Epi-mutations for spermatogenic defects by maternal exposure to di(2-ethylhexyl) phthalate
Figures
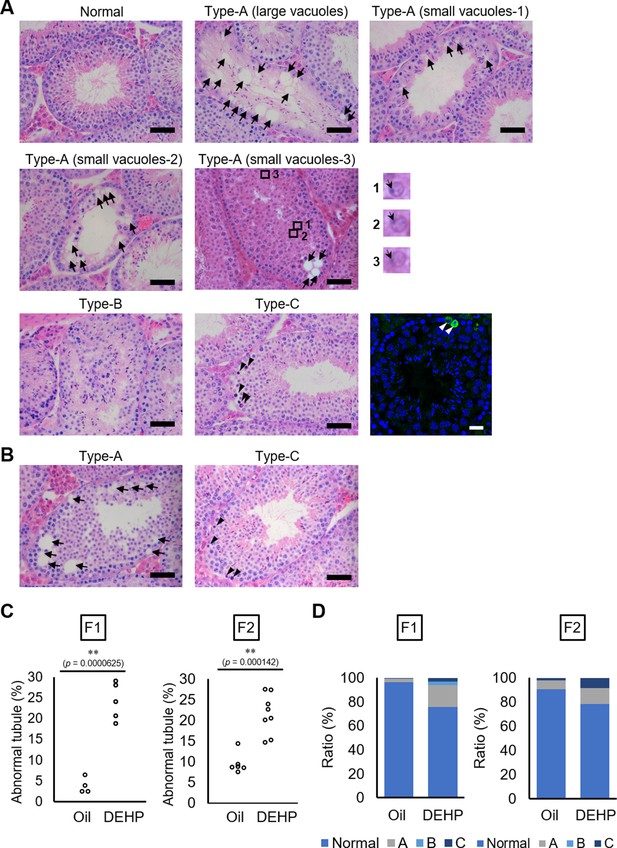
Histological analysis of testicular tubules of C57BL/6 offspring after prenatal di(2-ethylhexyl) phthalate (DEHP) exposure.
(A, B) Representative images of testicular tubules in F1 (A) and F2 (B). Abnormal testicular tubules were categorized as ‘Type A’ (presence of various size of vacuoles; indicated by arrows), ‘Type B’ (no lumen and complete loss of germ cell organization), or ‘Type C’ (presence of dead cells; indicated by arrowheads). Type A was further classified into three subtypes as described in Results. Enlarged images corresponding to the rectangular areas in Type A (small vacuoles-3) are shown. Arrows indicate nuclei. Active caspase-3 immunoreactive apoptotic cells (green) are indicated by arrowheads in a picture on the right of the Type C picture. (C) Ratios of abnormal tubules in F1 and F2 animals (F1 oil as vehicle control: n = 4; F1 DEHP: n = 5; F2 oil: n = 6, F2 DEHP: n = 8). (D) Ratios of abnormal tubule types in F1 and F2. **p<0.01 (unpaired two-sided Student's t-test). Scale bars: 50 μm.
-
Figure 1—source data 1
Numerical source data of abnormal testicular tubule rates (Figure 1C) and ratios of abnormal tubule types (Figure 1D), and data for Kolmogorov–Smirnov test for Figure 1C.
