalong to represent the PGS-BA × time interaction of the linear mixed models. Confidence intervals (CIs) represent 99.9% CI for the fit. XGB = boosting gradient as implemented in XGBoost.
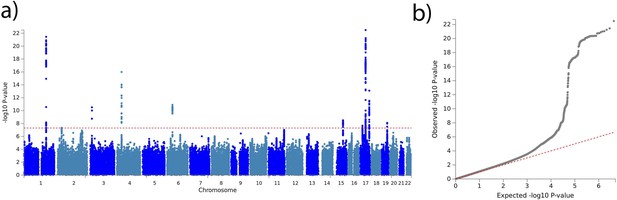
Brain age delta genome-wide association study (GWAS).
(a) Manhattan plot of the GWAS results for the test set on brain age delta (38,163 individuals). The horizontal line represents the threshold for genome-wide significance. (b) Quantile-quantile (QQ) plot illustrating the deviation of the observed p-values from the null hypothesis.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Software, algorithm | R Project for Statistical Computing | https://www.r-project.org/ | RRID:SCR_001905 | Version 3.6.3 |
| Software, algorithm | FreeSurfer | https://surfer.nmr.mgh.harvard.edu/ | RRID:SCR_001847 | Version 6.0 |
Additional files
-
Supplementary file 1
List of cortical brain features.
List of cortical features included in the brain age model and age variance explained in the UK Biobank and the Lifebrain training datasets. Vol = volume; GWC = gray-white matter contrast; Cth = cortical thickness.
- https://cdn.elifesciences.org/articles/69995/elife-69995-supp1-v1.docx
-
Supplementary file 2
List of subcortical brain features.
List of subcortical features included in the brain age model and age variance explained in the UK Biobank and the Lifebrain training datasets. Vol = volume; Int = intensity; hemi = hemisphere.
- https://cdn.elifesciences.org/articles/69995/elife-69995-supp2-v1.docx
-
Supplementary file 3
Sociodemographics.
Main sample descriptives for the training and test datasets. Obs = mean number of observations per participant (SD). Follow-up = mean time (years) between the first and the last MRI observation (SD). For the test datasets, age and age range refer to age at baseline. *AIBL does not belong to the Lifebrain consortium but was included to enrich the replication sample.
- https://cdn.elifesciences.org/articles/69995/elife-69995-supp3-v1.docx
-
Supplementary file 4
Relationship between brain age delta and change in brain features.
Long. change = longitudinal change in the raw neuroimaging features (mean change [log10(p)]). PC1 load = feature loadings on the first component of longitudinal change. Deltacross = relationship between cross-sectional brain age delta and feature change (r2 [log10(p)]). Deltalong = relationship between longitudinal brain age delta and feature change (r2 [log10(p)]). GWC = gray-white matter contrast. Cth = cortical thickness. Bil = bilateral. Subc = subcortical. n = 1372 and 1500 for the UK Biobank and the Lifebrain datasets. |N| = 365 and 372 features in the UK Biobank and the Lifebrain datasets. XGB = boosting gradient as implemented in XGBoost.
- https://cdn.elifesciences.org/articles/69995/elife-69995-supp4-v1.docx
-
Supplementary file 5
Contact information.
Contact information and ethical comittees for the different cohorts.
- https://cdn.elifesciences.org/articles/69995/elife-69995-supp5-v1.docx
-
Supplementary file 6
Data acquisition parameters.
Data acquisition parameters for the T1w sequences. *UK Biobank employed three scanners of the same model and with equivalent parameters (Cheadle, Reading, and Newcastle centers). **AIBL does not belong to the Lifebrain consortium but was included in the Lifebrain replication dataset.
- https://cdn.elifesciences.org/articles/69995/elife-69995-supp6-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69995/elife-69995-transrepform1-v1.pdf
-
Source code 1
Analysis Code.
- https://cdn.elifesciences.org/articles/69995/elife-69995-supp7-v1.zip





