BRAFV600E induces reversible mitotic arrest in human melanocytes via microRNA-mediated suppression of AURKB
Figures
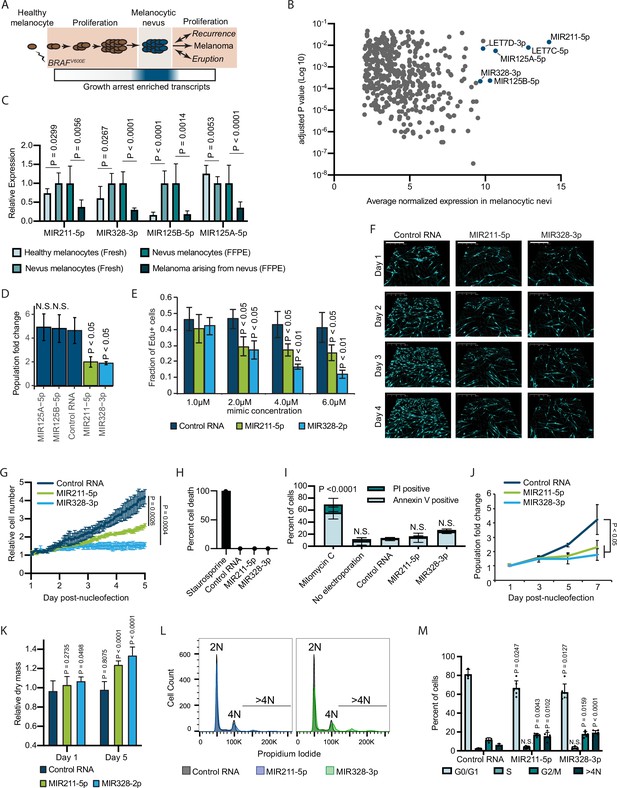
Melanocytic nevus enriched miRNAs induce arrest in primary human melanocytes.
(A) Schematic of hypothesized plastic nature of BRAFV600E- and nevus-associated proliferation arrest. Expression of the predicted nevus enriched transcriptional program is highlighted in blue. (B) Scatter plot depicting gene expression combining publicly available sequencing data sets totaling 14 melanoma-arising-from-nevus specimens, dissected into benign and malignant portions (28 total samples). Plotted are the adjusted p values (DESeq2) comparing matched nevus and melanoma samples against average normalized expression in nevus portion. Only genes exhibiting both high expression and significant differential expression in nevus portions are plotted (see Materials and methods). The full list of genes is included as Supplementary file 1. (C) Left two columns, the relative expression of indicated miRNAs in freshly isolated human melanocytes from BRAFV600E melanocytic nevi (n=6) compared to BRAFWT healthy skin melanocytes (n=8). Right two columns, the relative expression of indicated miRNAs in melanocytes dissected from FFPE nevus (n=7) or melanoma (n=7) specimens. Data represent mean and standard deviation of normalized RNA sequencing read counts relative to nevus-derived samples. (D) Mean and standard deviation for population fold change over 7 days of melanocytes nucleofected with indicated miRNA mimics compared to non-targeting control (Control RNA) (n=3). (E) Mean and standard deviation for fraction of EdU positive cells 4 days after nucleofection with indicated concentrations of miRNA mimics (n=3). P values indicate comparisons to concentration matched control. (F) Representative QPI images of melanocytes nucleofected with indicated miRNA mimics. Pixel color indicates low (black), mid (blue), or high (red) optical density. Scale bar=200 µm. (G) Mean and standard deviation for relative QPI-derived cell number over time (n=3). (H) Mean and standard deviation for QPI-derived cell death counts as percentage of total cells imaged on day 1 (n=3). Cells treated with 0.1 µM staurosporine were included as a control (n=1) for induction of cell death. (I) Mean and standard deviation for percent Annexin V/PI positive cells nucleofected with indicated miRNA mimics (n=3). Cells treated with 5 µg/ml mitomycin C were included as a control (n=3) for induction of apoptosis. (J) Mean and standard deviation for relative cell number over time (n=3). (K) Mean and standard deviation for relative QPI-derived dry mass per cell (n=3). (L) Representative histograms of DNA content profiling via propidium iodide incorporation measured by flow cytometry 5 days post-nucleofection of miRNA mimics. (M) Mean and standard deviation for percent of cells in indicated phases of cell cycle based upon profiles as in (K) (n=3–6, individual datapoints shown). P values for D-L calculated by unpaired t-tests comparing experimental to Control RNA. N.S, not significant (p≥0.05).