- https://cdn.elifesciences.org/articles/70322/elife-70322-fig1-data1-v1.xlsx
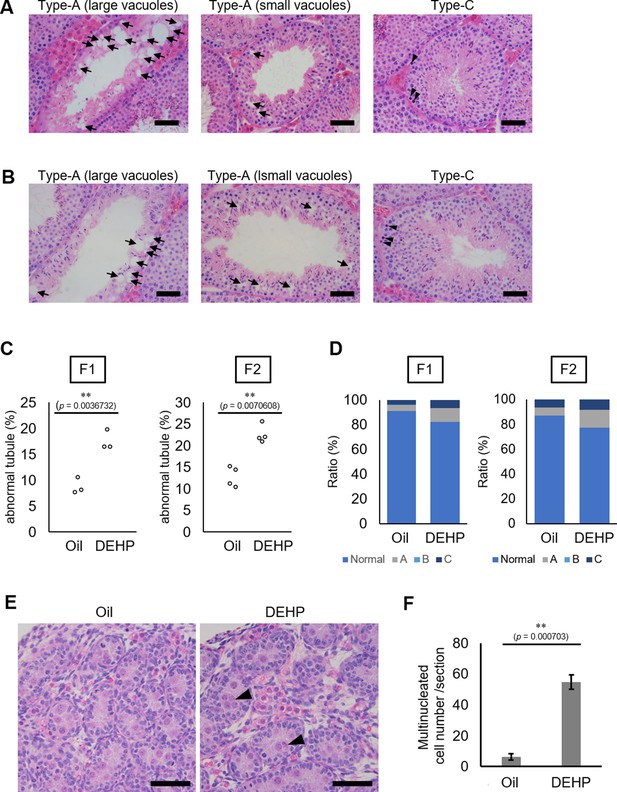
Histological analysis of the testicular tubules of prenatally di(2-ethylhexyl) phthalate (DEHP)-exposed offspring from C57BL/6 females mated with Oct4-deltaPE-GFP transgenic males.
(A, B) Representative images of testicular tubules in F1 (A) and F2 (B). Abnormal testicular tubules were categorized as ‘Type A’ (presence of various size of vacuoles; indicated by arrows) and ‘Type C’ (presence of dead cells; indicated by arrowheads). ‘Type B’ (no lumen and complete loss of germ cell organization) abnormal tubules were very rare in F1 and absent in F2. Arrowheads indicate apoptotic cells. (C) Ratios of abnormal tubules in F1 and F2 testes (F1: n = 3; F2: n = 4). (D) Ratios of abnormal tubule types in F1 and F2. (E) Representative histology of testis from oil- or DEHP-treated E19.5 embryos. Multinucleated cells are indicated by arrowheads. (F) Quantification of the multinucleated cell number per section. Values are plotted as mean ± SEM of all serial sections from one testis in each of three independent embryos. **p<0.01 (unpaired two-sided Student's t-test). Scale bars: 50 μm.
-
Figure 1—figure supplement 1—source data 1
Numerical source data of abnormal testicular tubule rates (Figure 1—figure supplement 1C), ratios of abnormal tubule types (Figure 1—figure supplement 1D), and multinucleated cells (Figure 1—figure supplement 1F), and data for Kolmogorov–Smirnov test for Figure 1—figure supplement 1C, F.
- https://cdn.elifesciences.org/articles/70322/elife-70322-fig1-figsupp1-data1-v1.xlsx
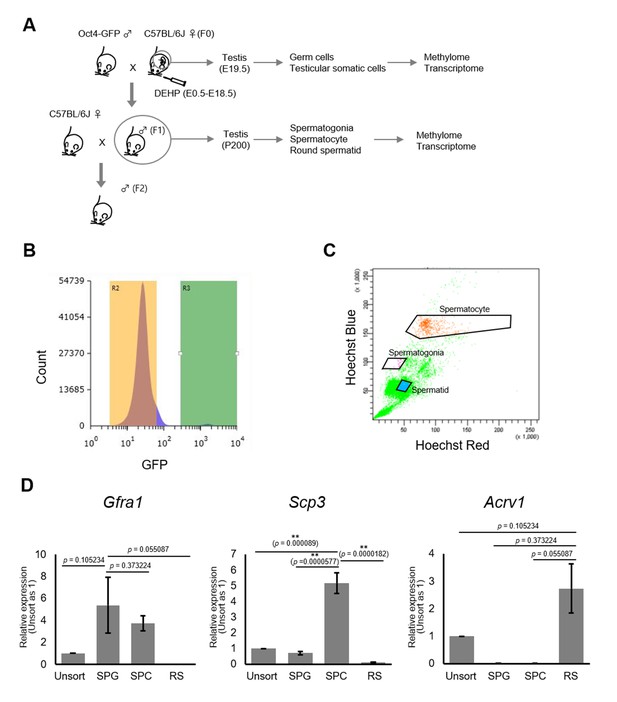
Purification of germ cells from E19.5 and adult testis.
