The enteric pathogen Cryptosporidium parvum exports proteins into the cytosol of the infected host cell
Figures
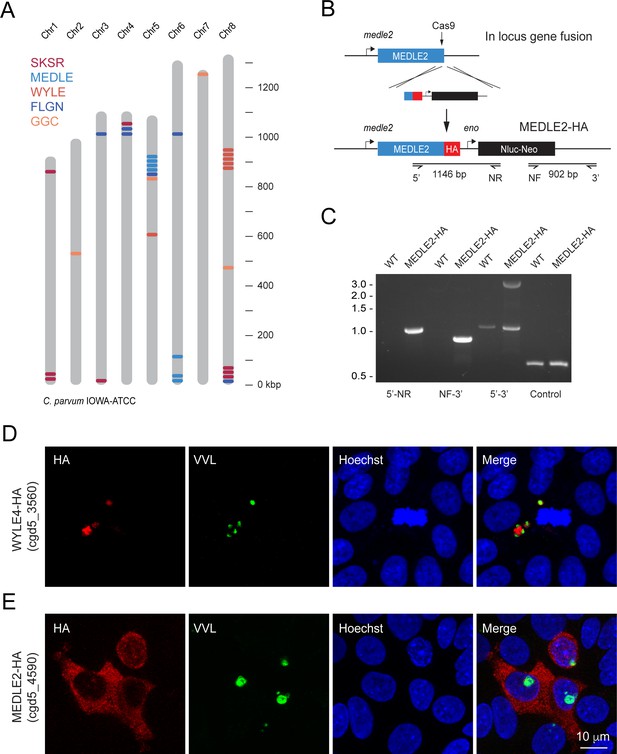
MEDLE2 is exported to the host cell cytoplasm.
(A) Schematic overview of the chromosomal location for polymorphic gene families in the C. parvum genome. (B) Map of the MEDLE2 locus targeted in C. parvum for insertion of a 3× hemagglutinin (HA) epitope tag, a nanoluciferase reporter gene (Nluc), and neomycin phosphotransferase selection marker (Neo). (C) PCR mapping of the MEDLE2 locus using genomic DNA from wild type (WT) and transgenic (MEDLE2-HA) sporozoites, corresponding primer pairs are shown in (B), and thymidine kinase (TK) gene used as a control. Note the presence of two bands in the 5′–3′ amplification, indicating the presence of a transgene (3081 bp) and persistence of an unmodified copy (1174 bp), suggesting multiple copies of MEDLE2 in the C. parvum genome; also see Figure 1—figure supplement 2. (D, E) HCT-8 cultures were infected with WYLE4-HA (D) or MEDLE2-HA (E) transgenic parasites and fixed after 24 hr for immunofluorescence assay (IFA). Red, antibody to HA; green, Vicia villosa lectin stain, VVL (Gut and Nelson, 1999); blue, Hoechst DNA dye. Additional genes targeted and the localizations of their products are summarized in Table 1 and Figure 1—figure supplements 1 and 3.
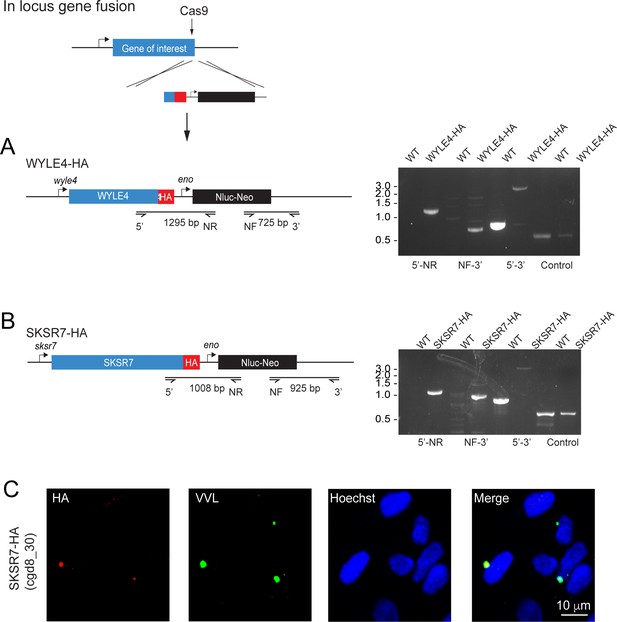
Additional secretory proteins tested in this study.
(A–C) Schematic depicting the generation of in-locus gene fusion transgenic parasite strains for WYLE4 (A) and SKSR7 (B). In each case, the C terminus of the gene was targeted for integration of a repair construct containing a hemagglutinin (HA) epitope tag, a nanoluciferase reporter gene (Nluc), and a neomycin phosphotransferase (Neo) selectable marker. Individual primer pairs used to map integration in wild type (WT) and transgenic strains are shown. (C) SKSR7-HA does not localize to the host cell; rather, the protein (red) exhibits expression in the parasite (green).
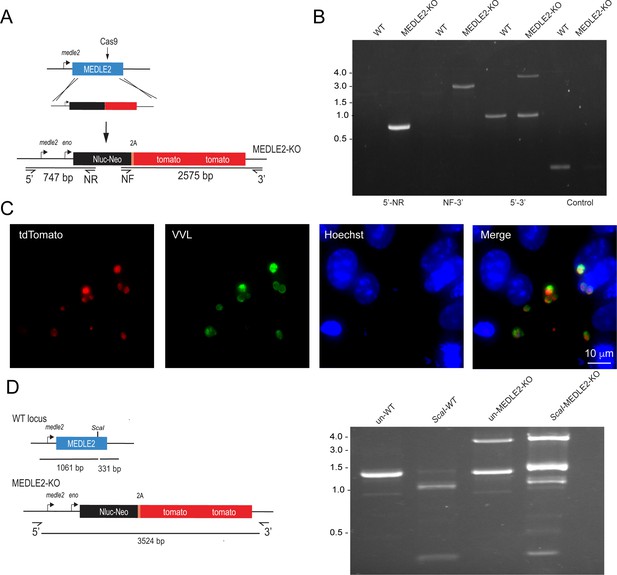
Knockout of MEDLE2 reveals multiple copies of the gene in the genome.
(A) Schematic representation for the strategy used to generate a MEDLE2 KO line, in which the entire locus of MEDLE2 is replaced with a nanoluciferase reporter gene (Nluc) and the neomycin phosphotransferase (Neo) selection marker fused to a 2A peptide, and tdTomato, such that the parasites express a red fluorescent protein in their cytoplasm. The solid black arrow indicates the position of the Cas9-induced double-stranded break in the middle of the gene. Note that this is a different guide from the one used for C-terminal tagging. (B) PCR mapping modification of the MEDLE2 locus using genomic DNA from wild type (WT) and transgenic (MEDLE2 KO) sporozoites using the primer pairs shown in (A) and the thymidine kinase (TK) gene as a control. Note the persistence of a WT band (1392 bp) in the 5′–3′ amplification product, despite the presence of the transgene (3524 bp). (C) HCT-8 cultures were fixed 24 hr after being infected with MEDLE2 KO transgenic parasites. MEDLE2 KO parasites exhibit red fluorescence in their cytoplasm as expected (red, tdTomato, parasite cytoplasm; green, parasites VVL; blue, Hoechst). (D) The full gene PCR products from WT (1392 bp) and MEDLE2 KO parasites (3524 bp) were used for restriction digest with ScaI. A single ScaI restriction site is found in the C terminus of WT MEDLE2; however, integration of the repair cassette disrupts this site. ScaI digested WT PCR product results in two digest products: 331 bp and 1061 bp. Undigested MEDLE2 KO full gene product has the expected 3524 bp fragment, as well as a persisting 1392 bp WT band. ScaI digested MEDLE2 KO shows the 3525 bp repair cassette resistant to ScaI digest, as well as the 331 bp and 1061 bp fragments produced from digest of the unmodified MEDLE2 locus. As a result, there are multiple copies of MEDLE2 in the genome and we have only targeted one for knockout.