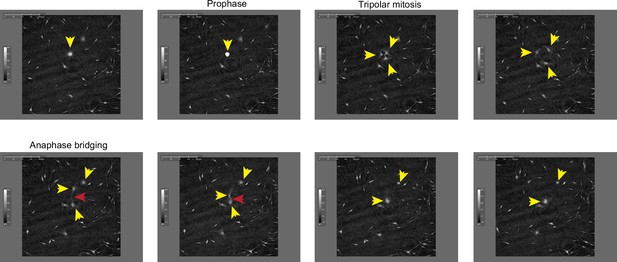
Still frames from Video 1 documenting representative cytokinesis failure in primary melanocytes transduced with MIR211-5p.
Yellow arrowheads follow a single cell and its progeny through a tripolar mitosis. Red arrowhead follows an anaphase bridge connecting two daughter cells that then remerge.
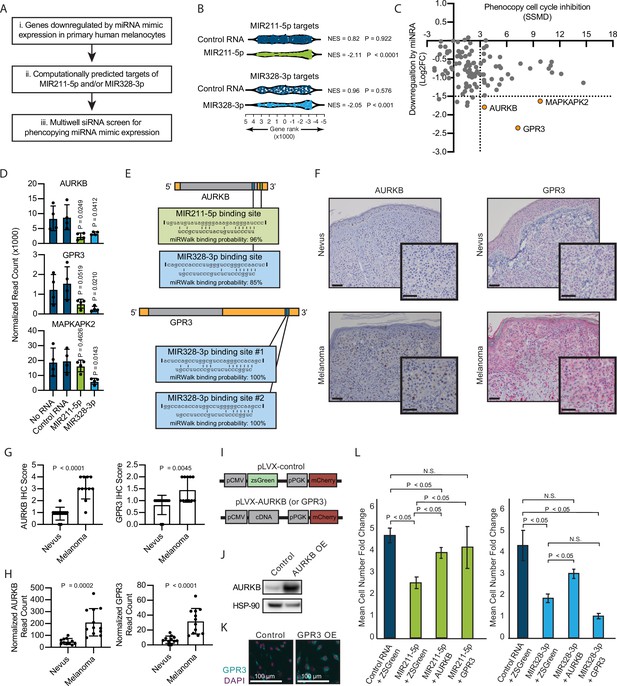
Inhibition of AURKB by MIR211-3p and MIR328-5p restricts proliferation in nevi.