(A) The bleeding schema of the experiments. (B) A representative FACS histogram of E19.5 testis. GFP-positive germ cells in the R3 gate (green color), and GFP-negative testicular somatic cells in the R2 gate (yellow color), were collected. (C) A representative FACS plot of F1 testicular cells stained with Hoechst 33342. Gates for spermatogonia, spermatocytes, and round spermatids are indicated. (D) Relative expression of stage-specific germ cell marker genes in F1 sorted cells was determined by RT-qPCR and consisted of the following: Gfra1 for spermatogonia, Scp3 for spermatocytes, and Acrv1 for spermatids. SPG: spermatogonia; SPC: spermatocytes; RS: round spermatids. Values were plotted as mean ± SEM of each cell sample from six individuals of F1. Statistics for data in (D) was unpaired two-sided Student's t-test.
-
Figure 2—source data 1
Numerical source data for relative gene expression of Gfra1, Scp1, and Acrv1, and data for Kolmogorov–Smirnov test for Figure 2D.
SPG: spermatogonia; SPC: spermatocytes; RS: round spermatids.
- https://cdn.elifesciences.org/articles/70322/elife-70322-fig2-data1-v1.xlsx
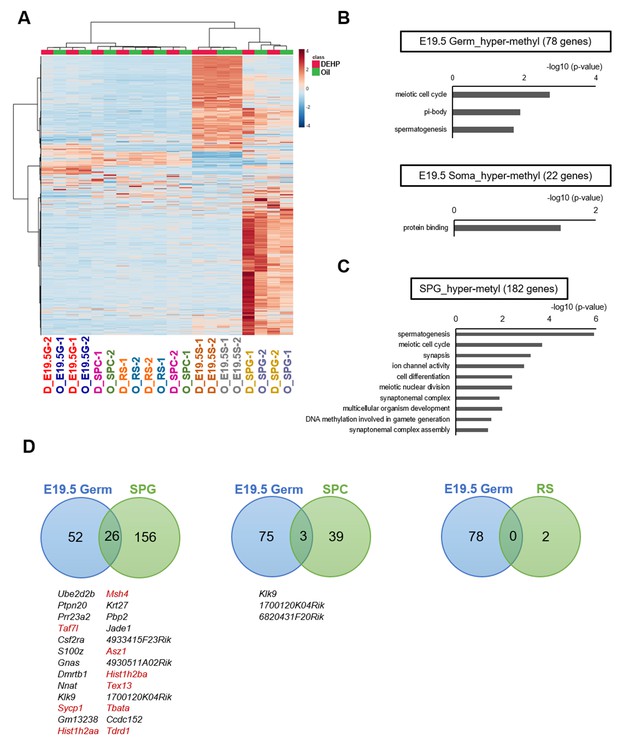
Methylome of testicular cells of F1 after maternal exposure to di(2-ethylhexyl) phthalate (DEHP).
(A) Heatmap of promoter DNA methylation levels in the testicular cell populations prenatally exposed to oil as vehicle control (O) or DEHP (D) (clustering distance: Euclidean; clustering method: Ward). Each colored cell on the map corresponds to normalized value of methylation percentage. Reddish and bluish colors represent relatively hyper- and hypomethylation, respectively. E19.5G: E19.5 germ cell; E19.5S: E19.5 testicular somatic cells; SPG: spermatogonia; SPC: spermatocytes. (B, C) Functional annotation chart of hypermethylated (more than 5% increase in DEHP-treated samples compared to oil-treated samples) genes in E19.5 germ cells (Germ) and testicular somatic cells (Soma) (B) and in spermatogonia (C) using DAVID. Statistically significant (p<0.05) GO terms with BH-corrected p-values are shown. Threshold was FDR < 0.05. (D) Venn diagram analysis of the hypermethylated genes in E19.5 germ cells, SPG, SPC, or round spermatids (RS). Genes that are hypermethylated in E19.5 germ cell and SPG or SPC are listed, and spermatogenesis-related genes are shown in red.
-
Figure 3—source data 1
Analytical codes of reduced representation bisulfite sequencing for Figure 3.