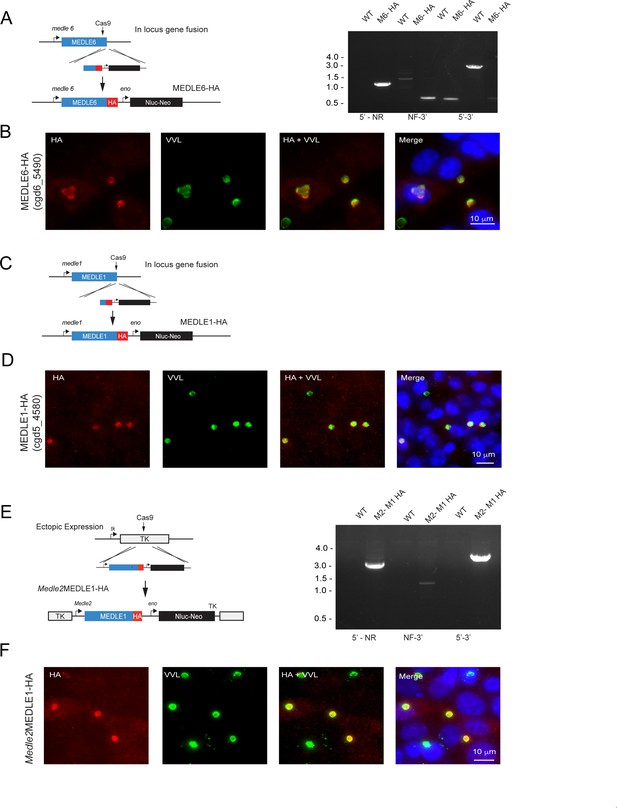
Other members of the MEDLE gene family are exported to the host cell.
(A) Schematic representation depicting the generation of a MEDLE6-HA transgenic parasite line, in which the endogenous locus of MEDLE6 (cgd6_5490) is a hemagglutinin (HA) epitope tagged at the C terminus. Proper integration at the desired locus was confirmed using PCR mapping with gDNA isolated from MEDLE6 transgenic parasites (M6) and a wild type control (WT). (B) MEDLE6-HA parasites were used to infect HCT-8 cells for an immunofluorescence assay. Cells were fixed every 12 hr and stained for immunofluorescence assay (IFA). Shown as a representative image, at 24 hr post infection, MEDLE6 (red) localizes in/around the parasite (green), as well as slightly in the host cell. Host cell expression is more apparent in multiply infected cells. (C) The MEDLE1 (cgd5_4580) locus was targeted for integration of an HA epitope tag at the C terminus. (D) MEDLE1-HA parasites were used to infect HCT-8 cells for a time-course infection, and IFA was performed on cells fixed every 12 hr. Shown as a representative image, at 12 hr post infection, MEDLE1 (red) localizes in/around the parasite (green), as well as at very low levels in the host cell. (E) Schematic representation for the strategy used to engineer a MEDLE1 overexpression line. The MEDLE2 promoter was used to drive expression of an ectopic copy of MEDLE1-HA expressed in the TK locus. Proper integration was assessed using PCR mapping with gDNA isolated from Medle2MEDLE1-HA (M2- M1 HA) and WT control (WT) parasites. (F) M2-M1 HA parasites were used to infect HCT-8 cells for IFA. At 24 hr post infection, MEDLE1-HA (red) can be seen in/around the parasite (green), as well as in the host cell when expression is driven by the MEDLE2 promoter.
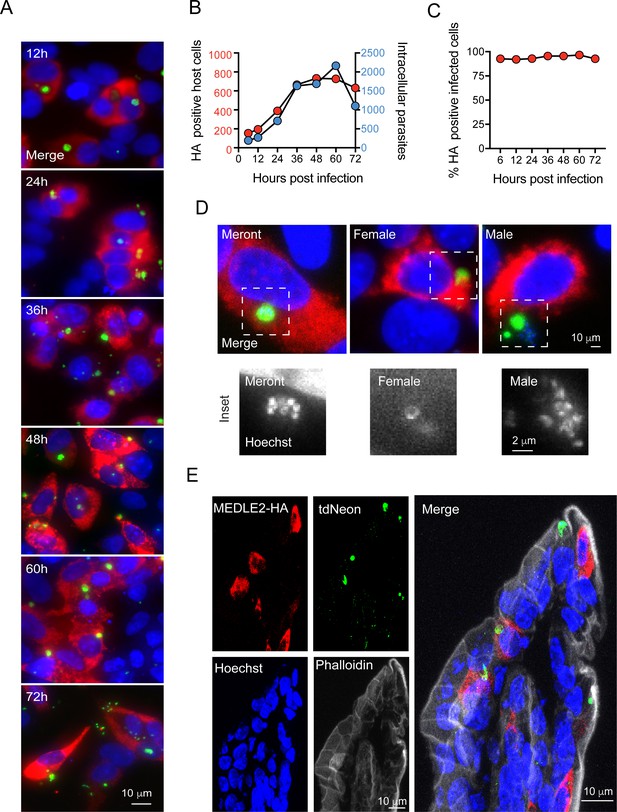
Infected cells express MEDLE2-HA across the parasite life cycle.
(A) 200,000 MEDLE2-HA-tdNeon transgenic parasites were used to infect HCT-8 cells and fixed at intervals across a 72 hr time period. Data shown are representative images from triplicate coverslips processed for immunofluorescence assay (IFA). Red, hemagglutinin (HA)-tagged protein; green, parasites (mNeon); blue, Hoechst. (B, C) Quantification of MEDLE2-expressing cells (red) versus intracellular parasites (blue) for 3695 host cells evaluated across a 72 hr time course. 20 fields of view quantified using ImageJ to identify host cells and parasites (B). The percentage of cell exhibiting MEDLE2-HA and mNeon staining is constant across the time course with a cumulative 94 ± 1.83% (mean ± SD) (C). (D) HCT-8 cultures infected with MEDLE2-HA parasites were fixed for IFA at 48 hr when sexual life stages were present. Cells were stained with stage-specific antibodies for female (COWP1) and male (α- tubulin), demonstrating MEDLE2 is exported across the parasite life cycle. Red, HA-tagged protein; green, parasites (stage-specific antibody); blue, Hoechst. (E) IFA of cryosections from the small intestine of Ifng-/- mice infected with MEDLE2-HA-tdNeon C. parvum (images representative of samples from three mice). Red, HA-tagged protein; green, parasites (tdNeon); blue, Hoechst; gray, Phalloidin (actin).
-
Figure 2—source data 1
Numerical data used for the quantification of HA positive host cells and the intracellular parasites.
- https://cdn.elifesciences.org/articles/70451/elife-70451-fig2-data1-v2.xlsx
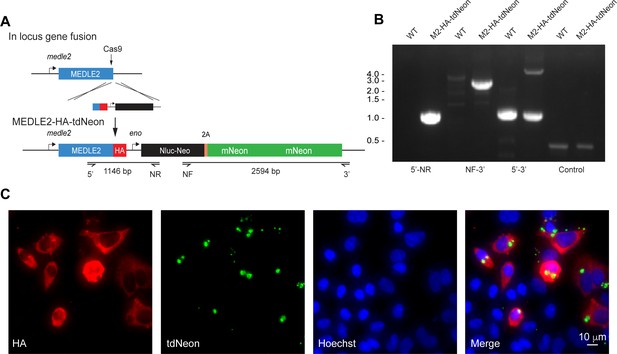
Construction of a MEDLE2-HA cytoplasmic tdNeon reporter parasite.