(A) Schematic of experimental and computational pipeline for identifying targets of MIR211-3p and/or MIR328-3p responsible for proliferation arrest. (B) GSEA analysis comparing predicted MIR211-5p and MIR328-3p target mRNAs to changes in gene expression after nucleofection with indicated mimics compared to nucleofection control. Supplementary file 3 provides full differentially expressed gene lists. (C) Plot of siRNA screen results for phenocopying proliferation arrest versus log2 fold change in expression after miRNA mimic nucleofection. Strictly standardized mean difference (SSMD) reports the magnitude and significance of inhibition of Edu incorporation (n=3, see Supplementary file 4 for list of genes and screen results). Dotted lines indicate cutoff values for further evaluation. Orange points indicate genes meeting cutoff criteria. (D) Mean and standard deviation for expression of indicated genes after nucleofection of indicated mimics (n=4). P values calculated by unpaired t-tests compared to no RNA. (E) Predicted binding sites and computed binding probabilities of MIR211-5p and MIR328-3p in AURKB and GPR3 transcripts. (F) Representative images of immunohistochemical staining for AURKB (brown chromagen) and GPR3 (red chromagen) expression in FFPE samples of nevi and melanoma. Scale bars=50 µm. (G) Mean and standard deviation for immunohistochemical staining scores as in (F) for cohorts of 11 nevi and 11 melanomas. P values calculated by unpaired t-tests. (H) Mean and standard deviation for read counts for cohorts of 11 nevi and 12 melanomas. P values calculated by unpaired t-tests. (I) Design of pLVX vectors expressing either AURKB or GPR3. (J) Western blot of AURKB expression in human melanocytes with or without lentiviral AURKB overexpression. HSP90 is the loading control. Source data provided as Supplementary files (Figure 3—source data 1). (K) Representative photomicrographs (20×) of immunofluorescence for GPR3 (green) or (DAPI) (purple) in human melanocytes with GPR3 lentiviral overexpression (GPR3 OE) or without (Control) overexpression. (L) Mean and standard deviation for cell number fold change over 7 days after nucleofection with indicated miRNA mimics in melanocytes transduced with lentiviral constructs expressing zsGreen, AURKB, or GPR3 (n=6 melanocyte preps, type 2 t test, N.S., not significant (p≥0.05)). P values calculated by unpaired t-tests. GSEA, gene set enrichment analysis; NES, normalized enrichment score; p, nominal p value.
-
Figure 2—source data 1
Zip files containing raw and annotated images of Western blots.
- https://cdn.elifesciences.org/articles/70385/elife-70385-fig2-data1-v2.zip
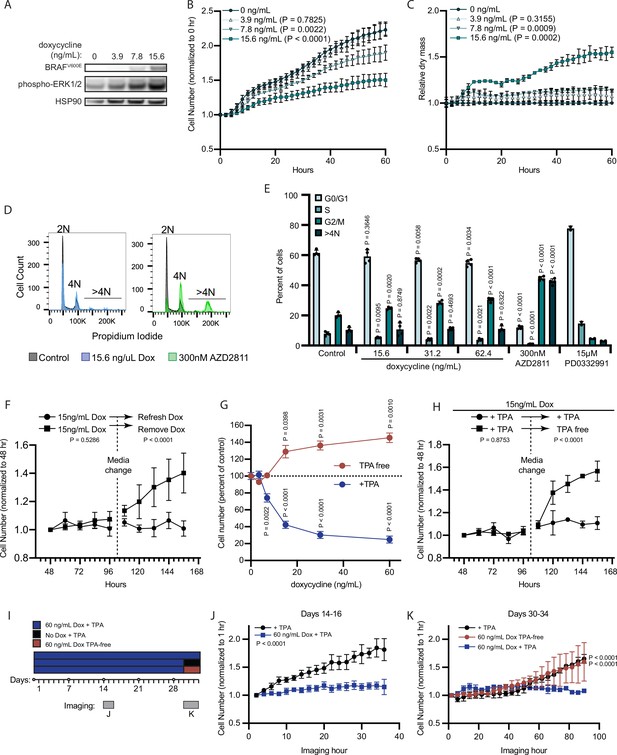
BRAFV600E induces a reversible and conditional arrest in human melanocytes.