- https://cdn.elifesciences.org/articles/70322/elife-70322-fig3-data1-v1.docx
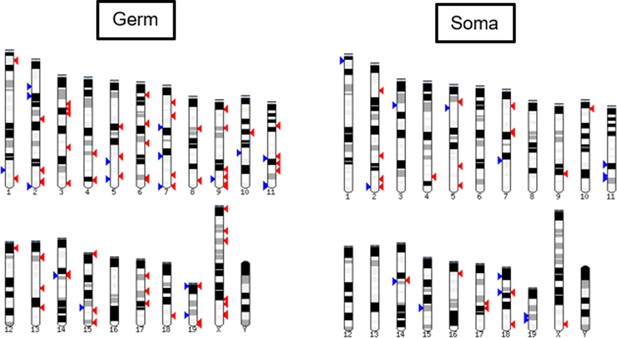
Chromosomal positions of differentially promoter-methylated genes in E19.5 germ and testicular somatic cells.
Red arrowhead: more than 5% increase in methylation in the di(2-ethylhexyl) phthalate (DEHP) group compared to the oil group; blue arrowhead: more than 5% decrease in methylation in the DEHP group compared to the oil group.
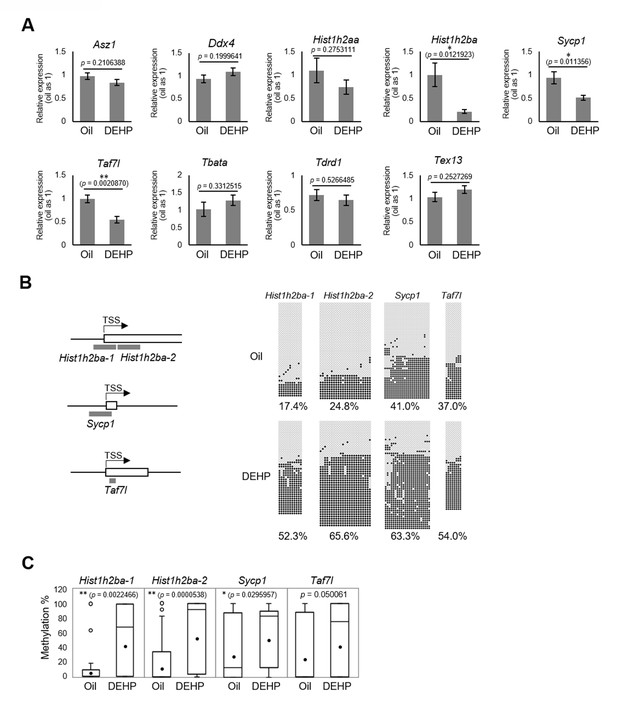
Gene expression and promoter methylation of spermatogenesis-related genes hypermethylated in both E19.5 germ cells and F1 spermatogonia following prenatal di(2-ethylhexyl) phthalate (DEHP) exposure.
(A) Relative mRNA levels in prenatally oil- or DEHP-treated adult F1 spermatogonia, as determined by RT-qPCR. Values of oil group are defined as 1.0. Values are plotted as mean ± SEM for samples obtained from six individuals. (B) The regions detected in bisulfite sequencing of Hist1h2ba, Sycp1, and Taf7l are indicated in gray bars in the left panel. TSS: transcription start site. Boxes indicate the first exon. Methylation status of these regions in F1 spermatogonia obtained from four individuals is indicated in the right panel. Methylated and unmethylated CpGs are presented as closed circles and open circles, respectively. The percentage of methylated CpGs is indicated. (C) Box-whisker plots of the CpG methylation levels shown in (B). The lines inside the boxes show the medians. The whiskers indicate the minimum and maximum values. Open and closed circles indicate outliers and mean value, respectively. *p<0.05, **p<0.01 (unpaired two-sided Student's t-test in A, Mann–Whitney U test in C).
-
Figure 4—source data 1
Numerical source data for relative gene expression (Figure 4A) and DNA methylation status (Figure 4D), and data for Kolmogorov–Smirnov test for Figure 4A.
