Structural characterization of NrnC identifies unifying features of dinucleotidases
Figures
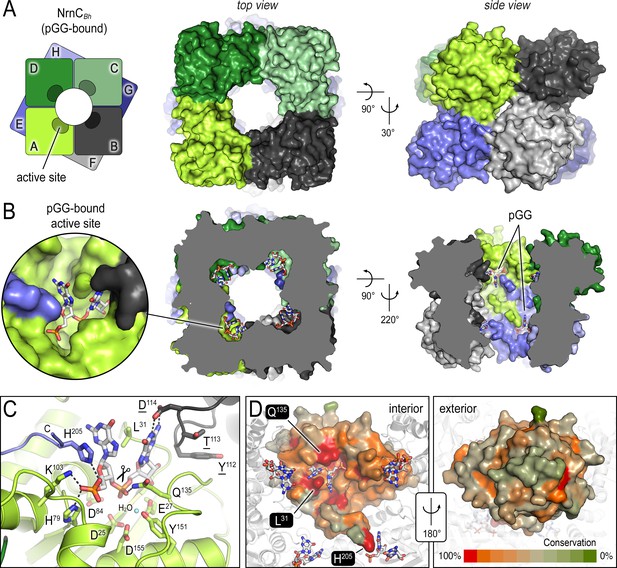
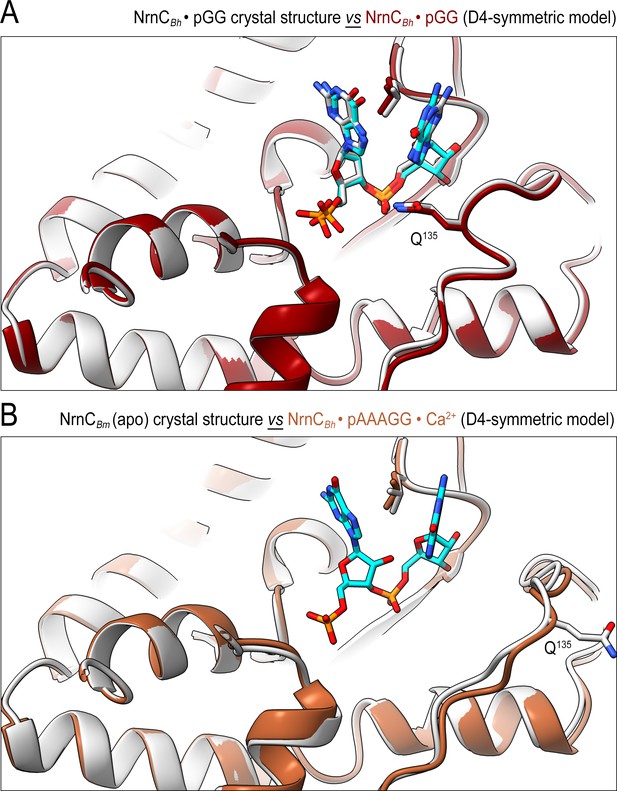
The crystal structures of B. henselae nano-RNase C (NrnC) bound to pGG reveals motifs defining substrate specificity.
(A) The octameric assembly. NrnCBh is shown as surface representation in two views. Each monomer is shown in a distinct color. The cartoon illustrates the stacking of the two tetrameric NrnC rings that form the octamer with a central, round opening. (B) Active-site position. Each monomer contributes one active site, here bound to the substrate pGG, facing toward NrnC’s central pore. Each active site includes a C-terminal tail of a subunit from an adjacent ring. (C) Substrate coordination. The catalytic DEDDy motif and residues coordinating each moiety of pGG contacts are shown as sticks, with carbon residues colored according to monomer identity. Residue Y151 coordinates water molecule near the scissile bond. (D) Conservation mapping on a surface representation of a NrnC monomer. Conservation scores were calculated based on a multisequence alignments (MUSCLE; Edgar, 2004) of NrnC homologs identified using a sequence search on the EggNOG resource, version 5.0.0 (Huerta-Cepas et al., 2019) and the sequence of NrnCBh as the input. Outliers were identified based on sequence length and non-consensus insertions, resulting in a final collection of 560 sequences of putative NrnC orthologs. The two views, separated by a 180° rotation, show the cavity-facing (interior, left) and outer-facing (exterior, right) surface regions.
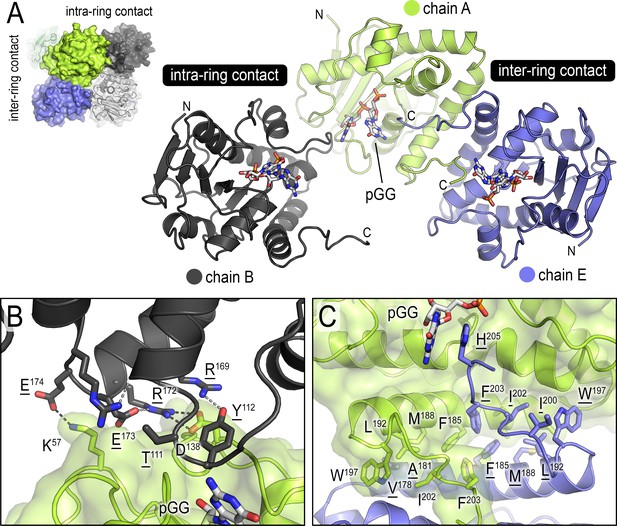
Inter- and intra-ring contacts in the NrnCBh octamer.
(A) Overview of three pGG-bound NrnCBh monomers. The monomers are shown in cartoon representation The chain colored in dark gray forms an intra-ring contact with the central, green-colored chain, whereas the chain colored in blue forms a representative inter-ring contact within the octameric nano-RNase C (NrnC). (B) Detailed intra-ring interface. (C) Detailed inter-ring interface. Residues contributing direct interactions between monomers are shown as sticks. Representative hydrogen bonds are shown as dashed lines.
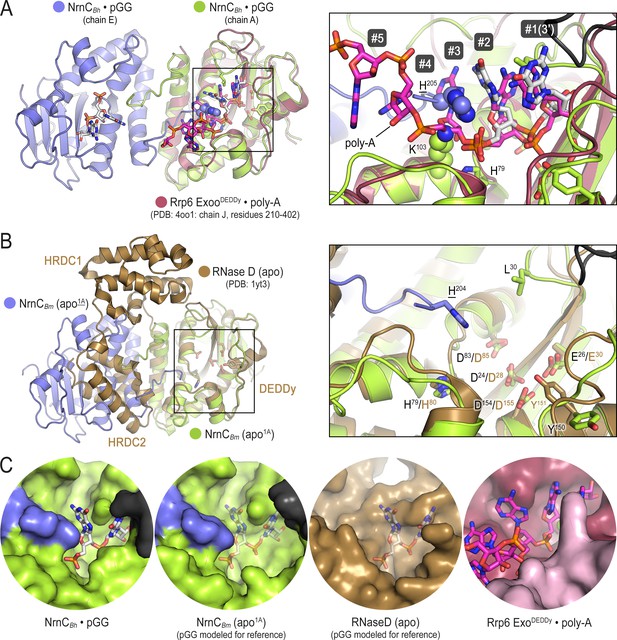
Comparison of nano-RNase C (NrnC) to structurally related proteins reveals the constricted nature of NrnC’s active site.
(A) A pGG-bound NrnCBh inter-ring dimer (slate and green chains, with pGG carbon atoms shown in white) was superimposed on a structure of the exosome’s Rrp6 exonuclease (purple chain) bound to poly-A (pink carbon atoms; PDB:4oo1; Wasmuth et al., 2014). The inset shows a detailed view of the superimposed active sites. Numbering refers to the residues in the substrate. Phosphate cap residues of NrnC that block the path of poly-A substrate are shown as spheres. The black protein chain in top-right corner stems from an adjacent intra-ring monomer. (B) A substrate-free NrnCBm inter-ring dimer (slate and green chains) was superimposed on a structure of apo-RNase D (Zuo et al., 2005), highlighting conservation of the catalytic DEDDy motif and differences in regions around NrnC’s L-wedge and phosphate cap (inset). (C) Surface views of the active sites of NrnC, RNaseD, and Rrp6 accentuate the constraint of the NrnC active site. Translucent pGG represents modeled substrate as opposed to co-crystallized substrate.
