Structural basis for membrane recruitment of ATG16L1 by WIPI2 in autophagy
Figures
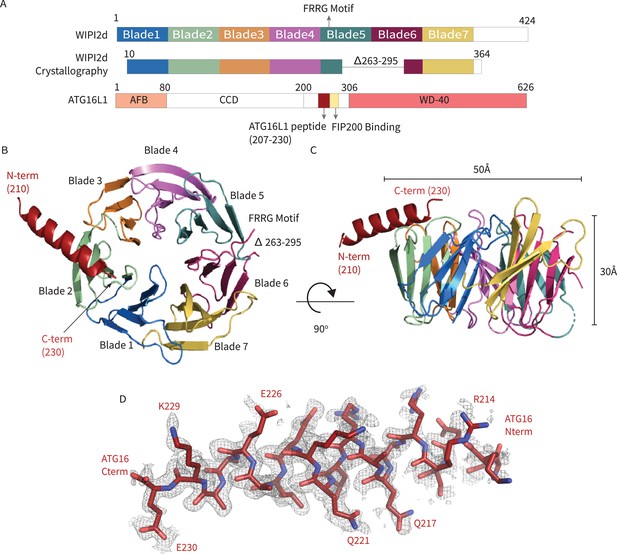
WIPI2d:ATG16L1 W2IR structure.
Structure of WIPI2d bound to ATG16L1 W2IR. (A) Annotated WIPI2d and ATG16L1 domain schematics. WIPI2d construct for crystallography is shown and W2IR from ATG16L1. (B, C) The ribbon diagram of the WIPI2d complex with ATG16L1 W2IR from the (B) bottom and (C) side views. Each blade is colored in accordance with (A). (D) Composite omit map of ATG16L1 W2IR. Modeled ATG16L1 is shown as red carton and the composite omit 2mFo-DFc map contoured at 1σ is shown in gray.
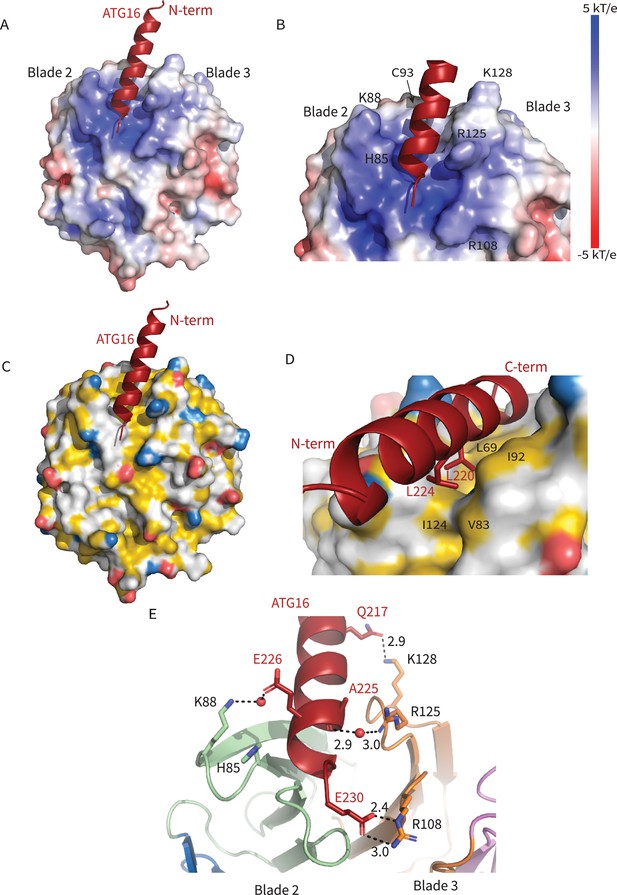
Interactions at the interface.
Analysis of the Interface. (A) Overall electrostatic surface and (B) closer view of electrostatic surface with ATG16 W2IR shown as a cartoon and key residues labeled. (C) Overall hydrophobic surface of WIPI2d and (D) closer view of the hydrophobic interface with key residues labeled where yellow represents hydrophobic regions. (E) A cartoon and stick representation of hydrogen bonds between ATG16 and WIPI2d shown as black dotted lines with distances noted and key residues shown as sticks.