(A) Schematic representation of the strategy to derive reporter parasite line in which MEDLE2 is 3× hemagglutinin (HA) epitope tagged and the parasite cytoplasm expresses a tandem mNeon green tag (tdNeon). The solid black arrow indicates the position of the Cas9-induced double-stranded break at the C terminus of the gene, which is the same guide used in Figure 1B to generate the MEDLE2-HA transgenic parasites. (B) PCR mapping modification of the MEDLE2 locus using genomic DNA from wild type (WT) and transgenic (MEDLE2-HA-tdNeon) sporozoites using the primer pairs shown in (A) and the thymidine kinase (TK) gene as a control. Note the presence of both a 1174 bp WT gene and a 4557 bp transgene. (C) HCT-8 cultures were infected with and MEDLE2-HA-tdNeon transgenic parasites and fixed at 24 hr. Red, HA-tagged protein; green, parasites (tdNeon); blue, Hoechst.
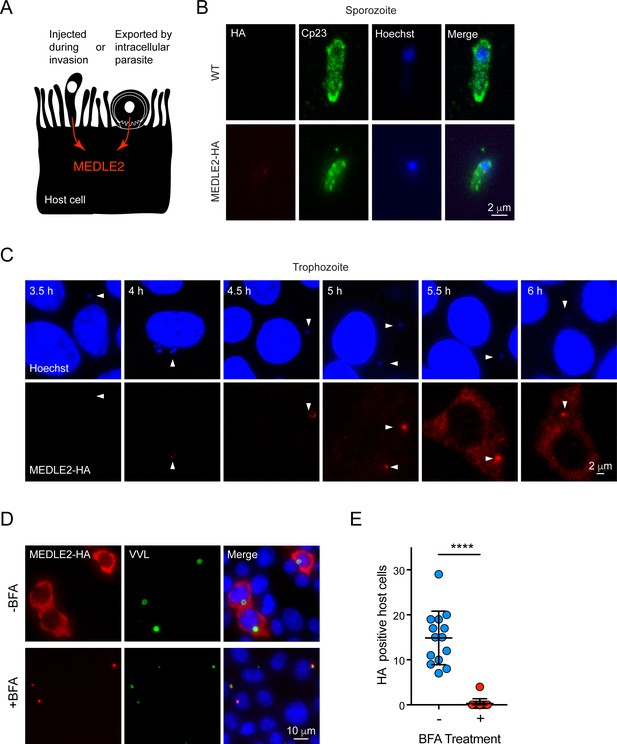
MEDLE2 is expressed by trophozoites and passes through the secretory pathway.
(A) Schematic representation of hypothetical patterns of MEDLE2 export in C. parvum. (B) Immunofluorescence assay (IFA) of wild type (WT) and MEDLE2-HA sporozoites fixed on poly-L-lysine-treated coverslips. We note that MEDLE2-HA is not observed in sporozoites. Red, hemagglutinin (HA)-tagged protein; green, sporozoite antigen Cp23; blue, Hoechst. (C) HCT-8 cells infected with MEDLE2-HA parasites were fixed in 30 min increments and processed for IFA. Data shown are representative images from a time-course bridging 3 hr (no observed MEDLE2-HA) and 6 hr (MEDLE2-HA abundant in host cell). White arrowheads denote parasite nuclei. Red, HA-tagged protein; blue, Hoechst. (D) MEDLE2-HA parasites were excysted and used to infect HCT-8 and after 3 hr media were supplemented with brefeldin A (BFA) (10 µg/mL). 10 hr post infection, cells were fixed and processed for IFA. Red, HA-tagged protein; green, parasites (VVL); blue, Hoechst (D). (E) The impact of BFA treatment on MEDLE2-HA export was quantified showing a significant reduction in MEDLE2 export when comparing BFA-treated (red) and untreated cells (blue) (n = 191 untreated, n = 98 treated; mean ± SEM; p<0.0001; unpaired t test with Welch’s correction).
-
Figure 3—source data 1
Numerical data used for the quantification of HA positive host cells in the absence and presence of Brefeldin A (BFA).
- https://cdn.elifesciences.org/articles/70451/elife-70451-fig3-data1-v2.xlsx
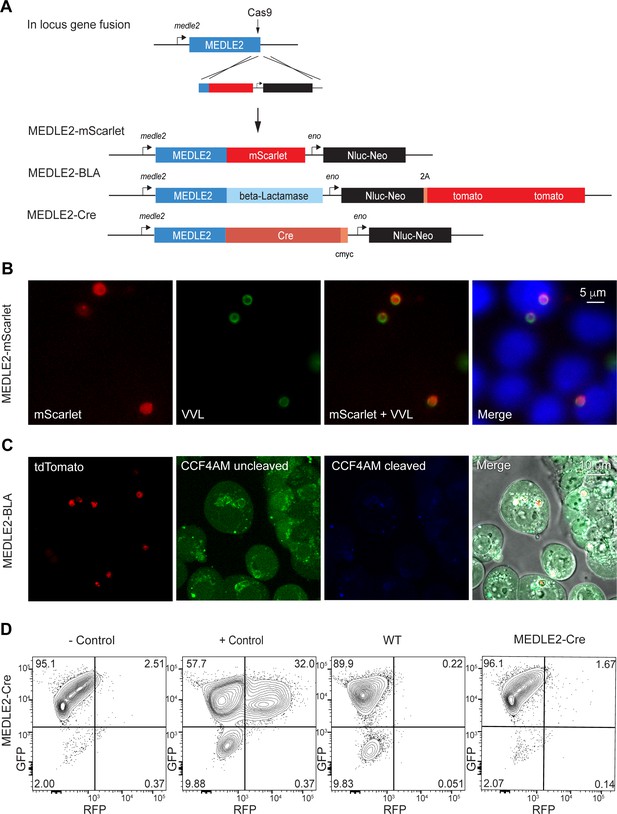
Ordered reporters disrupt MEDLE2 export.
(A) Schematic map of the MEDLE2 locus targeted for insertion of three different reporter genes (mScarlet, beta-lactamase, or Cre recombinase), nanoluciferase (Nluc), and the selection marker (Neo). The guide RNA and flanking sequences used here were the same as those employed to generate MEDLE2-HA transgenic parasites (see Figure 2—figure supplement 1, Figure 4—figure supplement 1 for more detail). (B) MEDLE2-mScarlet parasites were used to infect HCT-8 cells and fixed for immunofluorescence assay (IFA) across a time course. Data shown are from 10 hr post infection, which is representative of the MEDLE2 localization observed at all time points. Red, Medle2-mScarlet; green, parasites (VVL); blue, Hoechst. (C) HCT-8 cells were infected with MEDLE2-BLA C. parvum for 24 hr before incubation with the CCF4-AM beta-lactamase substrate and visualization by live microscopy. This experiment was repeated three times. Red, parasites (tdTomato); green, uncleaved CC4F-AM; blue, cleaved CCF4-AM; gray, DIC. We attribute lack of CCF4-AM cleavage to failure of MEDLE2-BLA to export (Figure 4—figure supplement 1). (D) MEDLE2-Cre parasites were used to infect loxGFP/RFP color switch HCT-8 cells ((Figure 4—figure supplement 2) for schematic representation). After 48 hr, cells were subjected to flow cytometry. Live, single cells were gated based upon forward and side scatter, and green fluorescence (GFP) and red fluorescence (RFP) were measured to detect Cre recombinase activity. Despite robust infection, MEDLE2-Cre-infected cultures did not express RFP (Figure 4—figure supplement 2) compared to the positive control that was transiently transfected to express Cre recombinase.