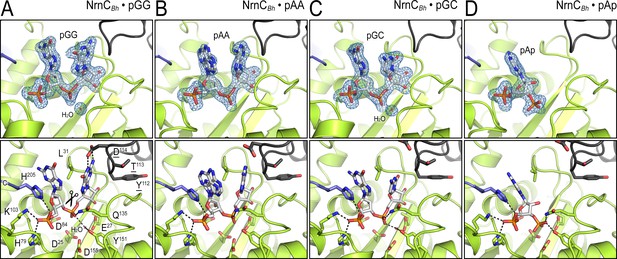
Structural comparison of NrnCBh bound to various ribonucleotides.
(A) Diribonucleotide pGG-bound NrnCBh, identical to the structure shown in Figure 1. (B) Diribonucleotide pAA-bound NrnCBh. (C) Diribonucleotide pGC-bound NrnCBh. (D) Adenosine-3′,5′-bisphosphate (pAp)-bound NrnCBh. The top row of images shows nano-RNase C (NrnC) as a cartoon representation with the nucleotide substrate represented as sticks with carbon atoms colored white. Polder omit maps for each substrate are shown as blue mesh. The bottom row shows detailed views of the active site residues contacting each ribonucleotide.
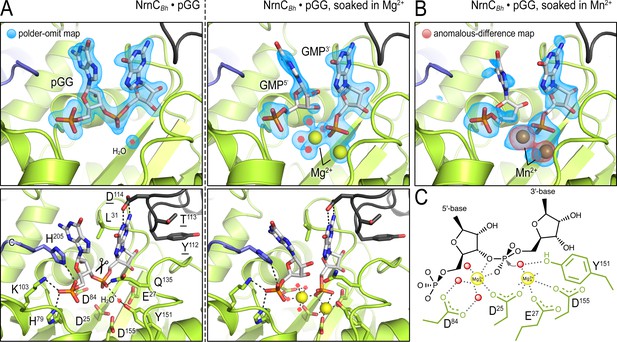
In crystallo catalysis indicates a two-metal mechanism of NrnCBh activity.
(A) Active sites of NrnCBh•pGG, before and after soaking crystals in a solution containing Mg2+ prior to data collection. (B) Active site of NrnCBh•pGG, after soaking crystals in a solution containing Mn2+ prior to data collection. Top panels show polder omit maps highlighting nucleotide and metal density. The red density in (B) represents an anomalous-difference map calculated from data collected at the Mn2+ absorption edge. The bottom panels show specific active site contacts between protein, nucleotide, ions, and water molecules. (C) Schematic overview of two-metal coordination at the active site of nano-RNase C (NrnC).
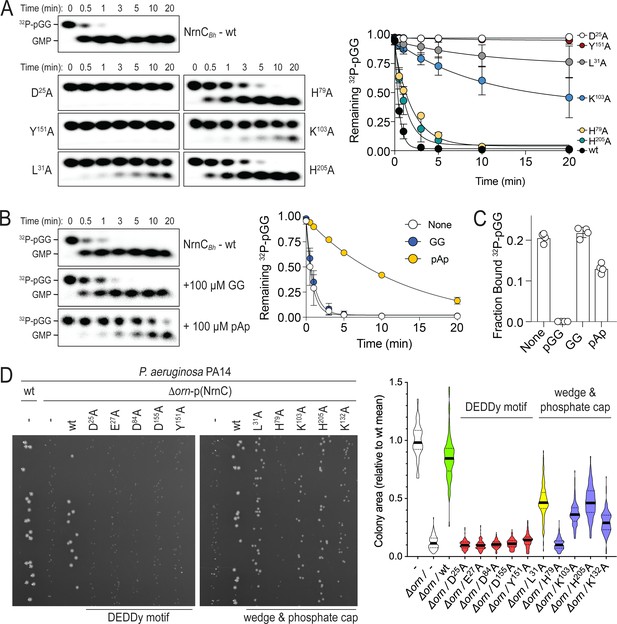
Phosphate cap and L-wedge contribute to nano-RNase C’s (NrnC’s) diribonucleotidase activity.
(A) In vitro enzyme activity. Degradation of 32P-pGG (1 μM total) by purified wild-type NrnCBh or variants with alanine substitutions (5 nM) at the indicated sites was assessed. Samples were stopped at the indicated times (min) and analyzed by denaturing 20% PAGE. Representative gels are shown (left). The graph (right) shows the means and SD of three independent experiments. (B) Effect of a dinucleotide lacking the 5′ phosphate (GG) and pAp on NrnC catalysis. pGG processing was assessed as in (A) but in the presence or absence of 100-fold excess (over 32P-pGG) GpG or pAp. Representative gels (left) and quantification from three independent experiments (right) are shown. Means and SD are plotted. (C) Competition binding studies. Fraction bound of 32P-pGpG to 200 nM purified NrnCBh in the presence of no competitor, 100 µM pGG, 100 µM GpG, or 100 µM pAp is plotted as individual data, means, and SD of four independent experiments. (D) Complementation of the small-colony phenotype of P. aeruginosa ∆orn by wild-type and mutant NrnCBh. Bacterial cultures were diluted and dripped on LB agar plates. After overnight incubation, representative images of the plates were taken (left). Experiments were performed in triplicate. Quantification of respective colony sizes is shown as violin plots (right).
-
Figure 2—source data 1
Source data for Figure 2A.
Original, unedited images, labeled overview, and quantification of wild-type and mutant NrnC activity using pGG as the substrate from three replicates.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig2-data1-v2.zip
-
Figure 2—source data 2
Source data for Figure 2B.
Original, unedited images, labeled overview, and quantification of nano-RNase C (NrnC) activity using pGG as the substrate in the presence or absence of GG or pAp from three replicates.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig2-data2-v2.zip
-
Figure 2—source data 3
Source data for Figure 2C.
Quantification of competition binding experiments using nano-RNase C (NrnC) and radiolabeled pGG in the presence or absence of unlabeled GG or pAp.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig2-data3-v2.zip
-
Figure 2—source data 4
Source data for Figure 2D.
Original, unedited images and labeled composite image of P. aeruginosa colony growth.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig2-data4-v2.zip
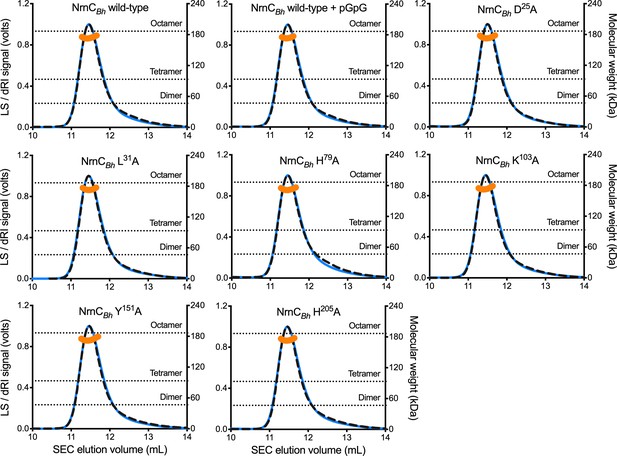
SEC-MALS of NrnCBh wild-type and mutant variants.
Molecular weight determination indicates that pGG binding does not impact oligomerization, and that purified NrnCBh point mutants remain octameric in solution. Absolute molecular weights of nano-RNase C (NrnC) are shown as orange data points across elution peaks plotted on the right axis. Theoretical oligomerization states are shown as dashed horizontal lines. 90°-light scattering: blue solid lines; refractive index signal: black dashed lines; plotted on left axis.