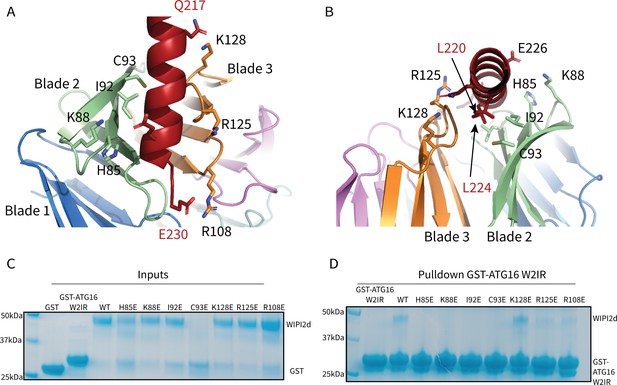
WIPI2d Interfacial mutants decrease ATG16L1 binding.
Key interacting residues shown as sticks in cartoon representation of WIPI2d:ATG16L1 interface shown from (A) the WIPI2d face or (B) down the ATG16L1 helix. (C) Inputs for the (D) Pull-down assays of mutant WIPI2d constructs and wild type with GST-ATG16L1 W2IR. GSH resin was used to pull-down GST-ATG16L1 W2IR from purified protein mixture. The pull-down results were performed in triplicates and visualized by SDS–PAGE and Coomassie blue staining.
-
Figure 3—source data 1
Uncropped SDS-PAGE gels for Figure 3.
Uncropped gel used in Figure 3C,D with lanes labeled similarly.
- https://cdn.elifesciences.org/articles/70372/elife-70372-fig3-data1-v3.zip
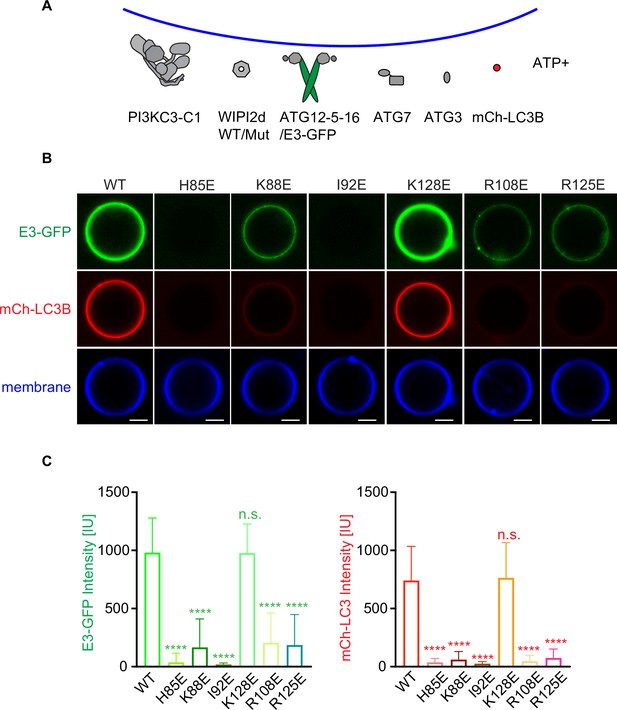
WIPI2d mutants disrupt E3 recruitment and LC3 lipidation on GUVs.
(A) The schematic drawing illustrates the reaction setting. Colors indicate fluorescent protein-fused components. Components in gray are not labeled but are present in the reaction. (B) Representative confocal images of GUVs showing E3 membrane binding and LC3B lipidation. PI3KC3-C1, WIPI2d WT or mutant, E3-GFP, ATG7, ATG3, mCherry-LC3B, and ATP/Mn2+ were incubated with GUVs (64.8% DOPC: 20% DOPE: 5% DOPS: 10% POPI: 0.2% Atto647 DOPE) at room temperature. Images taken at 30 min were shown. Scale bars, 10 µm. (C) Quantification of relative intensities of E3-GFP and mCherry-LC3B on GUV membranes in (A) (means ± SDs are shown; N = 40). p≥0.5: (ns); 0.01<p<0.05: (*); 0.001<p<0.01: (**); p<0.001 (***); p<0.0001 (****).