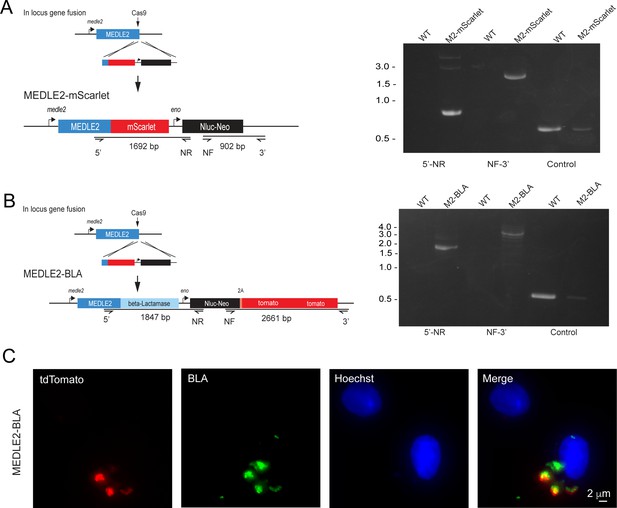
MEDLE2-BLA is not exported.
(A) Schematic representation of the strategy used to generate a MEDLE2-mScarlet parasite line, in which the MEDLE2 is C-terminally tagged with the fluorescent protein mScarlet, as well as engineered to express a nanoluciferase reporter gene (Nluc) and the neomycin phosphotransferase (Neo) selection marker. The solid black guide hit sequence is the same as used in Figure 1B to generate the MEDLE2-HA transgenic parasites. Integration PCR using MEDLE2-mScarlet and WT gDNA confirms proper integration. (B) The C terminus of MEDLE2 was targeted for insertion of a construct encoding beta-lactamase (BLA), a nanoluciferase reporter gene (Nluc) and the neomycin phosphotransferase (Neo) selection marker fused to a 2A peptide and a tdTomato reporter gene, using the same tagging strategy previously utilized for this locus. Integration PCR mapping the MEDLE2 locus using genomic DNA from wild type (WT) and transgenic (MEDLE2-BLA) sporozoites with the corresponding primer pairs shown in (A) and the thymidine kinase (TK) gene as a control shows the locus was successfully modified. (C) MEDLE2-BLA transgenic parasites were used to infect HCT-8 cells and fixed at 24 hr for immunofluorescence assay (IFA). Red, parasites (tdTomato); green, beta-lactamase (beta-lactamase antibody); blue, Hoechst.
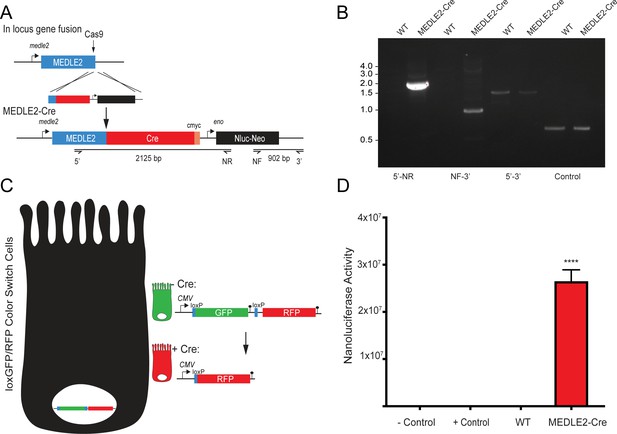
MEDLE2-Cre parasites infect loxGFP/RFP color switch cells.
(A) MEDLE2-Cre-expressing parasites were generated by targeting the MEDLE2 locus for insertion of a construct encoding Cre recombinase (Cre), a nanoluciferase reporter gene (Nluc), and the neomycin phosphotransferase (Neo) selection marker. The solid black arrow indicates the guide hit sequence, which is the same guide used in Figure 1B to generate the MEDLE2-HA transgenic parasites. (B) Integration PCR mapping the MEDLE2 locus using genomic DNA from wild type (WT) and transgenic (MEDLE2-Cre) sporozoites using the primer pairs shown in (A) and the thymidine kinase (TK) gene as a control. (C) Schematic of the LoxGFP/RFP color switch HCT-8. Introduction of Cre into these cells induces a color switch from green to red by recombinase-directed removal of the GFP coding sequence along with a stop codon preventing translation of RFP. (D) 1 × 106 transgenic MEDLE2-Cre oocyts were used to infect LoxGFP/RFP color switch HCT-8 cells for 48 hr before preparation of the sample for flow cytometry. 400,000 cells were removed from the sample preparation for nanoluciferase assay to determine infection in the culture. Mean nanoluciferase (relative luminescence) ± SEM is shown for three replicates (p<0.0001; one-way ANOVA with Dunnett’s multiple comparison test). Uninfected cells were used as a negative control, and Cre recombinase transfected cells were used as a positive control.
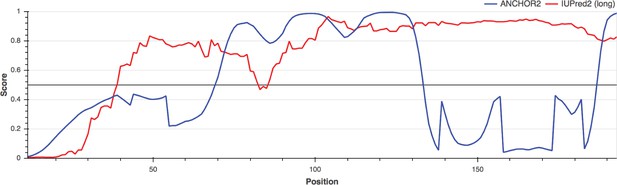
MEDLE2 is an intrinsically disordered protein.
MEDLE2 contains multiple single amino acid repeat regions and lacks a well-defined tertiary structure. The intrinsic disorder of MEDLE2 was assessed using IUPred2A (https://iupred2a.elte.hu/) and the resulting disordered plot is shown as greater than the threshold value of 0.5, represented by the horizontal black line.
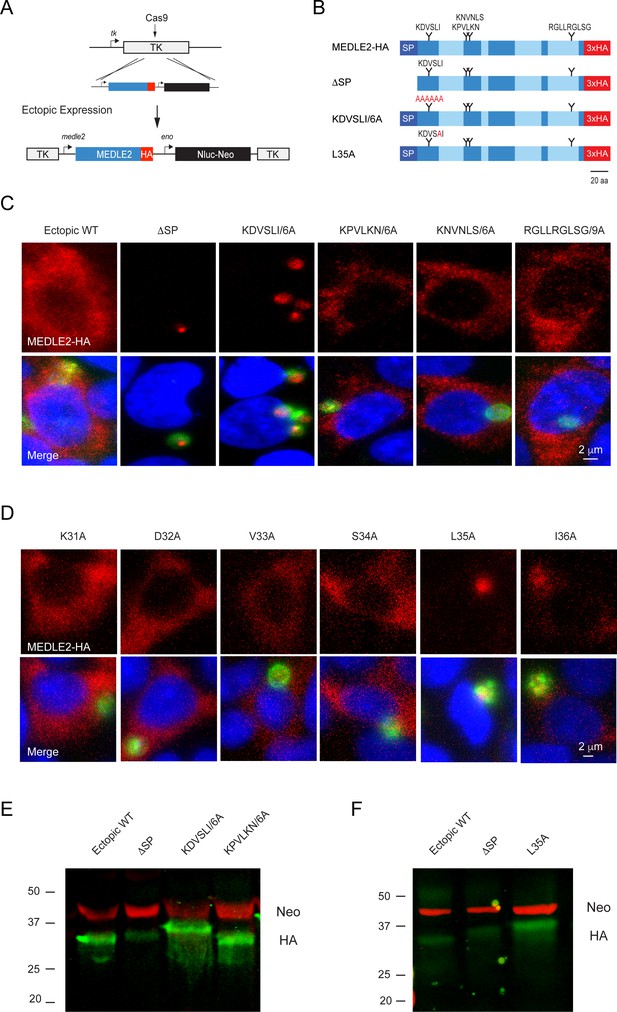
MEDLE2 contains a host-targeting motif that is processed during export.