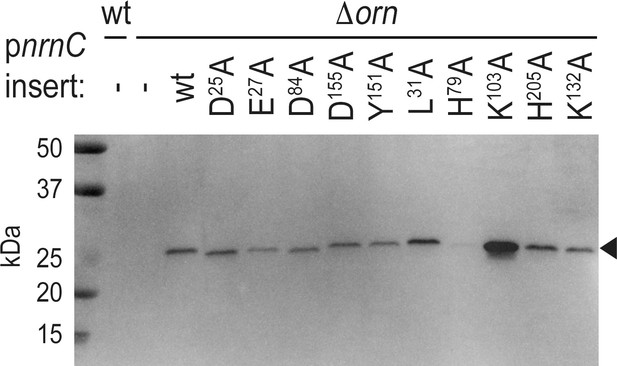
Expression of NrnCBh wild-type and mutant variants in P. aeruginosa ∆orn.
Cell lysates were analyzed by western blotting, detecting the C-terminal HA-tag in recombinantly expressed NrnCBh.
-
Figure 2—figure supplement 2—source data 1
Original and labeled, unedited western blot image.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig2-figsupp2-data1-v2.zip
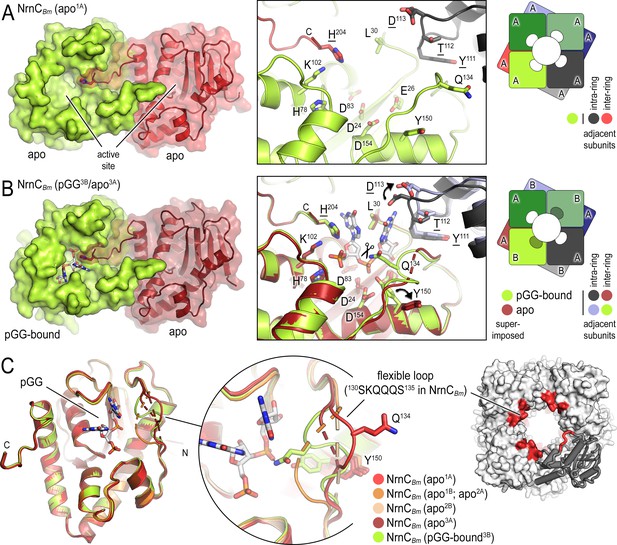
B. melitensis nano-RNase C (NrnC) crystal structures reveal a flexible loop that constraints the enzyme’s active site.
(A) Crystal structure of apo-NrnCBm. A crystallographic dimer as part of the octameric assembly is shown as surface presentation (left) and close-up of the active site (middle). The diagram (right) depicts the octamer and the spatial relationship of the monomers shown. (B) Crystal structure of NrnCBm with alternating substrate-bound and empty active sites. The close-up (middle) shows a superposition of the two monomers in the asymmetric unit, depicting their conformational difference and adjacent monomers, with intra- and inter-ring neighbors colored as shown in the diagram (right). (C) Superposition of four apo-NrnCBm conformations based on three independent crystal forms (comprising chains apo1A/apo1B for form 1; apo2A/apo2B for form 2, and apo3A/pGG-bound3B for form 3), compared to the pGG-bound conformation of the same protein shown in (B). The position of the flexible loop (red) in the NrnC octamer is shown (right panel).
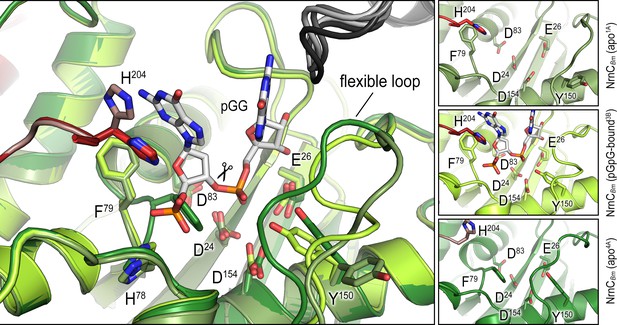
Overlay of an alternative crystallographic apo-NrnCBm state with the apo- and pGG-bound states observed in the crystal structure shown in Figure 3B.
The large panel shows three structures superimposed. The smaller panels on the right show each active site isolated with residues of interest labeled and shown as sticks.
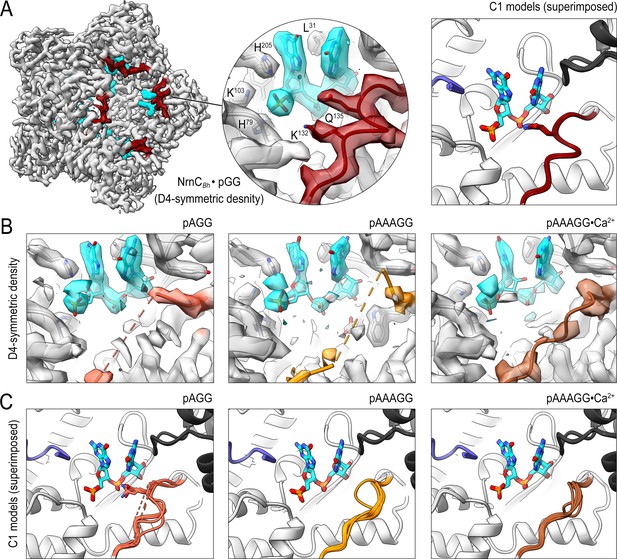
Cryo-electron microscopy (cryo-EM) structures of NrnCBh with 2-, 3-, 5-mer RNA substrates show substrate length-dependent active site conformations.
(A) Electron density map of a NrnCBh octamer in complex with pGG. D4 symmetry was applied during final map refinement. pGG molecule and density are colored cyan. The SKQQQS-containing loops (residues 130–137) are colored maroon. Superposition of all eight active sites from a reconstruction with C1 symmetry (right panel) shows consensus order in the loop when bound to pGG. (B) Active site images shown for NrnCBh incubated with 3-mer and 5-mer (with or without Ca2+) RNA substrates. Regions corresponding to those shown in (A) are shown in color, with light red (left panel), orange (middle panel), and brown (right panel) depicting the loop/loop density from structures determined with added pAGG, pAAAGG, and pAAAGG•Ca2+, respectively. D4-symmetric maps are shown. (C) Superposition of all eight active sites from octamer reconstructions based on respective C1-symmetric maps for each RNA substrate. Color scheme is as described in (B).
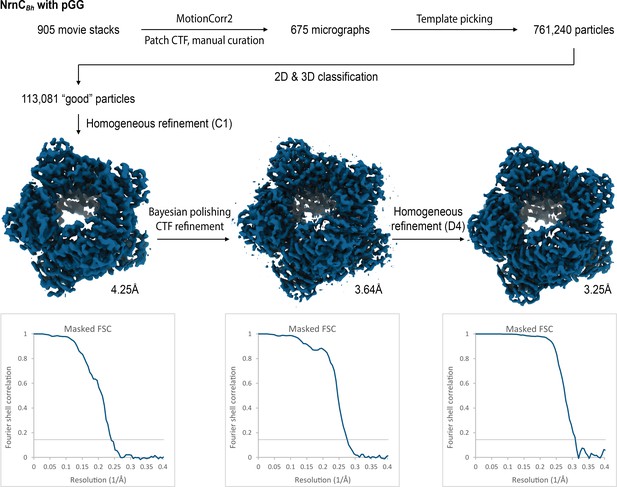
Cryo-electron microscopy (cryo-EM) workflow and resolution for NrnCBh•pGG.
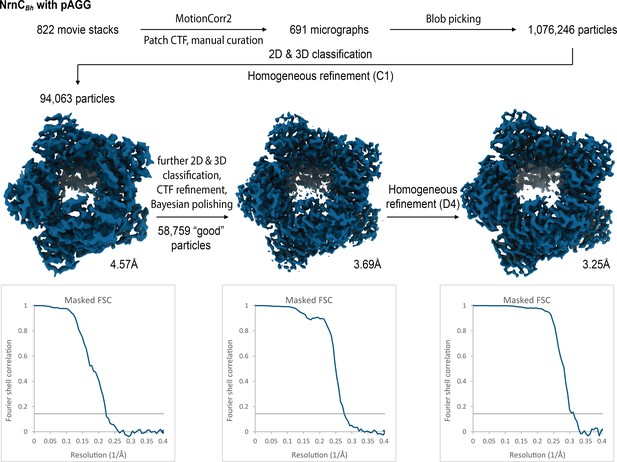
Cryo-electron microscopy (cryo-EM) workflow and resolution for NrnCBh•pAGG.