(A) Representative (of n=3) Western blot analysis of BRAFV600E, phospho-ERK1/2, and HSP90 (loading control) in diBRAFV600E melanocytes in response to increasing concentration of doxycycline. Source data provided in Supplementary files (Figure 3—source data 1). (B) Mean and standard deviation for relative QPI-derived cell number at indicated hours of diBRAFV600E melanocytes in response to increasing concentration of doxycycline (n=6). P values from unpaired t-tests compared to 0 ng/ml condition. (C) Mean and standard deviation for relative QPI-derived dry mass per cell of diBRAFV600E melanocytes in response to increasing concentration of doxycycline at indicated hours (n=6). P values from unpaired t-tests compared to 0 ng/ml condition. (D) Representative histograms of DNA content profiling via propidium iodide incorporation measured by flow cytometry 5 days after treatment with indicated molecules. AZD2811 included as a control for induced cytokinesis failure. (E) Mean and standard deviation for percent of cells in indicated phases of cell cycle after treatment with doxycycline based upon profiles as in (D) (n=4). AZD2811 (n=4) is a control for induced cytokinesis failure. PD0332991 (n=2) is a control for G0/G1 arrest. P values from unpaired t-tests compared to Control (no treatment) condition. (F) Mean and standard deviation for relative QPI-derived cell number of diBRAFV600E melanocytes at indicated hours after treatment doxycycline. Dotted line indicates change in media to either refresh doxycycline or add media without doxycycline (n=3). P values from unpaired t-tests comparing final timepoints. (G) Mean and standard deviation for cell number as percent of control of diBRAFV600E melanocytes cultured with or without TPA after 5 days of treatment with indicated concentrations of doxycycline. P values from unpaired t-tests compared to 0 ng/ml conditions. Baseline population doubling times were 3.2 days (+TPA) and 4.2 days (TPA-free). (H) Mean and standard deviation for relative QPI-derived cell number of diBRAFV600E melanocytes at indicated hours after treatment doxycycline. Dotted line indicates change in media to either refresh TPA-containing media or add media without TPA (n=3). P values from unpaired t-tests comparing final timepoints. (I) Schematic of experimental design testing the reversibility of growth arrest after 30 days exposure to high doxycycline (60 ng/ml). Days of live imaging represented in (J, K) are indicated by gray bars. (J) Mean and standard deviation for relative QPI-derived cell number of diBRAFV600E melanocytes at 14 days after addition of doxycycline (n=3). P values from unpaired t-tests comparing final timepoints. (K) Mean and standard deviation for relative QPI-derived cell number of diBRAFV600E melanocytes at 30 days after addition of doxycycline (n=3). Media was changed just prior to imaging to either refresh doxycycline (blue), change to +TPA media without doxycycline (black), or change to TPA-free media with doxycycline (red). P values from unpaired t-tests comparing final time points. QPI, quantitative phase imaging; TPA, tetradecanoylphorbol acetate.
-
Figure 3—source data 1
Zip files containing raw and annotated images of Western blots.
- https://cdn.elifesciences.org/articles/70385/elife-70385-fig3-data1-v2.zip
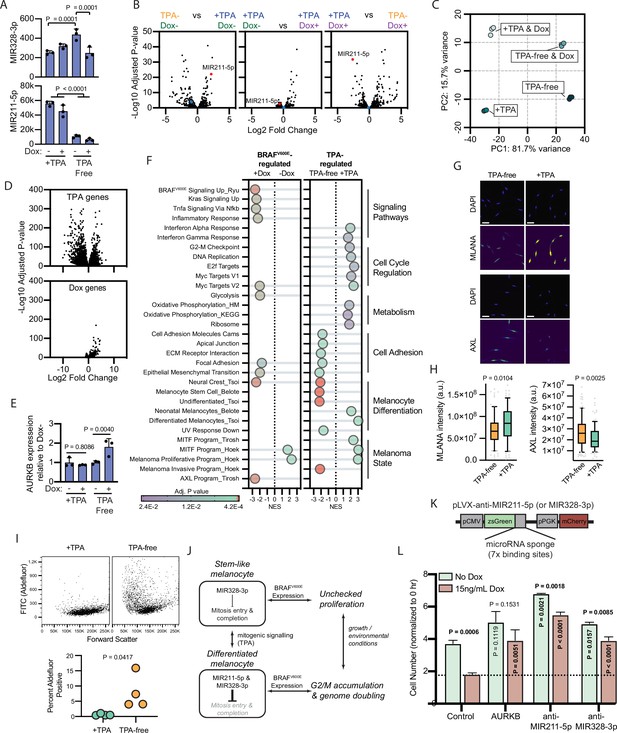
BRAFV600E-induced arrest is dependent on melanocyte growth conditions and differentiation state.