(A) Map showing the strategy used to engineer an ectopic copy of MEDLE2-HA in the thymidine kinase (TK) locus. Expression of an ectopic copy of MEDLE2-HA was driven by the MEDLE2 promoter. All point mutations were confirmed by Sanger sequencing (Figure 5—figure supplement 1). (B) Schematic representation of the MEDLE2 mutants generated using the strategy outlined in (A). The signal peptide (SP) is represented by dark blue, and low-complexity regions are shown in light blue. Candidate motifs targeted for mutagenesis are indicated with black triangles, and mutagenized amino acids are shown in red for two representative mutants. (C, D) Mutant parasites were used to infect HCT-8 cells and fixed for immunofluorescence assay (IFA) after 24 hr. For mutants shown in (C), the entire candidate motif was replaced with a matching number of alanine residues (e.g., KDVSLI/6A → AAAAAA). For mutants shown in (D), each individual amino acid in the KDVSLI sequence was changed to alanine. Red, hemagglutinin (HA)-tagged protein; green, parasites (VVL); blue, Hoechst. We note that SP and leucine 35 within the KDVSLI sequence are required for MEDLE2 export. (E, F) 5 × 106 transgenic oocysts were used to infect HCT-8 cells for 48 hr before preparation of whole-cell lysates. Proteins were separated by for SDS-PAGE and analyzed by western blot. The resulting blots for infections with whole motif mutants (E) and individual amino acid point mutants (F) are shown. Red, neomycin; green, HA. Note that when mutants are expressed in mammalian cells and not C. parvum the resulting proteins do not show any size differences (Figure 5—figure supplement 2).
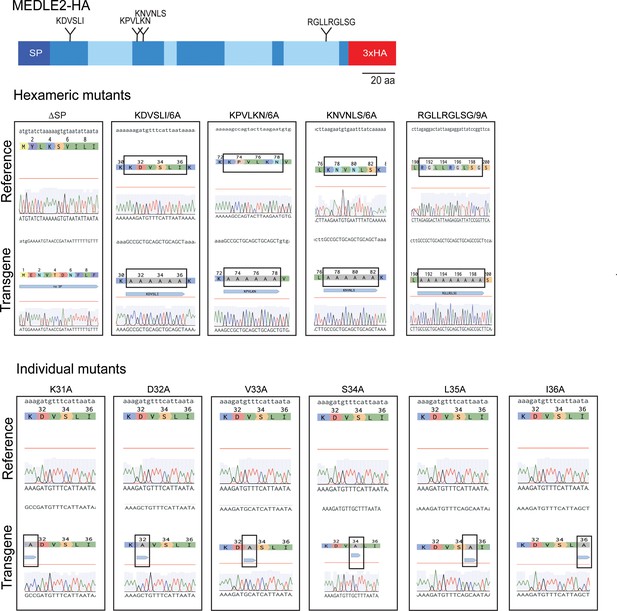
Sanger sequencing confirming the generation of MEDLE2 mutants.
100,000 sporozoites were used from each mutant strain for genomic DNA extraction. The resulting gDNA was used for PCR mapping of the thymidine kinase (TK) locus to verify the desired mutagenesis. 5′ TK–Nluc PCR products were used for TopoTA cloning, and the resulting colonies were grown and Sanger sequenced. Three colonies were sequenced for each strain using the M13 forward, M13 reverse, and an internal MEDLE2-specific primer to confirm targeted mutagenesis. The Benchling alignment of the Sanger sequencing result (transgene) to the reference sequence is shown. The black box highlights the mutation engineered in each strain.
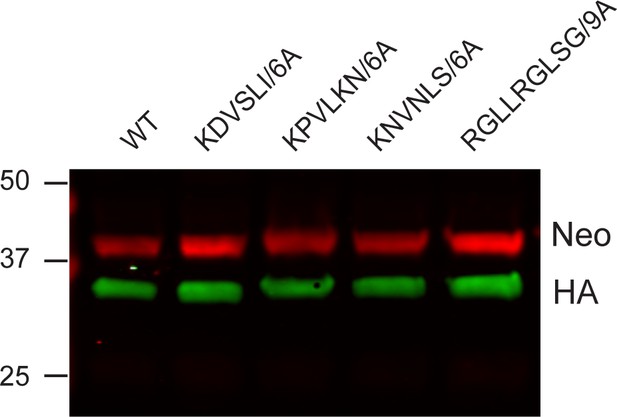
MEDLE2 mutants are of the same size as wild type (WT) MEDLE2 when expressed in HEK293T cells.
Plasmids encoding human codon-optimized MEDLE2 with the N-terminal signal peptide removed (aa 2–20) (WT) or encoding the desired point mutations (denoted residues were replaced with alanines) were transfected into HEK233T cells. After 24 hr, whole-cell lysates were prepared, and proteins separated by SDS-PAGE for western blot analysis. MEDLE2 mutants were indistinguishable in size from WT MEDLE2. Red, neomycin; green, hemagglutinin (HA).
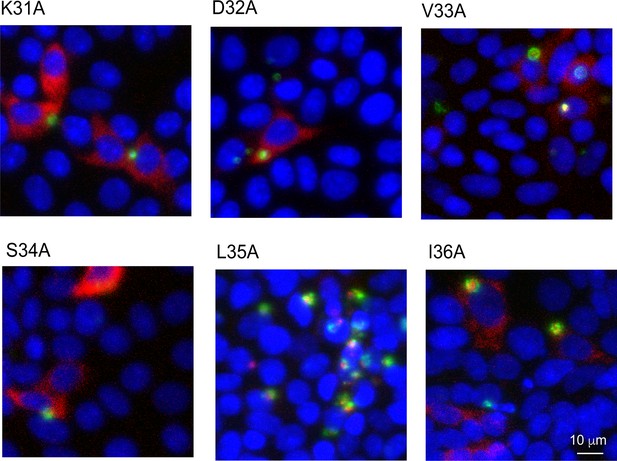
Uncropped images of panels shown in Figure 5D including infected and uninfected cells.
Transgenic C. parvum parasites with point mutations engineered in an ectopic copy of MEDLE2 expressed in the thymidine kinase (TK) locus were used to infect HCT-8 cells for 24 hr before fixation for immunofluorescence assay. L35A mutation results in accumulation of MEDLE2-HA (red) with the parasite (green).
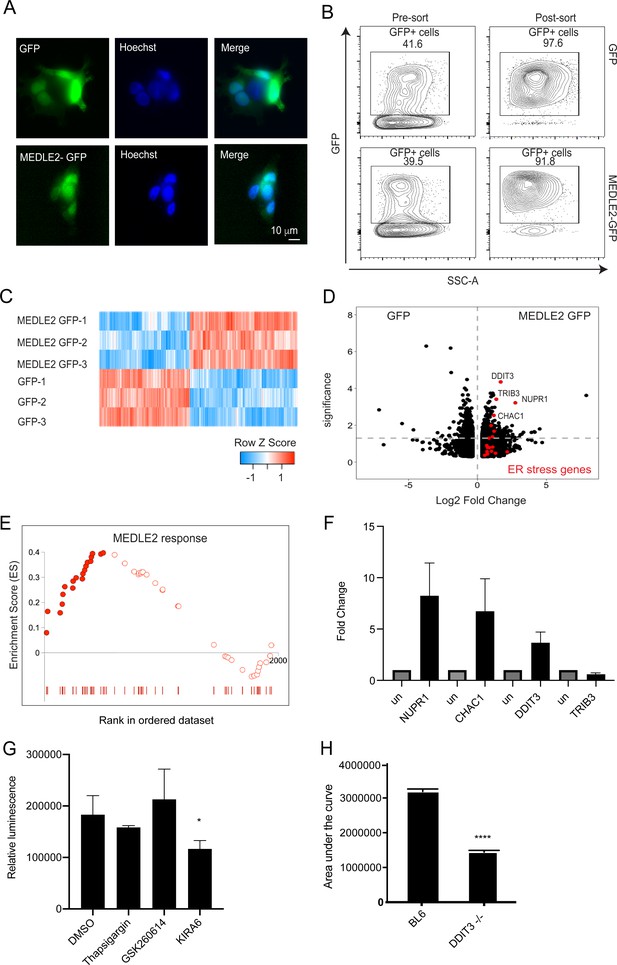
MEDLE2-expressing cells exhibit upregulation of genes involved in the unfolded protein response.