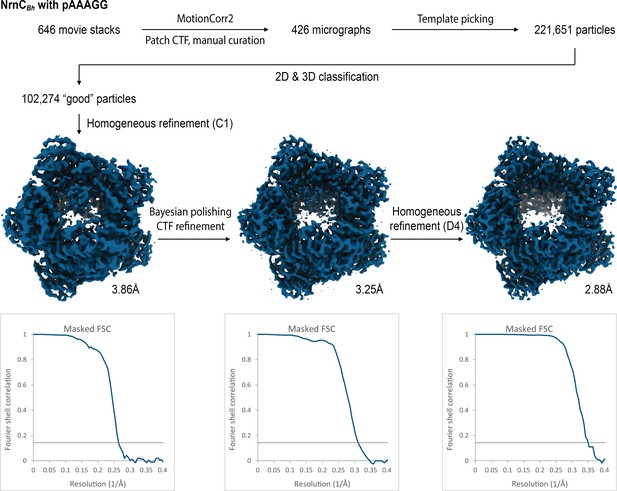
Cryo-electron microscopy (cryo-EM) workflow and resolution for NrnCBh•pAAAGG.
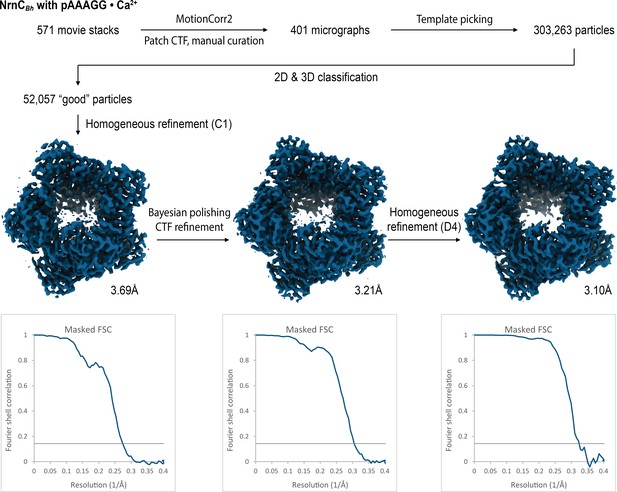
Cryo-electron microscopy (cryo-EM) workflow and resolution for NrnCBh•pAAAGG in the presence of Ca2+ ions.
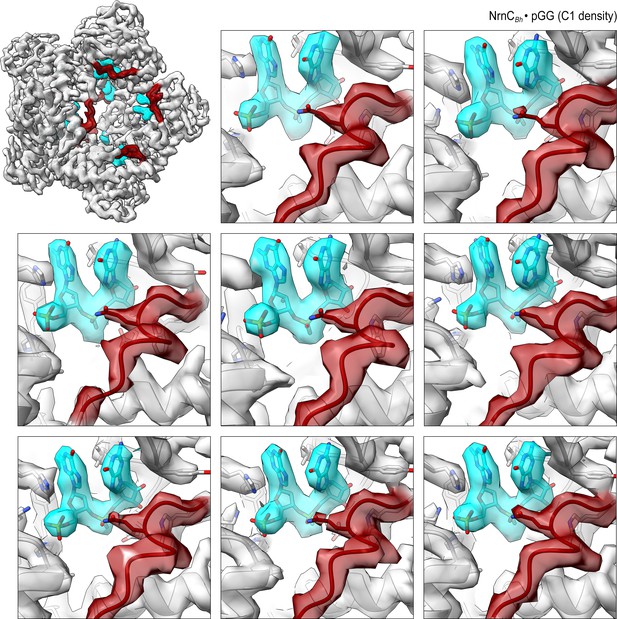
Overall and individual active site electron density of a NrnCBh•pGG octamer after refinement with C1 symmetry.
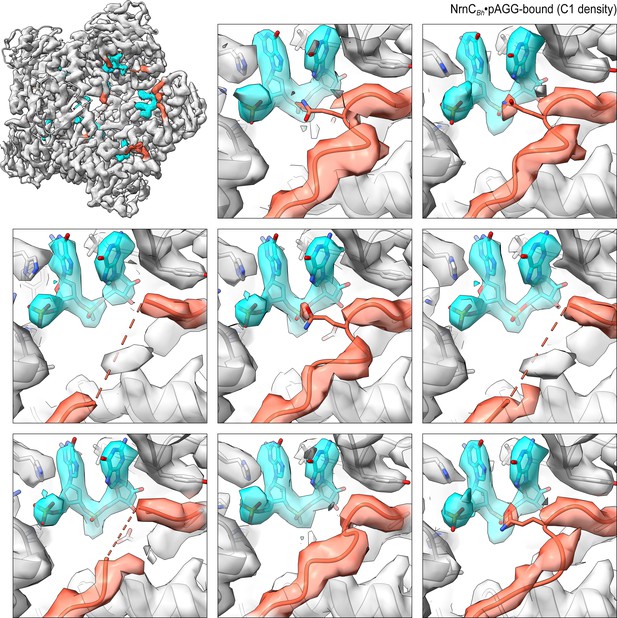
Overall and individual active site electron density of a NrnCBh•pAGG octamer after refinement with C1 symmetry.
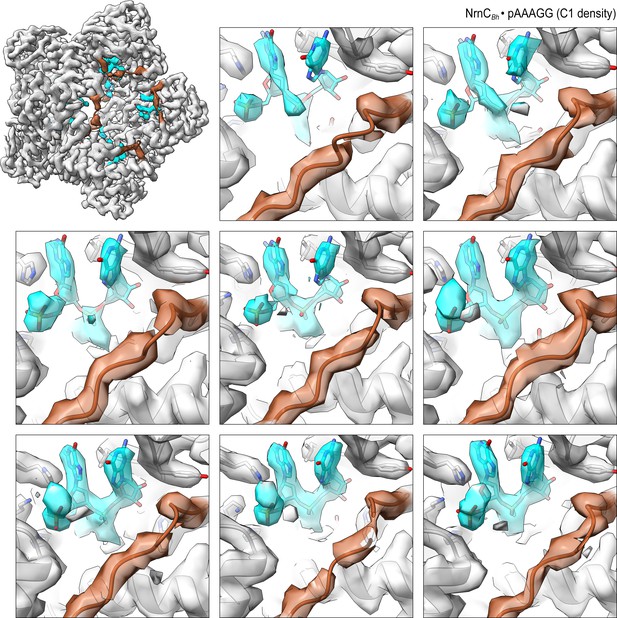
Overall and individual active site electron density of a NrnCBh•pAAAGG octamer after refinement with C1 symmetry.
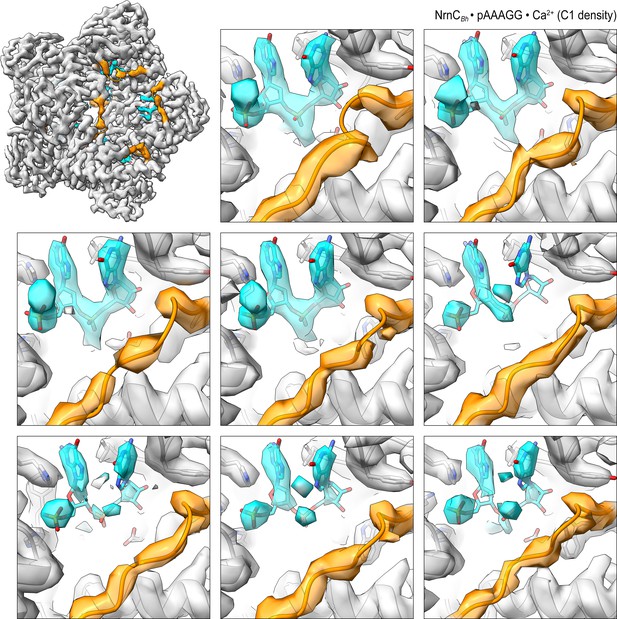
Overall and individual active site electron density of a NrnCBh•pAAAGG octamer in the presence of Ca2+ ions after refinement with C1 symmetry.