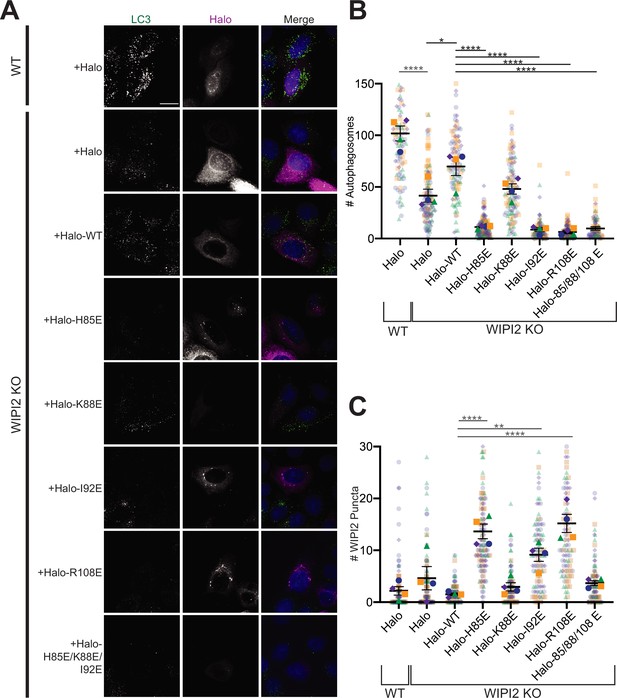
Altering the electrostatic interface of WIPI2 impairs starvation-induced autophagy in MEFs.
(A) Representative maximum projections of LC3 staining and either Halo or Halo-WIPI2 signal in WT or WIPI2 knockout (KO) cells (indicated on left) following 2 hr of starvation and 100 nM BafA treatment. Scale bar 15 µm. (B) Number of LC3-positive autophagosomes in either WT cells transfected with Halo, WIPI2 KO cells transfected with Halo, or WIPI2 KO cells transfected with the indicated Halo-tagged WIPI2 construct (labeled by mutation). (C) Number of discrete WIPI2 puncta under the conditions described in (B), measured using maximum projections of Halo-tag fluorescence. Experimental replicates are color-coded, with translucent dots representing individual measurements from each replicate and opaque dots, the corresponding arithmetic mean of that replicate. Error bars ± SEM; n = 4 independent experiments; *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 based on a one-way ANOVA with Tukey’s multiple comparisons between all conditions.
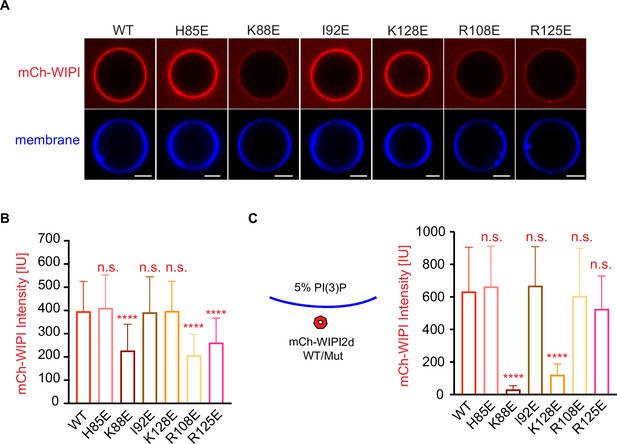
Reconstitution of membrane recruitment of WIPI2d mutants.
(A) Representative confocal images of GUVs showing membrane binding of mCherry-WIPI2d. PI3KC3-C1, mCherry-WIPI2d WT or mutant, E3-GFP were incubated with GUVs (64.8% DOPC: 20% DOPE: 5% DOPS: 10% POPI: 0.2% Atto647 DOPE) at room temperature. Images taken at 30 min were shown. Scale bars, 10 µm. (B) Quantification of relative intensities of mCherry-WIPI2d on GUV membranes in (A) membranes (means ± SDs are shown; N = 40). (C) Quantification of confocal images of GUVs (69.8% DOPC: 20% DOPE: 5% DOPS: 5% DOPI(3)P: 0.2% Atto647 DOPE) showing membrane binding of mCherry-WIPI2d. mCherry-WIPI2d WT or mutant were incubated with GUVs at room temperature for 30 min and then imaged. (Means ± SDs are shown; N = 40). p≥0.5: (ns); 0.01<p<0.05: (*); 0.001<p<0.01: (**); p<0.001 (***); p<0.0001 (****).