(A) HEK293T cells were transfected with plasmids encoding MEDLE2-GFP or GFP alone. After 24 hr, cells were fixed and processed for immunofluorescence assay (IFA). GFP is shown in green, Hoechst in blue. (B) 24 hr post transection, HEK293T cells were trypsinized and double sorted for live, GFP+ singlets directly into RNA lysis buffer and subjected to RNA sequencing. (C) Heat map depicting the differential gene expression between MEDLE2-GFP (top panel) and GFP control expressing cells (bottom panel). Upregulated gene expression is shown in red (row Z score > 0), while blue shows genes that are downregulated in expression (row Z score < 0). Expressing cells compared to GFP control cells. 413 transcripts showed upregulation in MEDLE2-GFP-expressing cells (right) and 487 genes had lower transcript abundance (left). The horizontal dashed line indicates p-value = 0.05. Gene set enrichment analysis (GSEA) performed on the 900 differentially expressed genes from the MEDLE2 transfection dataset identifies core enrichment of 20 genes that belong to ER stress response signaling pathways, which are indicated on the volcano plot in red. The most upregulated genes are identified by their gene ID. (E) The 234 genes with the greatest differential expression (p<0.01, log fold change absolute value > 1.5) were used to define a MEDLE2 gene set from the MEDLE2-GFP transfection dataset. This signature was used to perform GSEA using data from single-cell RNA sequencing on C. parvum-infected organoid-derived cultures, which showed enrichment of 51 genes with 22 genes in the core enrichment for the MEDLE2 response set highlighted in solid red. We note that we did not detect the MEDLE2 response signature in datasets from other enteric infections including rotavirus (Figure 6—figure supplement 1). (F) Ileal sections were removed from C. parvum-infected Ifng-/- mice and uninfected controls (each n = 3), and expression levels for the four differentially upregulated genes in the MEDLE2 response set (NUPR1, CHAC1, DDIT3, and TRIB3) were measured by qPCR. (G) HCT-8 cultures were pretreated for 2 hr with inhibitors (GSK2606414 and KIRA6) of ER stress signaling pathways prior to infection with 10,000 MEDLE2-HA parasites. After 24 hr, cells were lysed and nanoluciferase assay was performed as a measure of parasite growth. Inhibition of the IRE1 signaling pathway with KIRA6 significantly reduced parasite growth (one-way ANOVA, Dunnett’s multiple comparisons test p=0.0303; a representative experiment is shown [n = 6]). This experiment was repeated three times. (H) Ddit3-/- and C57BL/6J mice were treated with anti-mouse-IFN gamma antibody 1 day prior to infection with 10,000 MEDLE2-HA-tdNeon oocysts, and again at day 2 of infection. Fecal luminescence was determined by nanoluciferase activity to calculate the area under the curve for the duration of the infection. Ddit3-/- mice exhibited a 56% reduction in infection (1,416,227 ± 44,850; total peak area ± standard error) compared to control mice (3,189,123 ± 69,887; unpaired t test, p<0.001; n = 4 mice per group). One representative experiment is shown, which was repeated two more times, with a 54% reduction and no change in infection being observed.
-
Figure 6—source data 1
Source data for Figure 6.
- https://cdn.elifesciences.org/articles/70451/elife-70451-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Source data for Figure 6.
- https://cdn.elifesciences.org/articles/70451/elife-70451-fig6-data2-v2.xlsx
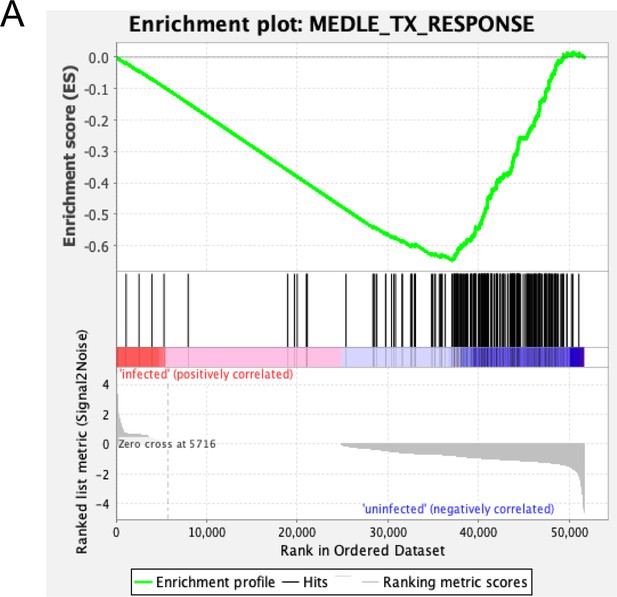
MEDLE2 response signature is absent from intestinal epithelial cells infected with human rotavirus.
Genes with the greatest differential expression (p<0.01, log fold change absolute value > 1.5) were used to define a MEDLE2 gene set from the MEDLE2-GFP transfected cells. RNA sequencing from small intestinal enteroid cultures infected with human rotavirus shows an absence of the MEDLE2 signature created from the MEDLE2 transfection dataset, evidenced by the negative enrichment score resulting from gene set enrichment analysis (GSEA).
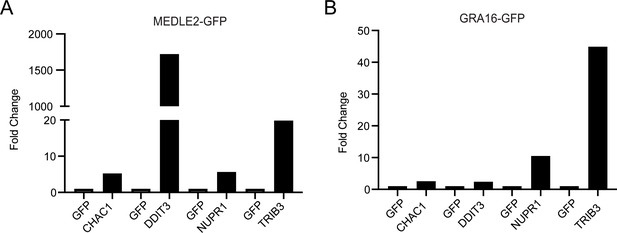
Transfection of HEK293T cells with T. gondii GRA16-GFP also results in ER stress.
HEK293T cells were transfected with plasmids encoding GFP-only, MEDLE2-GFP, and Tg. GRA16-GFP, RNA was extracted, and qPCR was performed to characterize the impact on ER stress response genes (CHAC1, DDIT3, NUPR1, and TRIB3). Introduction of disordered proteins, such as MEDLE2 (A) and GRA16 (B), both result in transcriptional changes in ER stress response genes.