The conformation of nano-RNase C (NrnC) bound to substrates with more than two bases resembles the crystallographic apo-state.
(A) Comparison of the crystal structure of NrnCBh-pGG with the corresponding cryo-electron microscopy (cryo-EM) structure shows agreement between the solution and crystalline state of the protein with a well-ordered conformation of the loop residues 130–137 engaging the substrate. (B) Comparison of the crystal structure of apo-NrnCBm with the cryo-EM structure of NrnCBm-pAAAGG. The superposition indicates that longer substrates may bind the active site but only the first full residues appear ordered, resulting in a conformation of nano-RNase C (NrnC) similar to the inactive state observed in the apo-state crystal structures.
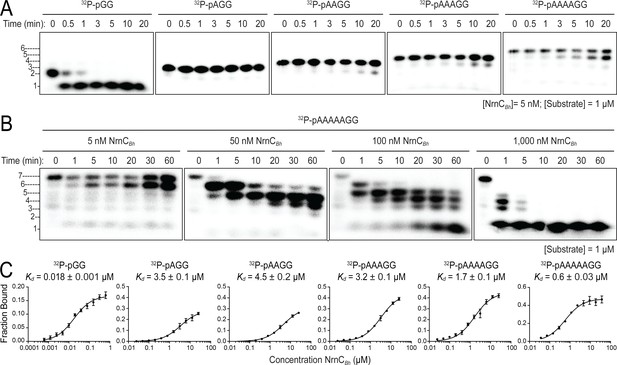
Nano-RNase C (NrnC) shows a strong preference for substrates with two residues in length.
(A, B). RNase assays. Experiments are similar to those in Figure 2 but were performed with radiolabeled substrates from 2 to 7 residues in length. Representative gels of at least two independent experiments are shown. In (B), enzyme concentration was varied from 5 to 1000 nM (1:200 to 1:1 enzyme:substrate ratio). Substrate length-dependent binding studies. (C) Affinity of NrnC for RNA with different lengths. Fraction bound of radiolabeled substrates of increasing length was assessed at different NrnC concentrations and is plotted as means and SD from three independent experiments.
-
Figure 5—source data 1
Source data for Figure 5A.
Original, unedited images and labeled composite overview of nano-RNase C (NrnC) activity against substrates with different length in three replicates.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig5-data1-v2.zip
-
Figure 5—source data 2
Source data for Figure 5B.
Original, unedited images and labeled composite overview of nano-RNase C (NrnC) activity against a 7-nucleotide RNA substrate at increasing enzyme concentration in three replicates.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig5-data2-v2.zip
-
Figure 5—source data 3
Source data for Figure 5C.
Quantification of binding studies using nano-RNase C (NrnC) and radiolabeled, single-stranded RNA with increasing length.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig5-data3-v2.zip
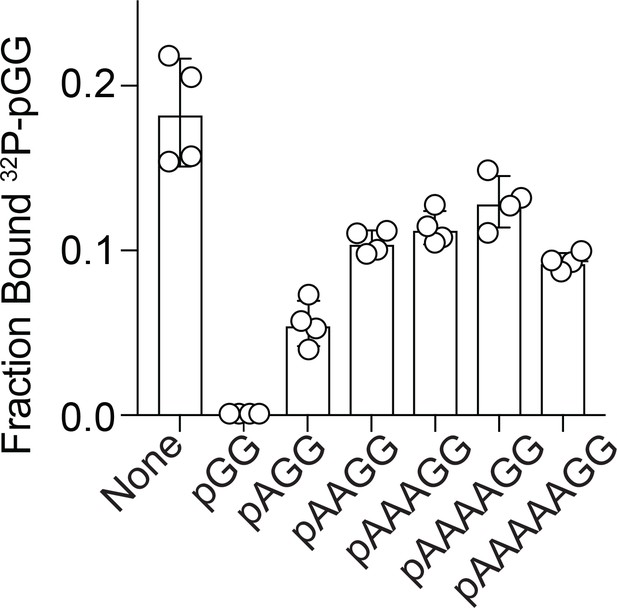
Competition binding studies.
Fraction bound of 32P-pGG to 200 nM purified NrnCBh in the presence of no competitor or 100 µM unlabeled RNA as indicated. Individual data, means, and SD of four independent experiments are plotted.
-
Figure 5—figure supplement 1—source data 1
Quantification of 32P-pGG binding to nano-RNase C (NrnC) in the presence of unlabeled RNA with increasing length (in three replicates).
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig5-figsupp1-data1-v2.zip
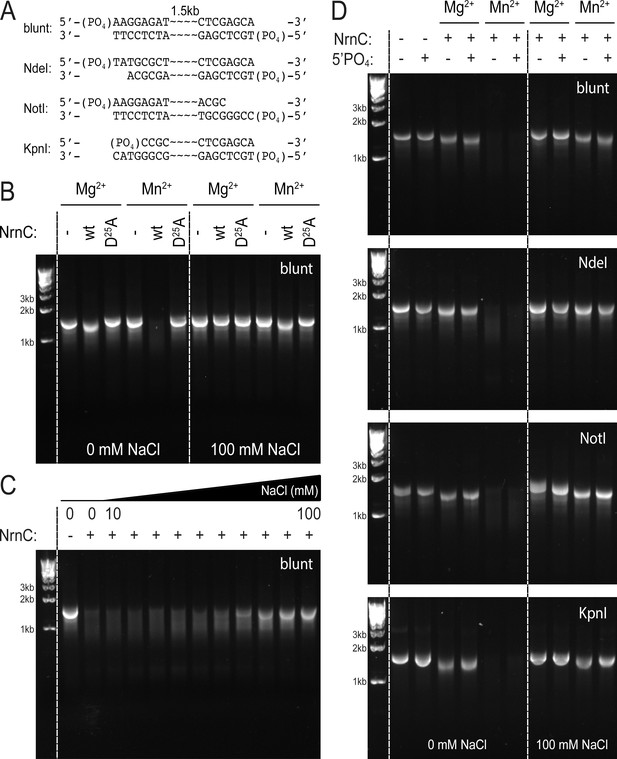
NrnCBh degrades long DNA fragments under distinct conditions.
(A) DNA fragments tested in this assay. (B) DNase activity of wild-type NrnCBh and a catalytically inactive mutant variant on blunt dsDNA in the presence of either Mg2+ or Mn2+ and in the absence or presence of NaCl. (C) NaCl titration on blunt dsDNA using wild-type NrnCBh. (D) Nano-RNase C (NrnC) activity on various dsDNA substrates with or without a 5′-PO4 and in the presence of Mg2+ of Mn2+. Representative agarose gels are shown from at least two independent experiments.
-
Figure 5—figure supplement 2—source data 1
Original, unedited agarose gel images and composite overview of nano-RNase C (NrnC) activity against 1.5 kb, double-stranded DNA substrates.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig5-figsupp2-data1-v2.zip
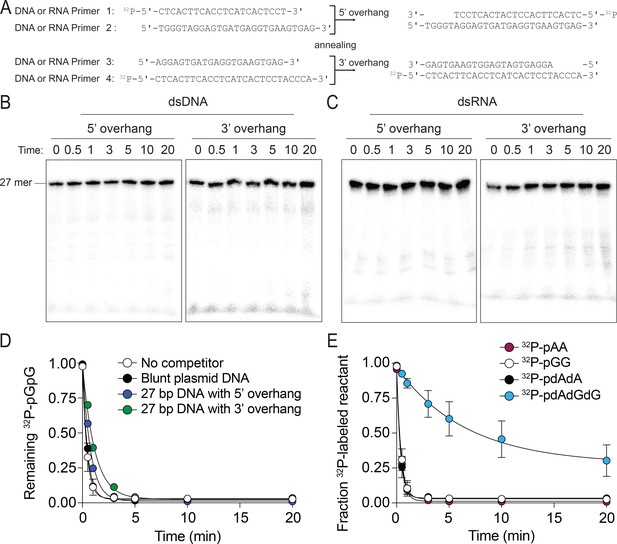
The preferred substrates of NrnCBh are diribonucleotides and deoxy-dinucleotides.