- https://cdn.elifesciences.org/articles/70322/elife-70322-fig4-data1-v1.xlsx
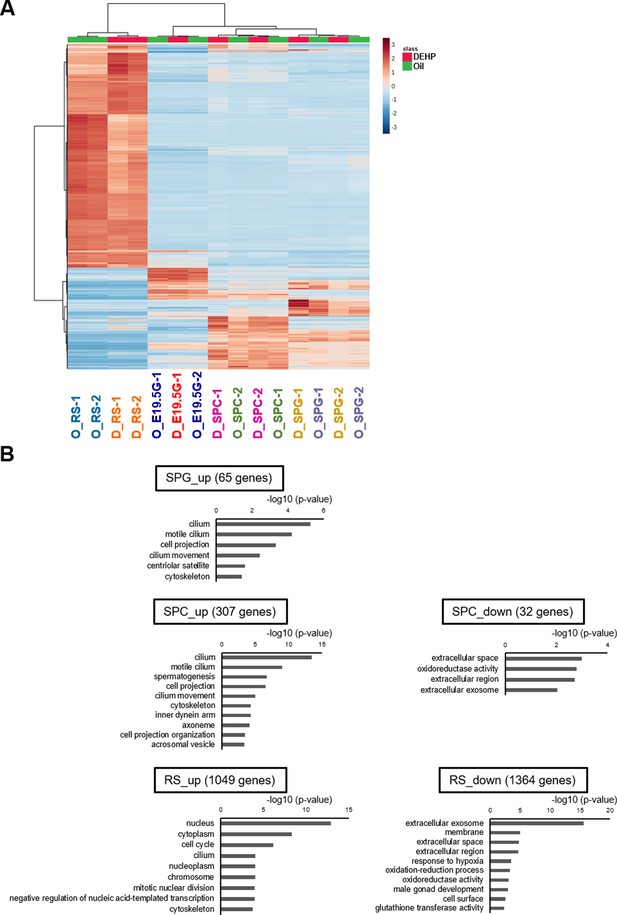
Transcriptome of testicular germ cells after prenatal di(2-ethylhexyl) phthalate (DEHP) exposure.
(A) A heatmap of TMM-normalized RNA-seq read count values in E19.5 and adult germ cell populations of F1 prenatally exposed to oil (O) or DEHP (D) (clustering distance: Euclidean; clustering method: Ward). Each colored cell on the map corresponds to normalized value of TMM-normalized read count. Reddish and bluish colors represent relatively up- and downregulation, respectively. (B) Functional annotation chart of genes exhibiting up- or downregulation (log2-fold-change > 1 or <−1, FDR < 0.05) following DEHP exposure (compared to oil exposure) in E19.5 germ cells (B) and F1 germ cell populations. Statistically significant (p<0.05) GO terms with BH-corrected p-values are shown. Threshold was FDR < 0.05. SPG: spermatogonia; SPC: spermatocytes; RS: round spermatids.
-
Figure 4—figure supplement 1—source data 1
Analytical codes of RNA-seq for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/70322/elife-70322-fig4-figsupp1-data1-v1.docx
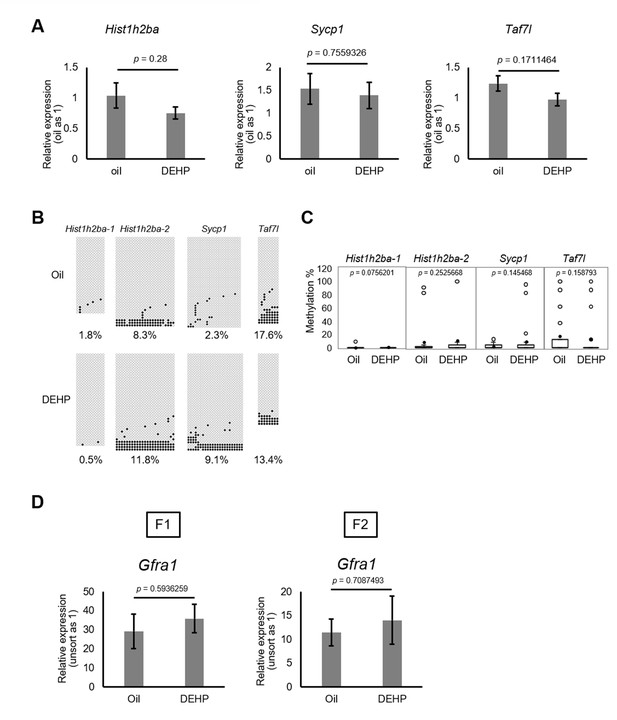
Gene expression and promoter methylation of Hist1h2ba, Sycp1, and Taf7l in F2 spermatogonia.