(A) Normalized read counts for MIR328-3p (top) and MIR211-5p (bottom) in TPA-containing (+TPA) or TPA-free media, with or without 15.6 ng/ml doxycycline (Dox). See Supplementary file 5 for individual values and exact p values. (B) Volcano plots depicting log2 fold change against adjusted p value (−log10) from differential expression (DE) analysis of small RNA sequencing of indicated comparisons. MIR211-5p labeled and shown in red. MIR328-3p shown in blue. (C) First and second principal components (PCs) separating diBRAFV600E melanocytes cultured in indicated conditions. (D) Volcano plots depicting log2 fold change against adjusted p value (−log10) from DE analysis of mRNA sequencing of all +TPA specimens versus all TPA-free specimens (top) or all Dox specimens versus all no-Dox specimens (bottom). (E) Normalized read counts for AURKB normalized to no Dox conditions (n=3). P values from unpaired t-tests. Sequencing data represented in (A–F) are from n=3 per condition analyzed with DESeq2. (F) Gene set enrichment analysis (GSEA) comparing DE gene sets from (D) to Molecular Signature Database Hallmark gene sets, KEGG Pathway gene sets and published signatures of melanocyte signaling and differentiation from Belote et al., 2021; Tsoi et al., 2018; Tirosh et al., 2016; Ryu et al., 2011; Hoek and Goding, 2010. All enrichments with adjusted p value<0.025 are depicted. See Supplementary file 6 for all associations. Normalized enrichment score (NES). Enrichment differences between pathways across the four conditions in (C) are depicted in Figure 4—figure supplement 1. (G) Representative photomicrographs of immunofluorescence for MLANA or AXL in human melanocytes cultured in +TPA or TPA-free conditions. White scale bar=30 µm. (H) Quantification of fluorescence intensity (arbitrary pixel intensity units, a.u.) from experiments represented in (G). Box (median, 25th and 75th percentiles) and whiskers (10th and 90th percentiles) of 84, 88, 113, and 96 cells, respectively. P values from unpaired t-tests. (I) Representative plots (top) and quantification (bottom, n=4) of relative aldehyde dehydrogenase activity of melanocytes grown in +TPA or TPA-free conditions. Normalized read counts for AURKB normalized to no Dox conditions (n=3). P values from unpaired t-tests. Sequencing data represented in (A–G) from n=3 per condition analyzed with DESeq2. (J) Schematic of model supported by transcriptomic analyses. TPA induces a more differentiated state in human melanocytes. Due to increased MIR211-5p and base-line MIR328-3p expression, AURKB expression is capped, resulting in G2/M accumulation upon BRAFV600E expression. (K) Design of pLVX vectors expressing microRNA sponges fused to zsGreen. (L) Mean and standard deviation for cell number as percent of time point 0 hr after 7 days of culture in +TPA media with or without 15.6 ng/ml Dox (n=3 per condition). Melanocytes were transduced with either pLVX-control, -AURKB, -anti-MIR211-5p, or -anti-MIR328-3p. P values from unpaired t-tests compared to matched no Dox condition (horizontal), Control no Dox condition (vertical in green bar), or Control Dox condition (vertical in red bar). TPA, tetradecanoylphorbol acetate.
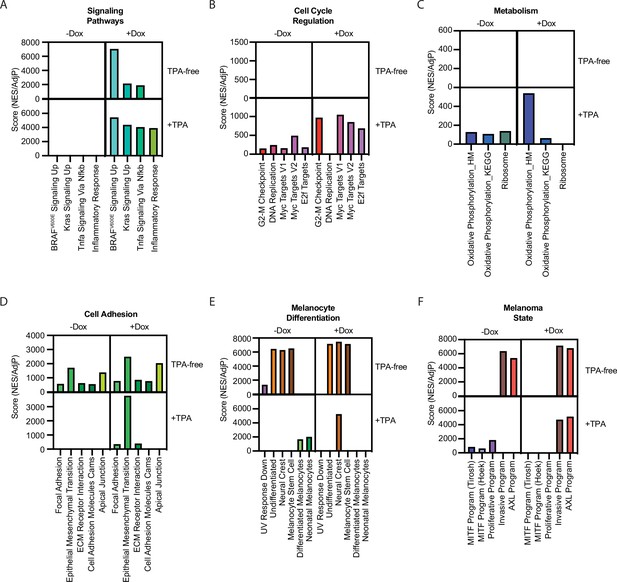
Summary of gene set enrichment for gene sets in Figure 1F across the four conditions in Figure 4C.