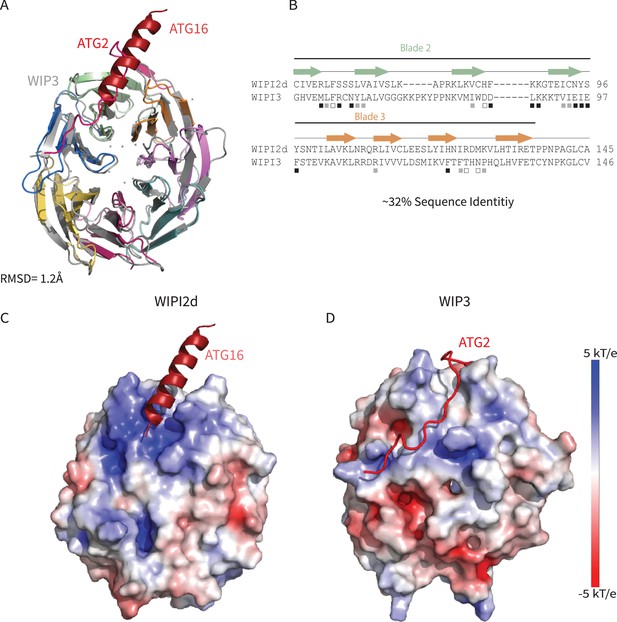
Comparing WIPI2d and WIPI3 structures and binding modes.
Comparison of WIPI2d and WIPI3. Alignment of WIPI2d and WIPI3 (A) structure and (B) sequence based on structures with W2IR residues denoted with white squares, W34IR with black, and from both with gray. Electrostatic surface comparison of (C) WIPI2d and (D) WIPI3.
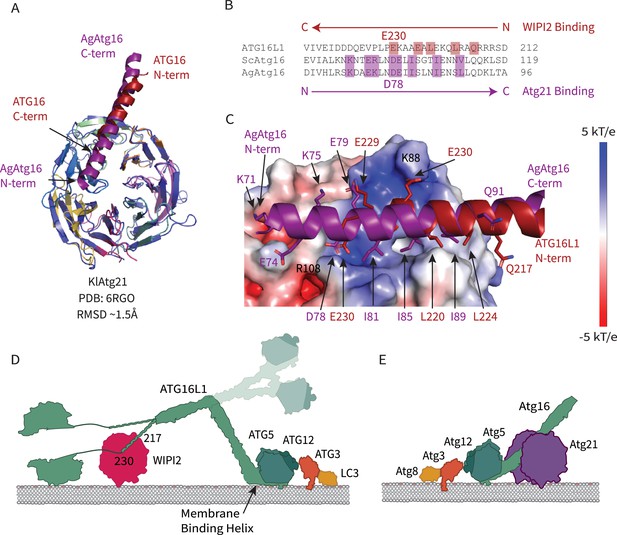
Comparison of WIPI2 and Atg21 binding to Atg16.
(A) Structural alignment of WIPI2 (PDB: 7MU2) and Atg21 (PDB: 6RGO; indigo) structures bound to Atg16. (B) Sequence alignment of Atg16 ß-propeller binding residues based on structure. Residues for WIPI2 and Atg21 binding are in highlighted in red and purple, respectively. (C) Electrostatic potential of Atg21 with overlay of AgAtg16 and ATG16L1 in purple and red, respectively. Key interacting residues are shown in sticks and labeled. Model of ATG12–5-16 performing LC3 lipidation on the autophagic membrane with (D) WIPI2 recruitment in humans with Helix one membrane binding is labeled (Lystad et al., 2019), and a secondary upward conformation is shown in faded colors versus (E) Atg21 recruitment in yeast.