(A) Relative mRNA levels determined by RT-qPCR in prenatal oil- or di(2-ethylhexyl) phthalate (DEHP)-treated adult F2 spermatogonia. Values are plotted as mean ± SEM of spermatogonia samples obtained from three individuals. (B) Methylation status of these regions determined by bisulfite sequencing in spermatogonia obtained from three individuals. Methylated and unmethylated CpGs are presented as closed circles and open circles, respectively. The percentage of methylated CpGs is indicated. (C) Box-whisker plots of the CpG methylation levels shown in (B). The lines inside the boxes show the medians. The whiskers indicate the minimum and maximum values. Open and closed circles indicate outliers and mean value, respectively. (D) Relative expression of Gfra1 in F1 and F2 sorted spermatogonia determined by RT-qPCR. Values were plotted as mean ± SEM of each cell sample from four individuals of F1 and three individuals of F2. Statistics for data in (A) and (D) was unpaired two-sided Student's t-test and that for (C )was Mann–Whitney U test.
-
Figure 4—figure supplement 2—source data 1
Numerical source data for relative gene expression Figure 4—figure supplement 2A, D and DNA methylation status (Figure 4—figure supplement 2C), and data for Kolmogorov–Smirnov test for Figure 4—figure supplement 2A, D. SPG: spermatogonia.
- https://cdn.elifesciences.org/articles/70322/elife-70322-fig4-figsupp2-data1-v1.xlsx
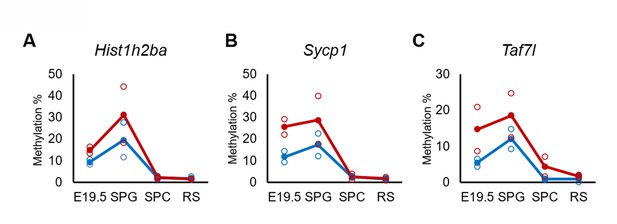
Changes in DNA methylation of the candidate epi-mutated genes in di(2-ethylhexyl) phthalate (DEHP) (red) and oil (blue)-treated E19.5 and adult F1 germ cell populations.
(A–C) Values of promoter methylation of each replicate (reduced representation bisulfite sequencing [RRBS], n = 2) of Hist1h2ba (A), Sycp1 (B), and Taf7l (C) are plotted by open red or blue circles, and means of two replicates are shown by closed circles.
-
Figure 5—source data 1
Numerical source data of DNA methylation level for Figure 5A–C.
SPG: spermatogonia; SPC: spermatocytes; RS: round spermatids.
- https://cdn.elifesciences.org/articles/70322/elife-70322-fig5-data1-v1.xlsx
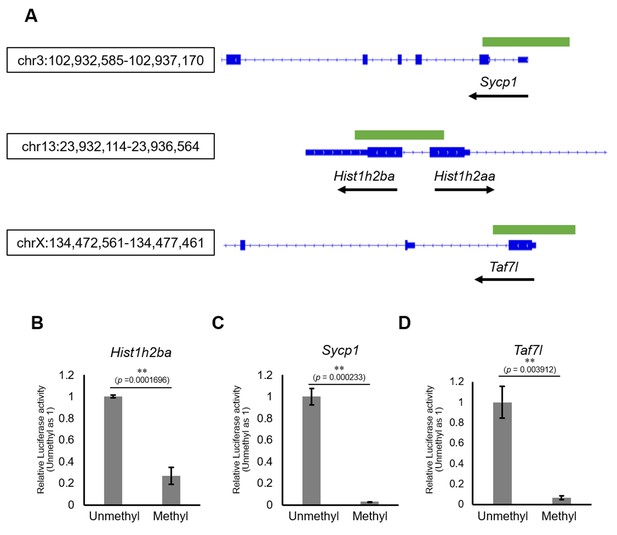
The effects on gene expression of promoter methylation in the candidate epi-mutated loci.