Pair-wise comparisons were conducted as in Figure 1F, comparing the +TPA Dox condition to other conditions. Scores were generated by dividing positive NES values by the AdjP value. For the +TPA Dox quadrant, scores were averaged when occurring in more than one pair-wise comparison. See Supplementary file 6 for complete information on all comparisons. NES, normalized enrichment score; TPA, tetradecanoylphorbol acetate.
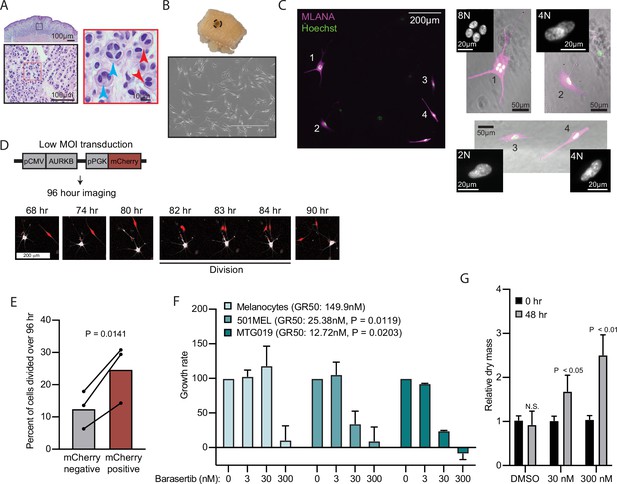
AURKB inhibition is critical for BRAFV600E-induced arrest of human nevi.
(A) Representative image of 15 H&E-stained melanocytic nevus. Border colors indicate two consecutive magnifications. Arrowheads indicate bi- (red) or multi- (blue) nucleation. (B) Example images of skin specimen containing a melanocytic nevus (circled) and phase-contrast microscopy of melanocytes isolated from nevus portion. Scale bar=400 µm. (C) Images of MLANA (purple) and Hoechst (green) co-staining of melanocytic nevus derived melanocytes in culture. Zoomed images show representative 2 N, 4 N, and 8 N cells. (D) Representative images of QPI coupled with fluorescence microscopy to identify adjacent mCherry-expressing and mCherry-negative melanocytes. Individual cells were identified at time zero, tracked, and monitored for division for 96 hr. (E) Mean and individual matched data points for the percent of n=3 mCherry-negative or mCherry-positive cells identified at time zero that divided over 96 hr. P value from paired t-test. (F) Mean and standard deviation for 48 hr growth rates of indicated cells treated with Barasertib. P values from unpaired t-tests comparing the 30 nM samples of melanoma lines to primary melanocytes (n=3). (G) Mean and standard deviation for relative QPI-derived dry mass per 501MEL cell at initial time point (black) compared to 48 hr treatment with Barasertib (gray) (n = 3). P values from unpaired t-tests. QPI, quantitative phase imaging.
Videos
Cytokinesis failure associated with MIR211-5p expression.