(A–C) NrnCBh does not degrade dsDNA and dsRNA. DNA or RNA oligonucleotide sequences used are shown in (A). Primers 1 and 2 were annealed to generate a 27-nucleotide DNA or RNA with a 5′ overhang and primers 3 and 4 were annealed to generate a 3′ overhang of (length of overhang: 5 nucleotides). NrnCBh activity on 5′ overhang or 3′ overhang of DNA(B) and RNA (C) in the presence of Mg2+ and 100 mM NaCl. These experiments were performed with 3.3 nM of 5′-radiolabeled DNA or RNA. Aliquots of each reaction were stopped at the indicated times and analyzed by 20% urea PAGE. (D) dsDNA is not an inhibitor of NrnCBh’s activity on diribonucleotides. The rate of 32P-pGG degradation by 5 nM NrnCBh was assayed over time with no competitor, plasmid DNA (30 nM) that was cut with StuI, or primers (1 µM) annealed to generate 5-nucleotide 5′ or 3′ overhangs (primers in panel A). Samples were stopped at the indicated time points and analyzed by thin-layer chromatography. Data are from triplicate independent experiments. (E) NrnCBh cleaves DNA deoxy-dinucleotides. Degradation of 32P-pGG, pAA, pdAdA, and pdAdGdG (3.3 nM) by 5 nM NrnCBh is shown. Samples were stopped at the indicated time points and analyzed by TLC. The graph shows quantification of triplicate independent experiments.
-
Figure 5—figure supplement 3—source data 1
Source data for Figure 5—figure supplement 3B.
Original, unedited images and labeled composite overview of nano-RNase C (NrnC) activity against double-stranded DNA oligonucleotides.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig5-figsupp3-data1-v2.zip
-
Figure 5—figure supplement 3—source data 2
Source data for Figure 5—figure supplement 3C.
Original, unedited images and labeled composite overview of nano-RNase C (NrnC) activity against double-stranded RNA oligonucleotides.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig5-figsupp3-data2-v2.zip
-
Figure 5—figure supplement 3—source data 3
Source data for Figure 5—figure supplement 3D and E.
Data quantification plotted in the graphs shown from three replicate experiments.
- https://cdn.elifesciences.org/articles/70146/elife-70146-fig5-figsupp3-data3-v2.zip
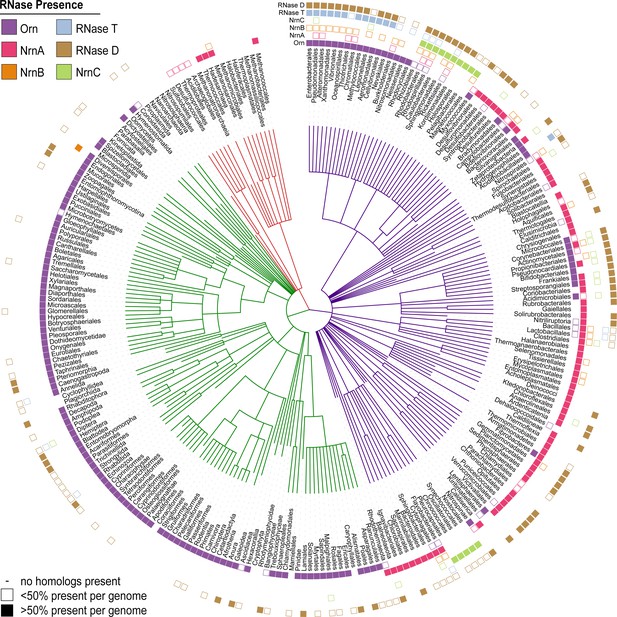
Presence of RNase homologs across sequenced organism classes.
Shown is a ‘Tree of Life’ with all taxonomic groups at the class level with at least one substantially complete proteome available in the dataset. The tree is based on the structure of the NCBI Taxonomy database, with bacterial taxa shown with purple lines, eukaryotic taxa shown with green lines, and archaeal taxa shown with red lines. The presence of each RNase homolog as a proportion of the total proteins in that taxonomic group is shown as either a filled square (>50% presence of a homolog per genome) or an empty square (<50% presence of a homolog per genome). Lack of a square indicates no homologs for that family were present in genomes of that class.
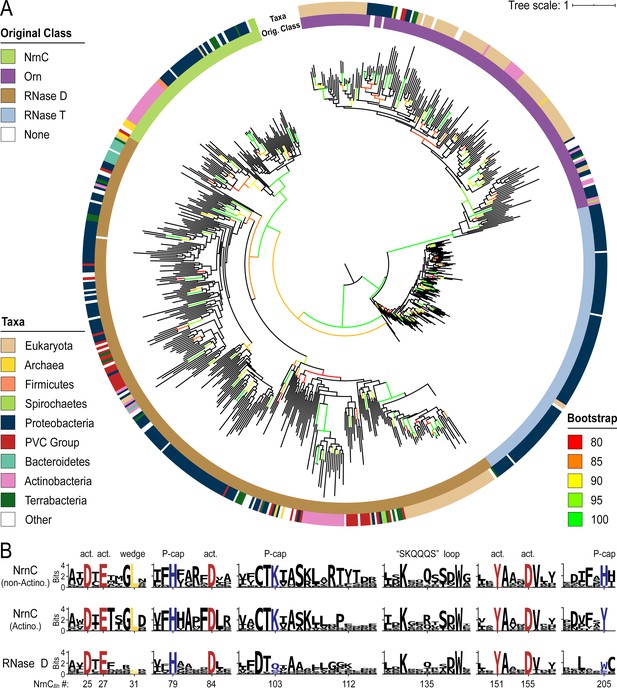
Phylogenetic tree of four DnaQ-fold RNase families.
(A) Phylogenetic tree of 669 representatives of the RNase T, RNase D, oligoribonuclease (Orn), and nano-RNase C (NrnC) families of RNase proteins. The inner ring represents the original classification of each sequence by HMM analysis. The outer ring represents the high-level taxonomic classification of the organism the protein is found in. The color of the branch represents the UFBoot bootstrap value, where black branches are <80%, red is 80%, orange is 85%, yellow is 90%, light green is 95%, and bright green is 100%. Bootstrap values > 90% indicate high-confidence splits. (B) Sequence logos of RNase D and NrnC subgroups. Sequence logos showing the relative entropy (information content) at selected positions in RNase D as well as the Actinobacterial and non-Actinobacterial subsets of NrnC. Sequence numbering is relative to Bartonella birtlessi NrnC (G4VUY7). Active site residues are shown in red, phosphate cap residues in dark blue, and the L-wedge in yellow.
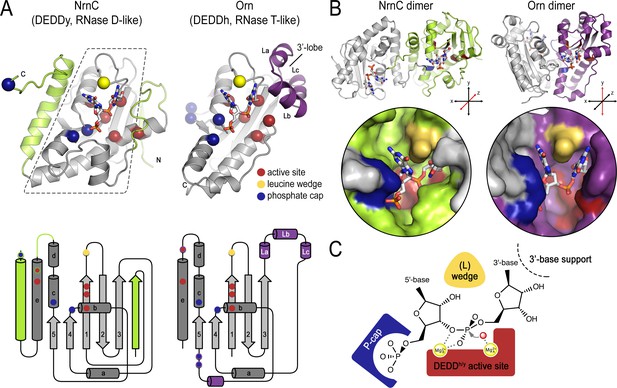
Structural comparison of nano-RNase C (NrnC) and oligoribonuclease (Orn).