Tables
Members of multigene families for which localization of protein product was initially attempted in this study.
| Gene family | Gene ID | Result |
|---|---|---|
| MEDLE | cgd5_4590 | Exported to host cell |
| FLGN | cgd4_4470 | Transgenic unsuccessful |
| GGC | cgd5_3570 | Transgenic unsuccessful |
| SKSR | cgd8_30 | Not in host cell |
| SKSR | cgd8_40 | Not in host cell |
| WYLE | cgd8_3560 | Not in host cell |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Cryptosporidium parvum) | MEDLE2 | Li et al., 2017 | cgd5_4590 | |
| Gene (C. parvum) | MEDLE1 | Fei et al., 2018 | cgd5_4580 | |
| Gene (C. parvum) | MEDLE6 | This paper | cgd6_5490 | Named according to the spatial localization in the genome similarly to MEDLE2 and MEDLE1 |
| Gene (C. parvum) | WYLE4 | This paper | cgd8_3560 | Named for the gene family and according to the spatial localization in the genome |
| Gene (C. parvum) | SKSR7 | This paper | Cgd8_30 | Named for the gene family and according to the spatial localization in the genome |
| Strain, strain background (C. parvum) | C. parvum oocysts, IOWAII strain (WT) | Bunchgrass | ||
| Strain, strain background (Mus musculus) | Ifng-/-, C57BL/6J | The Jackson Laboratory | Jax 002287; RRID:IMSR_JAX:002287 | |
| Strain, strain background (M. musculus) | Ddit3tm2.1Dron | The Jackson Laboratory | Jax 005530; RRID:IMSR_JAX:005530 | |
| Strain, strain background (M. musculus) | C57BL/6J | The Jackson Laboratory | Jax 000664; RRID:IMSR_JAX:000664 | |
| Genetic reagent (C. parvum) | MEDLE2-HA | This paper | cgd5_4590 modified | Stable transgenic parasite line expressing HA |
| Genetic reagent (C. parvum) | WYLE4-HA | This paper | cgd8_3570 modified | Stable transgenic parasite line expressing HA |
| Genetic reagent (C. parvum) | SKSR7-HA | This paper | cgd8_30 modified | Stable transgenic parasite line expressing HA |
| Genetic reagent (C. parvum) | MEDLE2 KO | This paper | Cgd5_4590 | Stable transgenic parasite line with one copy of MEDLE2 knocked out |
| Genetic reagent (C. parvum) | MEDLE1-HA | This paper | cgd5_4580 modified | Stable transgenic parasite line expressing HA |
| Genetic reagent (C. parvum) | MEDLE6-HA | This paper | cgd6_5490 modified | Stable transgenic parasite line expressing HA |
| Genetic reagent (C. parvum) | Medle2 MEDLE1-HA | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing HA |
| Genetic reagent (C. parvum) | MEDLE2-HA-tdNeon | This paper | cgd5_4590 modified | Stable transgenic parasite line expressing HA and tdNeon |
| Genetic reagent (C. parvum) | MEDLE2-mScarlet | This paper | cgd5_4590 modified | Stable transgenic parasite line expressing mScarlet |
| Genetic reagent (C. parvum) | MEDLE2-Bla-2A-tdTomato | This paper | cgd5_4590 modified | Stable transgenic parasite line expressing BLA and tdTomato |
| Genetic reagent (C. parvum) | MEDLE2-Cre | This paper | cgd5_4590 modified | Stable transgenic parasite line expressing Cre recombinase |
| Genetic reagent (C. parvum) | Ectopic MEDLE2-HA | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 |
| Genetic reagent (C. parvum) | ΔSP | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 (aa 21–209) |
| Genetic reagent (C. parvum) | KDVSLI/6A | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 with KDVSLI (aa 31–36) mutated to six alanines |
| Genetic reagent (C. parvum) | KPVLKN/6A | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 with KPVLKN (aa 73–78) mutated to six alanines |
| Genetic reagent (C. parvum) | KNVNLS/6A | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 with KDVSLI (aa 77–82) mutated to six alanines |
| Genetic reagent (C. parvum) | RGLLRGLSG/9A | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 with KDVSLI (aa 191–199) mutated to six alanines |
| Genetic reagent (C. parvum) | K31A | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 with K31 mutated to alanine |
| Genetic reagent (C. parvum) | D32A | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 with D32 mutated to alanine |
| Genetic reagent (C. parvum) | V33A | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 with V33 mutated to alanine |
| Genetic reagent (C. parvum) | S34A | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 with S34 mutated to alanine |
| Genetic reagent (C. parvum) | L35A | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 with L35 mutated to alanine |
| Genetic reagent (C. parvum) | I36A | This paper | cgd5_4440 modified | Stable transgenic parasite line expressing extra copy of MEDLE2 with I36 mutated to alanine |
| Cell line (human) | HCT-8 | ATCC | CCL-224; RRID:CVCL_2478 | |
| Cell line (human) | HEK293T | ATCC | CRL-3216; RRID:CVCL_0063 | |
| Cell line (Escherichia coli) | GC5 | Genesee Scientific | 42-653 | Electrocompetent cells |
| Cell line (E. coli) | One Shot Topo10, | Invitrogen | C404003 | Electrocompetent cells |
| Transfected construct (human) | loxP GFP/RFP color switch lentivirus | GenTarget Inc | Cat#: LVP460-Neo | Transfected construct (human) |
| Biological sample (M. musculus) | Isolated sections of ileum | Ifng-/- mice | Jax 002287; RRID:IMSR_ JAX:002287 | 6-week-old male mice |
| Antibody | Anti-HA (rat monoclonal) | MilliporeSigma | Cat# 11867431001; RRID:AB_390919 | IF (1:500), WB (1:500), IHC (1:500) |
| Antibody | Anti-Cp23 (mouse monoclonal) | LS Bio | Cat# LS-C137378; RRID:AB_10947007 | IF (1:100) |
| Antibody | Anti-alpha tubulin (mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat#12G10; RRID:AB_1157911 | IF (1:1000) |
| Antibody | Anti-COWP1 (rat monoclonal) | This paper, produced by GenScript | This paper | IF (1:100) |
| Antibody | Anti-neomycin phosphotransferase II (rabbit polyclonal) | MilliporeSigma | Millipore Cat# 06-747; RRID:AB_310234 | WB (1:1000) |
| Antibody | Anti-mouse IFN gamma | Bio X Cell | Clone: XMG1.2; Cat# BE0055; RRID:AB_1107694 | In vivo 100 µg |
| Antibody | Goat anti-rat polyclonal Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A-21213; RRID:AB_2535799 | IFA (1:500) |
| Antibody | Goat anti-mouse polyclonal Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11001; RRID:AB_2534069 | IFA (1:500) |
| Strain, strain background | Alexa Fluor 647 Phalloidin | Thermo Fisher Scientific | A22287; RRID:AB_2620155 | IFA (1:1000) |
| Antibody | IRDye 800CW goat anti-rat IgG | LI-COR | 926-32219; RRID:AB_1850025 | WB (1:10,000) |
| Antibody | IRDye 680RD goat anti-rabbit IgG | LI-COR | 926-68071; RRID:AB_2721181 | WB (1:10,000) |
| Recombinant DNA reagent | Cas9 cgd5_4590 (plasmid) | This paper | Guide targeting C terminus of MEDLE2 | |
| Recombinant DNA reagent | Cas9 cgd8_3560 (plasmid) | This paper | Guide targeting C terminus of WYLE4 | |
| Recombinant DNA reagent | Cas9 cgd8_30 (plasmid) | This paper | Guide targeting C terminus of SKSR7 | |
| Recombinant DNA reagent | Cas9 cgd5_4580 (plasmid) | This paper | Guide targeting C terminus of MEDLE1 | |
| Recombinant DNA reagent | Cas9 Cgd6_5490 (plasmid) | This paper | Guide targeting C terminus of MEDLE6 | |
| Recombinant DNA reagent | Cas9 Tk guide int (plasmid) | Tandel et al., 2019 | Guide targeting internal cgd5_4440 | |
| Recombinant DNA reagent | Cas9 MEDLE2 KO (plasmid) | This paper | Guide targeting internal MEDLE2 | |