Four day time-lapse quantitative phase imaging of cytokinesis failure in primary melanocytes transduced with MIR211-5p.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | BRAF | https://www.ncbi.nlm.nih.gov/gene | Gene ID: 673 | |
| Cell line (H. sapiens) | Epidermal melanocytes (adult normal, adult nevus, and neonatal normal) | Other | Prep specific | Non-immortalized primary lines, derived from fresh skin. |
| Cell line (H. sapiens) | 501MEL, human melanoma | Boris Bastian | RRID:vCVCL_4633 | |
| Cell line (H. sapiens) | HCIMel019, human melanoma | This paper | HCIMel019 | Patient-derived melanoma line. Available from the HCI PRR core. |
| Biological sample (H. sapiens) | FFPE melanocytic tumor specimens | UCSF Dermatopathology Biospecimen Archive | De-identified | |
| Antibody | Anti-HSP90 (rabbit polyclonal) | CST | 4,874 | Western (1:1000) |
| Antibody | Anti-BRAFV600E (mouse monoclonal) | Spring Bioscience Corp | E19292; RRID:AB_11203851 | Western (1:1000) |
| Antibody | Anti-phosphoERK1/2 (rabbit monoclonal) | CST | 4,970 | Western (1:1000) |
| Antibody | Anti-AURKB (rabbit polyclonal) | Abcam | ab2254, RRID:AB_302923 | Western (1:1000); IHC (1:400) |
| Antibody | Anti-GPR3 (mouse monoclonal) | Abnova | H00002827-M01, RRID:AB_425462 | IHC/IF (1:125) |
| Antibody | Anti-AXL (rabbit monoclonal) | Cell Signalling Technology | 8661, RRID:AB_11217435 | IF (1:250) |
| Antibody | Anti-MLANA (mouse monoclonal) | Abcam | ab731, RRID:AB_305836 | IF (1:100) |
| Recombinant DNA reagent | pTRIPZ-diBRAFV600E | Todd Ridky | gift | |
| recombinant DNA reagent | pLVX-AURKB | This paper | Addgene #153,316 | See Materials and Methods: Generation of Lentiviral vectors. Available from Addgene. |
| recombinant DNA reagent | pLVX-GPR3 | This paper | Addgene #153,317 | See Materials and Methods: Generation of Lentiviral vectors. Available from Addgene. |
| recombinant DNA reagent | pLVX-anti-MIR211-5p | This paper | Addgene #153,318 | See Materials and Methods: Generation of Lentiviral vectors. Available from Addgene. |
| recombinant DNA reagent | pLVX-anti-MIR328-3p | This paper | Addgene #153,319 | See Materials and Methods: Generation of Lentiviral vectors. Available from Addgene. |
| recombinant DNA reagent | pLVX-Che-zsGreen | This paper | Addgene #153,320 | See Materials and Methods: Generation of Lentiviral vectors. Available from Addgene. |
| sequence-based reagent | hsa-MIR211-5p | Dharmacon | C-300566-03-0005 | |
| sequence-based reagent | hsa-MIR328-3p | Dharmacon | C-300695-03-0005 | |
| sequence-based reagent | MIRControl 1 (Control RNA) | Dharmacon | CN-001000-01-05 | |
| commercial assay or kit | ALDEFLUOR kit | Stemcell Technologies | 01700 | |
| chemical compound, drug | barasertib | Selleckchem | A1147 | |
| chemical compound, drug | doxycycline | Sigma | D9891 |
Additional files
-
Supplementary file 1
Results of differential expression analyses performed on previously published databases of matched melanocytic nevi and melanoma-arising-from-nevi.
- https://cdn.elifesciences.org/articles/70385/elife-70385-supp1-v2.xlsx
-
Supplementary file 2
Normalized read counts from small RNA sequencing performed on melanocytes derived from healthy human skin and melanocytic nevi.
- https://cdn.elifesciences.org/articles/70385/elife-70385-supp2-v2.xlsx
-
Supplementary file 3
Results of differential expression analyses performed on primary melanocytes nucleofected with microRNA or control mimics.
- https://cdn.elifesciences.org/articles/70385/elife-70385-supp3-v2.xlsx
-
Supplementary file 4
Results from siRNA screen for EdU incorporation performed in primary melanocytes.
Mean and standard deviation of triplicate screens shown for each gene. Column D indicates the microRNA predicted to targeted the gene.
- https://cdn.elifesciences.org/articles/70385/elife-70385-supp4-v2.xlsx
-
Supplementary file 5
Results of differential expression analyses performed on primary melanocytes with or without BRAFV600E in+ TPA or TPA-free conditions.
- https://cdn.elifesciences.org/articles/70385/elife-70385-supp5-v2.xlsx
-
Supplementary file 6
Results of GSEA analyses performed on primary melanocytes with or without BRAFV600E in+ TPA or TPA-free conditions.
- https://cdn.elifesciences.org/articles/70385/elife-70385-supp6-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70385/elife-70385-transrepform1-v2.pdf