(A) Gene structures of Hist1h2ba, Sycp1, and Taf7l. Exons are indicated by blue bars, and the promoter regions (defined as ± 500 bp from transcription start site) used for firefly luciferase (Luc) assays are indicated by green bars. (B–D) The Luc activity of the reporter vector with either methylated or unmethylated promoters of Hist1h2ba (B), Sycp1 (C), and Taf7l (D) in HEK293T cells. Luc activity generated by the indicated vector was normalized to that generated by the Renilla phRL-TK vector. The Luc activity by the unmethylated vectors was defined as 1.0. Values are plotted as mean ± SEM of three independent experiments (n = 3). **p<0.01 (unpaired two-sided Student's t-test).
-
Figure 6—source data 1
Numerical source data for relative luciferase activity and data for Kolmogorov–Smirnov test for Figure 6B–D.
- https://cdn.elifesciences.org/articles/70322/elife-70322-fig6-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus, males) | Oct4-delta PE-GFP | PMID:10646797 | C57BL/6J background | |
| Cell line (Human) | HEK293T | ATCC RRID:CVCL_0063 | CRL-3216 | |
| Antibody | Anti-Cleaved Caspase-3 (rabbit polyclonal) | CST RRID:AB_2341188 | Cat# 9661 | (1:400) |
| Recombinant DNA reagent | pCpGL (plasmid) | PMID:17965610 | http://www.ag-rehli.de/materials.html | |
| Commercial assay or kit | EZ DNA Methylation-Gold Kit | Zymo Research | Cat# D5005 | |
| Commercial assay or kit | NEBNext Ultra II RNA Library Prep Kit for Illumina | BioLabs | Cat#E7770S | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay System | Promega | Cat#E1910 | |
| Chemical compound, drug | DEHP | Sigma | Cat#80030 | |
| Chemical compound, drug | Corn oil | Sigma | Cat#C8267 | |
| Software, algorithm | Bismark | PMID:21493656 | v0.10.1 | https://www.bioinformatics.babraham.ac.uk/projects/bismark/ |
| Software, algorithm | TopHat2 | PMID:23618408 | V2.1.0 | https://ccb.jhu.edu/software/tophat/index.shtml |
| Software, algorithm | Bowtie2 | PMID:22388286 | V2.2.6.0 | http://bowtie-bio.sourceforge.net/index.shtml |
| Software, algorithm | featureCounts | PMID:24227677 | Packaged in Subread v1.5.0-p2 | http://subread.sourceforge.net/ |
| Software, algorithm | edgeR | PMID:19910308 | V3.13 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
Additional files
-
Supplementary file 1
Methylation levels (%) of various genomic regions.
- https://cdn.elifesciences.org/articles/70322/elife-70322-supp1-v1.xlsx
-
Supplementary file 2
List of hyper- (DEHP-oil [%] > 5) and hypo- (DEHP-oil [%] < −5) methylated genes in E19.5 testicular germ cells and somatic cells for Figure 3.
- https://cdn.elifesciences.org/articles/70322/elife-70322-supp2-v1.xlsx
-
Supplementary file 3
List of hyper- (DEHP-oil [%] > 5) and hypo- (DEHP-oil [%] < −5) methylated genes in spermatogonia, spermatocyte, and round spermatid for Figure 3.
- https://cdn.elifesciences.org/articles/70322/elife-70322-supp3-v1.xlsx
-
Supplementary file 4
List of down- and upregulated genes in E19.5 testicular germ cells, and spermatogonia, spermatocyte, and round spermatid in adult testis (log2-fold-change > 1 or < −1, FDR < 0.05) for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/70322/elife-70322-supp4-v1.xlsx
-
Supplementary file 5
Primers used in this study (for Figure 2, Figure 4, Figure 6, and Figure 4—figure supplement 2).
- https://cdn.elifesciences.org/articles/70322/elife-70322-supp5-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70322/elife-70322-transrepform-v1.docx