| Recombinant DNA reagent | Lic HA (plasmid) | This paper | Crypto expression vector for HA tagging | |
| Recombinant DNA reagent | Lic tdTomato KO (plasmid) | This paper | Crypto expression vector for replacing gene KO with tdTomato | |
| Recombinant DNA reagent | Lic medle2-MEDLE1 HA (plasmid) | This paper | Crypto expression vector with medle2 promoter driving MEDLE1-HA expression | |
| Recombinant DNA reagent | Lic HA-2A- TdNeon (plasmid) | This paper | Crypto expression vector for HA tagging and cytoplasmic tdNeon | |
| Recombinant DNA reagent | Lic mScarlet (plasmid) | This paper | Crypto expression vector for mScarlet tagging | |
| Recombinant DNA reagent | Lic Bla-2A-TdTomato (plasmid) | This paper | Crypto expression vector for BLA tagging and cytoplasmic tdTomato | |
| Recombinant DNA reagent | Lic Cre (plasmid) | This paper | Crypto expression vector for Cre tagging | |
| Recombinant DNA reagent | Lic Extra MEDLE2-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA (MEDLE2 promoter) | |
| Recombinant DNA reagent | Lic ΔSP MEDLE2-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA (aa 21–209) | |
| Recombinant DNA reagent | Lic KDVSLI/6A-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA with KDVSLI (aa 31–36) mutated to six alanines | |
| Recombinant DNA reagent | Lic KPVLKN/6A-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA with KPVLKN (aa 73–78) mutated to six alanines | |
| Recombinant DNA reagent | Lic KNVNLS/6A-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA with KNVNLS (aa 77–82) mutated to six alanines | |
| Recombinant DNA reagent | Lic RGLLRGLSG/9A-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA with RGLLRGLS (aa 191–199) mutated to six alanines | |
| Recombinant DNA reagent | Lic K31A-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA with K31 mutated to alanine | |
| Recombinant DNA reagent | Lic D32A-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA with D32 mutated to alanine | |
| Recombinant DNA reagent | Lic V33A-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA with V33 mutated to alanine | |
| Recombinant DNA reagent | Lic S34A-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA with S34 mutated to alanine | |
| Recombinant DNA reagent | Lic L35A-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA with L35 mutated to alanine | |
| Recombinant DNA reagent | Lic I36A-HA (plasmid) | This paper | Crypto expression vector for extra copy of MEDLE2-HA with I36 mutated to alanine | |
| Recombinant DNA reagent | mEGFP-Lifeact-7 (plasmid) | Addgene | # 54610 | Used as a mammalian expression vector to clone codon optimized MEDLE2 into |
| Recombinant DNA reagent | GFP-only | This paper | Removed Lifeact domain from Addgene plasmid #54610 for a GFP-only control plasmid | |
| Recombinant DNA reagent | Recod MEDLE2-GFP (plasmid) | This paper | Human codon optimized MEDLE2 (aa 21–209) with a GFP tag | |
| Recombinant DNA reagent | GRA16-GFP | This paper | T. gondii GRA16 (aa 24–505) with GFP tag. | |
| Recombinant DNA reagent | Recod MEDLE2-HA (plasmid) | This paper | Human codon optimized MEDLE2 (aa 21–209) with a HA tag. | |
| Recombinant DNA reagent | Recod KDVSLI/6A-HA (plasmid) | This paper | Human codon optimized MEDLE2 (aa 21–209) with a HA tag and KDVSLI (aa 31–36) mutated to six alanines | |
| Recombinant DNA reagent | Recod KPVLKN/6A-HA (plasmid) | This paper | Human codon optimized MEDLE2 (aa 21–209) with a HA tag and KPVLKN (aa 73–78) mutated to six alanines | |
| Recombinant DNA reagent | Recod KNVNLS/6A-HA (plasmid) | This paper | Human codon optimized MEDLE2 (aa 21–209) with a HA tag and KNVNLS (aa 77–82) mutated to six alanines | |
| Recombinant DNA reagent | Recod RGLLRGLSG/9A-HA (plasmid) | This paper | Human codon optimized MEDLE2 (aa 21–209) with a HA tag and RGLLRGLS (aa 191–199) mutated to six alanines | |
| Sequence-based reagent | Recodonized MEDLE2 | Integrated DNA Technologies | MEDLE2 (aa 21–209) codon optimized for human expressionSee Supplementary file 1 for sequence | |
| Sequence-based reagent | PCR primers | This paper | Please see Supplementary file 1 | |
| Commercial assay or kit | DNeasy Blood & Tissue Kit | QIAGEN | Cat# 69504 | |
| Commercial assay or kit | ZymoPureII Plasmid Maxiprep Kit | Zymo Research | Cat# 11-555B | |
| Commercial assay or kit | Nano-Glo Luciferase Assay System | Promega | Cat# N1130 | |
| Commercial assay or kit | SF Cell Line 4D X Kit L | Lonza | Cat# V4XC-2024 | |
| Commercial assay or kit | LiveBLAzer FRET-B/G Loading Kit | Thermo Fisher | Cat# K1095 | |
| Commercial assay or kit | ZeroBlunt TopoTA Kit | Invitrogen | Cat# 450245 | |
| Commercial assay or kit | RNeasy Microkit | QIAGEN | Cat# 74004 | |
| Commercial assay or kit | SMART cDNA synthesis kit | Takara | Cat# 635040 | |
| Commercial assay or kit | Nextera XT DNA Library Prep Kit | Illumina | Cat# FC-131-1096 | |
| Commercial assay or kit | RNeasy MiniKit | QIAGEN | Cat# 74104 | |
| Commercial assay or kit | QIAshredder | QIAGEN | Cat# 79656 | |
| Commercial assay or kit | SuperScript First Strand Synthesis kit | Thermo Fisher | Cat#18091050 | |
| Commercial assay or kit | Lipofectamine 3000 | Thermo Fisher | Cat# L3000015 | |
| Chemical compound, drug | Paromomycin | Gemini | Cat# 400-155P | Used 16 g/L water |
| Chemical compound, drug | Brefeldin A (BFA) | BioLegend | Cat# 420601 | Used 10 µg/mL |
| Chemical compound, drug | Thapsigargin | MedChemExpress | HY-13433 | Used 1 µM |
| Chemical compound, drug | GSK2606414 | MedChemExpress | HY-18072 | Used 30 nm |
| Chemical compound, drug | KIRA6 | MedChemExpress | HY-19708 | Used 500 nm |
| Software, algorithm | Prism 8 | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | ImageJ | Fiji | RRID:SCR_003070 | |
| Software, algorithm | FlowJo v10, LLC | TreeStar | RRID:SCR_008520 | |
| Software, algorithm | Kallisto v0.44.0 | BioConductor (Bray et al., 2016) | Pachter Lab | |
| Software, algorithm | Limma-Voom | BioConductor (Law et al., 2014; Ritchie et al., 2015) | ||
| Software, algorithm | Bioconductor tximport | BioConductor (Robinson et al., 2010) | DOI: 10.18129/B9.bioc.tximport | |
| Software, algorithm | Molecular Signatures Database (MSigDB) | UC San Diego and Broad Institute (Mootha et al., 2003; Subramanian et al., 2005) | https://www.gsea-msigdb.org/gsea/msigdb | |
| Software, algorithm | CryptoDB | VEuPathDB | cryptodb.org | |
| Other | Fluorescin Vicia villosa lectin stain | Vector Labs | Cat# FL-1231-2 | IF (1:1000) |
| Other | DAPI stain | Invitrogen | Cat# D1306 | Flow cytometry (1 µg/mL) |
| Other | Hoechst 33342 | Thermo Fisher | Cat# H3570 | IF (1:10,000) |
| Other | Alexa Fluor 647 Phalloidin | Thermo Fisher | Cat# A22287 | IF (1:1000) |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70451/elife-70451-transrepform1-v2.docx
-
Supplementary file 1
Primer sequences used for this study.
- https://cdn.elifesciences.org/articles/70451/elife-70451-supp1-v2.xlsx
-
Source code 1
Supplemental code detailing the R packages used for analysis of the MEDLE2 transfection RNAsequencing dataset.
- https://cdn.elifesciences.org/articles/70451/elife-70451-supp2-v2.zip