(A) Fold topology. pGG-bound NrnC and Orn monomers are shown in a similar orientation as cartoons (top) or schematic topology diagrams (bottom). Conserved catalytic core elements are colored in gray. NrnC and Orn-specific features are colored in green and purple, respectively. Other color codes mark the positions of the DEDDy/h motif (red spheres), L-wedge (yellow sphere), and phosphate cap residues (dark blue spheres). (B) Comparison of dimer units of NrnC and Orn (top) with close-ups of the composite active sites of the enzymes (bottom). An NrnC monomer is colored green and an Orn monomer is colored purple, with adjacent monomers in the biological assemblies colored in light gray. Specific residues are colored as in (A). Coordinate systems indicate the twofold symmetry axis of the enzyme dimers, with the colored monomers shown in a similar orientation. (C) Structurally and functionally conserved features common among NrnC- and Orn-type diribonucleotidases.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Pseudomonas aeruginosa) | PA14 | Rahme et al., 1995; PMID:7604262 | ||
| Strain, strain background (P. aeruginosa) | PA14 ∆orn | This study | ||
| Strain, strain background (Escherichia coli) | Stellar cells | Takara/Clontech | ||
| Strain, strain background (E. coli) | BL21(DE3) | New England Biolabs | ||
| Recombinant DNA reagent | pEX-Gn-∆orn (plasmid) | This study | ||
| Recombinant DNA reagent | pJN105 (plasmid) | Newman and Fuqua, 1999; PMID:10023058 | ||
| Recombinant DNA reagent | pJHA (plasmid) | Kim et al., 2019; PMID:31225796 | ||
| Sequence-based reagent | ACAGATTGGTGGATC CATGACCGAAATTCGCG TGCATCAGGGCGATCTGC CGAACCTGGATAACTAT CGCATTGATG CGGTGGCG GTGGATACCG AAACCCTGG GCCTGCAGCC GCATCGCGAT CGCCTGTGCG TGGTGCAGCTG AGCAGCGGCGA TGGCACCGC GGATGTGATTCA GATTGCGAA AGGCCAGAAAA GCGCGCCGAA CCTGGTGCGCC TGCTGAGCG ATCGCGATATT ACCAAAATTTTT CATTTTGGCCGC TTTGATCTG GCGATTCTGGCG CATACCTTTG GCGTGATGCCG GATGTGGTGT TTTGCACCAAAAT TGCGAGCAA ACTGACCCGCAC CTATACCGATCGC CATGGCCTGAAAG AAATTTGCGG CGAACTGCTGAAC GTGAACATTAG CAAACAGCAGCAG AGCAGCGATTG GGCGGCGGAAAC CCTGAGCCGCG CGCAGATTGAATAT GCGGCGAGCG ATGTGCTGTATCTG CATCGCCTGAA AGATATTTTTGAAG AACGCCTGAAA CGCGAAGAACGCG AAAGCGTGGCG AAAGCGTGCTTTC AGTTTCTGCCGA TGCGCGCGAACC TGGATCTGCTGG GCTGGAGCGAAATTG ATATTTTTGCG CATAGCTAAGCGGC CGCACTCGAGCA (DNA fragment) | Geneart | BH02530 | Custom DNA fragment for Bartonella henselae NrnC, cloned in to His6-SUMO-pET28 |
| Sequence-based reagent | ACAGATTGGTG GATCCATGACCA TTCGCTTTCATC GCAACGATCT GCCGAACCTGGA TAACTATCAGG TGGATGCGGTG GCGATTGATAC CGAAACCCTGG GCCTGAACCCGC ATCGCGATCGCC TGTGCGTGGTG CAGATTAGCCCG GGCGATGGCAC CGCGGATGTGA TTCAGATTGAAGC GGGCCAGAAAAAA GCGCCGAACC TGGTGAAACTGC TGAAAGATCGC AGCATTACCAAAA TTTTTCATTTTG GCCGCTTTGATC TGGCGGTGCTG GCGCATGCGTTT GGCACCATGCC GCAGCCGGTGTT TTGCACCAAAAT TGCGAGCAAACTG ACCCGCACCT ATACCGATCGCCAT GGCCTGAAAG AAATTTGCAGCGA ACTGCTGGATG TGAGCATTAGCAA ACAGCAGCAG AGCAGCGATTGGG CGGCGGAAG TGCTGAGCCAGG CGCAGCTGGAA TATGCGGCGAG CGATGTGCTGTAT CTGCATCGCCTG AAAGCGGTGCT GGAACAGCGCCT GGAACGCGAT GGCCGCACCAAA CAGGCGGAAGC GTGCTTTAAATTT CTGCCGACCCG CAGCGAACTGGA TCTGATGGGCTG GGCGGAAAGCGA TATTTTTGCGCAT AGCTAAGCGGCCGCACTC GAGCA (DNA fragment) | Geneart | BMEI1828 | Custom DNA fragment for Brucella melitensis NrnC, cloned in to His6-SUMO-pET28 |
| Recombinant DNA reagent | His6-SUMO-pET28- NrnCBh (plasmid) | This study | Cloned from custom DNA fragment | |
| Recombinant DNA reagent | His6-SUMO-pET28- NrnCBm (plasmid) | This study | Cloned from custom DNA fragment | |
| Recombinant DNA reagent | pJHA-NrnCBh (plasmid) | This study | NrnCBh cloned into pJHA for expression in P. aeruginosa | |
| Recombinant DNA reagent | pJHA-NrnCBm (plasmid) | This study | NrnCBm cloned into pJHA for expression in P. aeruginosa | |
| Recombinant DNA reagent | pJHA-NrnCBh D25A (plasmid) | This study | Product of site-directed mutagenesis of pJHA-NrnCBh | |
| Recombinant DNA reagent | pJHA-NrnCBh E27A (plasmid) | This study | Product of site-directed mutagenesis of pJHA-NrnCBh | |
| Recombinant DNA reagent | pJHA-NrnCBh D84A (plasmid) | This study | Product of site-directed mutagenesis of pJHA-NrnCBh | |
| Recombinant DNA reagent | pJHA-NrnCBh D155A (plasmid) | This study | Product of site-directed mutagenesis of pJHA-NrnCBh | |
| Recombinant DNA reagent | pJHA-NrnCBh Y151A (plasmid) | This study | Product of site-directed mutagenesis of pJHA-NrnCBh | |
| Recombinant DNA reagent | pJHA-NrnCBh L31A (plasmid) | This study | Product of site-directed mutagenesis of pJHA-NrnCBh | |
| Recombinant DNA reagent | pJHA-NrnCBh H79A (plasmid) | This study | Product of site-directed mutagenesis of pJHA-NrnCBh | |
| Recombinant DNA reagent | pJHA-NrnCBh K103A (plasmid) | This study | Product of site-directed mutagenesis of pJHA-NrnCBh | |
| Recombinant DNA reagent | pJHA-NrnCBh H205A (plasmid) | This study | Product of site-directed mutagenesis of pJHA-NrnCBh | |
| Recombinant DNA reagent | pJHA-NrnCBh K132A (plasmid) | This study | Product of site-directed mutagenesis of pJHA-NrnCBh | |
| Sequence-based reagent | TTTGGGCTAGCCA TATGACCGAAATT CGTGTTCATCAGGG | Life Technologies | NrnCBh_ infusionprimer_F | Primer for cloning NrnCBh into pJHA |
| Sequence-based reagent | GCTCAAGCTTGAAT TCGCTGTGTGCAA AGATATCAATTTCG | Life Technologies | NrnCBh_ infusionprimer_R | Primer for cloning NrnCBh into pJHA |
| Sequence-based reagent | TTTGGGCTAGCCATATGACCATTCGTTTTCATCGTAATGATC | Life Technologies | NrnCBm_ infusionprimer_F | Primer for cloning NrnCBm into pJHA |
| Sequence-based reagent | GCTCAAGCTTGAATTCGCTATGTGCAAAAATATCGCTTTC | Life Technologies | NrnCBm_ infusionprimer_R | Primer for cloning NrnCBm into pJHA |
| Sequence-based reagent | Acccagtgtttcggtagc aacggcaactgcatc | Life Technologies | NrnCBh D25A_a | Primer for site directed mutagenesis |