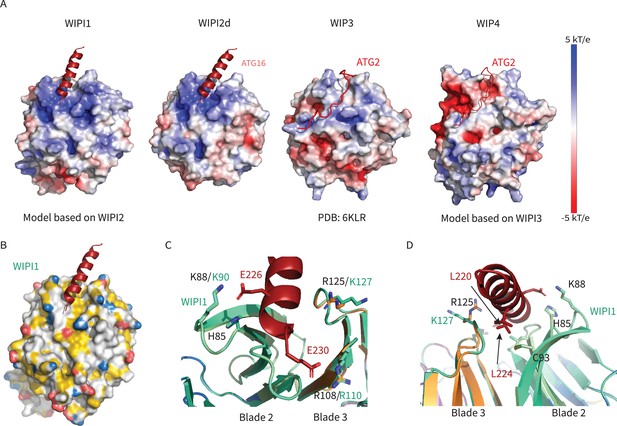
WIPI1-4 comparison.
Comparison of electrostatic surface potential of (A) WIPI1-4. (B) Hydrophobic surface of WIPI1 with predicted ATG16L1 W2IR shown as cartoon. (C, D) Alignment of WIPI2d crystal structure and WIPI1 homology structure with WIPI1 shown as light green and key residues labeled in the same color as structure.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HeLa human epithelial cell line | ATCC | CCL-2 | Authenticated by STR profiling; tested negative for mycoplasma |
| Cell line (Homo sapiens) | WIPI2-KO cells: HeLa cell line gene-edited to knockout WIPI2 expression | Fischer et al., 2020 | ||
| Recombinant DNA reagent | pHTC HaloTag | Promega | G7711 | |
| Recombinant DNA reagent | Halo-WIPI2-WT (Homo sapiens) plasmid for transfection | Stavoe et al., 2019 | Addgene 175025 | Available from Addgene |
| Recombinant DNA reagent | Halo-WIPI2-H85E (Homo sapiens) plasmid for transfection | Modified from Halo-WIPI2-WT in Stavoe et al., 2019 | Addgene 175027 | Available from Addgene |
| Recombinant DNA reagent | Halo-WIPI2-K88E (Homo sapiens) plasmid for transfection | Modified from Halo-WIPI2-WT in Stavoe et al., 2019 | Addgene 175028 | Available from Addgene |
| Recombinant DNA reagent | Halo-WIPI2-I92E(Homo sapiens) plasmid for transfection | Modified from Halo-WIPI2-WT in Stavoe et al., 2019 | Addgene 175029 | Available from Addgene |
| Recombinant DNA reagent | Halo-WIPI2-R108E (Homo sapiens) plasmid for transfection | Stavoe et al., 2019 | Addgene 176004 | Available from Addgene |
| Recombinant DNA reagent | Halo-WIPI2-H85/K88/I92E (Homo sapiens) plasmid for transfection | Modified from Halo-WIPI2-WT in Stavoe et al., 2019 | Addgene 175033 | Available from Addgene |
| Antibody | Anti-LC3B, (Rabbit polyclonal) primary antibody | Abcam | Cat.#ab48394 | IF (1:1000) |
| Antibody | Anti-Rabbit AlexaFluor488, (Goat polyclonal) secondary antibody | ThermoFisher | Cat.#A11034 | IF (1:1000) |
| Chemical compound, drug | TMRDirect Halo Ligand | Promega | Cat.#G2991 | 37.5 nM final concentration |
| Software, algorithm | FIJI | PMID:22743772 | ||
| Software, algorithm | Ilastik | PMID:31570887 | ||
| Software, algorithm | Adobe Illustrator 2021 | Adobe | ||
| Software, algorithm | Prism 9 | GraphPad | ||
| Other | 35 mm #1.5 glass bottom imaging dishes | MatTek | Cat.# P35G-1.5–20 C | |
| Other | EBSS | ThermoFisher | Cat.# 24010043 | |
| Cell line (Homo sapiens) | HEK GnTi | ATCC | CRL-3022 | |
| Recombinant DNA reagent | pCAG-WIPI2d-cs-TEV | Fracchiolla et al., 2020 | Addgene 171419 | |
| Recombinant DNA reagent | pCAG-WIPI2d10-364Δ263–295-cs-TEV | This paper | Addgene 171830 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-WIPI2dH85E-cs-TEV | This paper | Addgene 171831 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-WIPI2dK88E-cs-TEV | This paper | Addgene 171832 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-WIPI2dI92E-cs-TEV | This paper | Addgene 171833 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-WIPI2dC93E-cs-TEV | This paper | Addgene 171834 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-WIPI2dR108E-cs-TEV | This paper | Addgene 171835 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-WIPI2dR125E-cs-TEV | This paper | Addgene 171836 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-WIPI2dK128E-cs-TEV | This paper | Addgene 171837 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-mcherry-WIPI2d-cs-TEV | Fracchiolla et al., 2020 | Addgene 178912 | |
| Recombinant DNA reagent | pCAG-mcherry-WIPI2dH85E-cs-TEV | This paper | Addgene 171838 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-mcherry-WIPI2dK88E-cs-TEV | This paper | Addgene 171839 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-mcherry-WIPI2dI92E-cs-TEV | This paper | Addgene 171840 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-mcherry-WIPI2dC93E-cs-TEV | This paper | Addgene 171841 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-mcherry-WIPI2dR108E-cs-TEV | This paper | Addgene 171842 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-mcherry-WIPI2dR125E-cs-TEV | This paper | Addgene 171843 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pCAG-mcherry-WIPI2dK128E-cs-TEV | This paper | Addgene 171844 | Materials and methods section: Plasmids |
| Recombinant DNA reagent | pLEXm-GST-TEV-ATG14 | Addgene 99329 | ||
| Recombinant DNA reagent | pCAG-TSF-TEV-BECN1 | Addgene 99328 | ||
| Recombinant DNA reagent | pCAG-TSF-TEV-VPS34 | Addgene 99327 | ||
| Recombinant DNA reagent | pCAG-VPS15 | Stjepanovic et al., 2017 | Addgene 99326 | |
| Recombinant DNA reagent | pGBdest-ATG12-10xHis-TEV-ATG5-10xHis-TEVcs-ATG16L1-GFP-TEVcs-StrepII, ATG7, ATG10 | Fracchiolla et al., 2020 | Addgene 169077 | |
| Recombinant DNA reagent | pFast BacHT(B)–6xHis-TEV-ATG7 | Fracchiolla et al., 2020 | ||
| Recombinant DNA reagent | pET Duet-1-6xHis-TEV-ATG3 | Fracchiolla et al., 2020 | Addgene 169079 | |
| Recombinant DNA reagent | pET Duet-1-6xHis-TEV-mCherry-LC3B-Gly(∆5 C) | Zaffagnini et al., 2018 | Addgene 169168 | |
| Other | 96–2 well,INTELLI-PLATE (original) tray | Molecular Dimensions, Maumee, OH | ||
| Other | Greiner pre-greased 24 well Combo Plate (SBS format) with lid | Molecular Dimensions, Maumee, OH | ||
| Software, algorithm | Nikon Elements microscope imaging software 4.60 | Nikon Corporation, Tokyo, Japan | https://www.nikoninstruments.com/Products/Software/NIS-Elements-Advanced-Research/NIS-Elements-Viewer | |
| Other | Glutathione Sepharose 4B GST-tagged protein purification resin | GE healthcare, Chicago, IL | Cat# 17075605 | |
| Other | Strep-Tactin Superflow high capacity 50 % suspension | IBA Lifesciences,Göttingen, Germany | Cat# 2-1208-010 | |
| Software, algorithm | phenix.refine | PMID:20124702, 22505256, 31588918 | RRID:SCR_016736 | |
| Software, algorithm | XDS | PMID:20124692 | RRID:SCR_015652 | |
| Software, algorithm | POINTLESS | PMID:21460446 | RRID:SCR_014218 | |
| Software, algorithm | PHASER | PMID:19461840 | RRID:SCR_014219 | |
| Software, algorithm | PyMol | PyMol (pymol.org) | RRID:SCR_000305 | |
| Software, algorithm | APBS | PMID:11517324 | RRID:SCR_008387 | |
| Software, algorithm | SWISS-MODEL | PMID:29788355 | RRID:SCR_018123 | |
| Software, algorithm | Coot | PMID:20383002 | RRID:SCR_014222 | |
Additional files
-
Supplementary file 1
Data collection and refinement statistics.
- https://cdn.elifesciences.org/articles/70372/elife-70372-supp1-v3.docx
-
Supplementary file 2
Oligos used for cloning.
- https://cdn.elifesciences.org/articles/70372/elife-70372-supp2-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70372/elife-70372-transrepform1-v3.docx