| Sequence-based reagent | Gatgcagttgccgttgct accgaaacactgggt | Life Technologies | NrnCBh D25A_b | Primer for site directed mutagenesis |
| Sequence-based reagent | Gttgccgttgataccgca acactgggtctgcag | Life Technologies | NrnCBh E27A_a | Primer for site directed mutagenesis |
| Sequence-based reagent | Ctgcagacccagtgttgc ggtatcaacggcaac | Life Technologies | NrnCBh E27A_b | Primer for site directed mutagenesis |
| Sequence-based reagent | Cgatgcggctgcgcac ccagtgtttcggtatcaacg | Life Technologies | NrnCBh L31A_a | Primer for site directed mutagenesis |
| Sequence-based reagent | Cgttgataccgaaaca ctgggtgcgcagc cgcatcg | Life Technologies | NrnCBh L31A_b | Primer for site directed mutagenesis |
| Sequence-based reagent | Agatcgaaacgacca aaggcaaagattttg gtaatatcacgatcgc | Life Technologies | NrnCBh H79A_a | Primer for site directed mutagenesis |
| Sequence-based reagent | Gcgatcgtgatattacc aaaatctttgcctttg gtcgtttcgatct | Life Technologies | NrnCBh H79A_b | Primer for site directed mutagenesis |
| Sequence-based reagent | Gtgccagaattgccag agcgaaacgac caaagtga | Life Technologies | NrnCBh D84A_a | Primer for site directed mutagenesis |
| Sequence-based reagent | Tcactttggtcgtttcgct ctggcaattctggcac | Life Technologies | NrnCBh D84A_b | Primer for site directed mutagenesis |
| Sequence-based reagent | Cgggtcagtttgcttgc aattgcggtacaa aaaacaacatccgg | Life Technologies | NrnCBh K103A_a | Primer for site directed mutagenesis |
| Sequence-based reagent | Ccggatgttgttttttgtacc gcaattgcaagca aactgacccg | Life Technologies | NrnCBh K103A_b | Primer for site directed mutagenesis |
| Sequence-based reagent | Acatcacttgctgcagct tcaatctgtgcac ggctcagg | Life Technologies | NrnCBh Y151A_a | Primer for site directed mutagenesis |
| Sequence-based reagent | Cctgagccgtgcacag attgaagctgca gcaagtgatgt | Life Technologies | NrnCBh Y151A_b | Primer for site directed mutagenesis |
| Sequence-based reagent | Cggtgcagatacaga acagcacttgctgcatattcaa | Life Technologies | NrnCBh D155A_a | Primer for site directed mutagenesis |
| Sequence-based reagent | Ttgaatatgcagcaag tgctgttctgtatctgcaccg | Life Technologies | NrnCBh D155A_b | Primer for site directed mutagenesis |
| Sequence-based reagent | Gctcaagcttgaattc gctggctgcaa agatatcaatttcgc | Life Technologies | NrnCBh H205A_a_pJHA | Primer for site directed mutagenesis |
| Sequence-based reagent | Gcgaaattgatatctttgc agccagcgaattcaagcttgagc | Life Technologies | NrnCBh H205A_b_pJHA | Primer for site directed mutagenesis |
| Sequence-based reagent | Gtgcggccgcttagctgg ctgcaaagatatcaatttcgct | Life Technologies | NrnCBh H205A_a_SUMO | Primer for site directed mutagenesis |
| Sequence-based reagent | Agcgaaattgatatcttt gcagccagctaa gcggccgcac | Life Technologies | NrnCBh H205A_b_SUMO | Primer for site directed mutagenesis |
| Sequence-based reagent | 5′-GG-3′ (RNA primer) | Sigma | ||
| Sequence-based reagent | 5′-AGG-3′ (RNA primer) | Sigma | ||
| Sequence-based reagent | 5′-AAGG-3′ (RNA primer) | Sigma | ||
| Sequence-based reagent | 5′-AAAGG-3′ (RNA primer) | Sigma | ||
| Sequence-based reagent | 5′-AAAAGG-3′ (RNA primer) | Sigma | ||
| Sequence-based reagent | 5′-AAAAAGG-3′ (RNA primer) | Sigma | ||
| Sequence-based reagent | 5′-pGG-3′ (RNA primer) | Biolog’ catalog number P023-01 | ||
| Sequence-based reagent | 5′-pAA-3′ (RNA primer) | Biolog’ catalog number P033-01 | ||
| Sequence-based reagent | 5′-pGC-3′ (RNA primer) | GE Healthcare Dharmacon | ||
| Chemical compound, drug | 5′-pAp-3′ (RNA primer) | Sigma | Cat# A5763 | |
| Sequence-based reagent | Various RNA and DNA oligonucleotides | IDT | ||
| Antibody | Anti-HA (rabbit polyclonal) | Takara | Cat# 631207 | (1:100) |
| Antibody | Anti-HA−agarose (mouse monoclonal, clone HA-7) | Sigma | Cat# A2095; RRID:AB_257974 | (10 µl) |
| Antibody | Anti-rabbit (donkey polyclonal, HRP-conjugated) | Cytiva | Cat# NA934 | (1:5000) |
| Software, algorithm | Prism | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | XDS Program Package | Kabsch, 2010; PMID:20124693 | RRID:SCR_015652 | Distributed through SBGrid |
| Software, algorithm | Pointless | Evans, 2006; PMID:16369096 | RRID:SCR_014218 | Distributed through SBGrid |
| Software, algorithm | Scala | Evans, 2006; PMID:16369096 | Distributed through SBGrid | |
| Software, algorithm | Phenix | Adams et al., 2010; PMID:20124702 | RRID:SCR_014224 | Distributed through SBGrid |
| Software, algorithm | Coot | Emsley et al., 2010; PMID:20383002 | RRID:SCR_014222 | Distributed through SBGrid |
| Software, algorithm | MrBump | Keegan et al., 2018;PMID:29533225 | Distributed through SBGrid | |
| Software, algorithm | PyMOL | Schrödinger | RRID:SCR_000305 | Distributed through SBGrid |
| Software, algorithm | UCSF ChimeraX | Pettersen et al., 2021; PMID:32881101 | RRID:SCR_015872 | Distributed through SBGrid |
| Software, algorithm | cryoSPARC | Punjani et al., 2017; PMID:28165473 | RRID:SCR_016501 | |
| Software, algorithm | RELION | Zivanov et al., 2018; PMID:30412051 | RRID:SCR_016274 | |
| Software, algorithm | GCTF | Zhang, 2016; PMID:26592709 | RRID:SCR_016500 | |
| Software, algorithm | iTOL | Letunic and Bork, 2019; PMID:30931475 | RRID:SCR_018174 | |
| Software, algorithm | TCoffee | Notredame et al., 2000; PMID:10964570 | RRID:SCR_019024 | |
| Software, algorithm | Hmmer | Eddy, 2011; PMID:22039361 | RRID:SCR_005305 | |
| Software, algorithm | MAFFT | Katoh et al., 2005; PMID:15661851 | RRID:SCR_011811 | |
| Software, algorithm | SnakeMake | Köster and Rahmann, 2018; PMID:29788404 | RRID:SCR_003475 |
Additional files
-
Supplementary file 1
X-ray data collection and refinement statistics.
- https://cdn.elifesciences.org/articles/70146/elife-70146-supp1-v2.docx
-
Supplementary file 2
Cryo-electron microscopy model validation statistics.
- https://cdn.elifesciences.org/articles/70146/elife-70146-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70146/elife-70146-transrepform1-v2.docx





