MACF1 controls skeletal muscle function through the microtubule-dependent localization of extra-synaptic myonuclei and mitochondria biogenesis
Figures
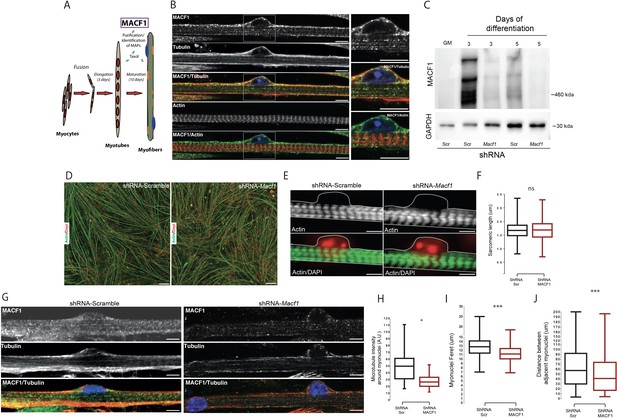
MACF1 is an essential regulator of myonuclei positioning in mature myofibers.
(A) Scheme presentation of the sequential steps to obtain immature myotubes and mature myofibers from primary mouse myoblasts. Taxol stabilized microtubules proteome was collected either from WT elongated myotubes or from mature contractile myofibers. For downregulating experiments, siRNA/shRNA were transfected in early steps of differentiation just before myotubes formation. (B) Confocal immunofluorescence images (63×) presenting the organization of MACF1 (green) and total alpha tubulin or F-actin (red) in WT mature mouse primary myofibers. Scale bar = 10 µm. (C) Western blot analysis of MACF1 protein expression (main two isoforms at 830 and 630 kD) in total protein extracts of primary cells at proliferation cycle (GM for growth media) and in 3- or 5 days post-differentiation myotubes or myofibers. Cells were treated either with a scramble shRNA or a pool of four shRNAs targeting Macf1. GAPDH was used as loading control. (D) Immunofluorescence staining (10×) of F-actin (green) and myonuclei (red) in primary myofibers treated either with scramble shRNA (left panel) or with a pool of four distinct shRNAs targeting Macf1 (right panel) after 13 days of differentiation. Scale Bar = 50 µm. (E) Representative immunofluorescence microscopy images (63×) of F-actin (green) and myonuclei (red) in primary myofibers treated with scramble shRNA (left panel) or with a pool of four distinct shRNAs targeting Macf1 (right panel) after 13 days of differentiation. Scale Bar = 10 µm. (F) Sarcomere length measured on the immunofluorescence images of F-actin from 13 days mature primary myofibers treated with either scramble shRNA or a pool of four distinct shRNAs targeting Macf1. Data were obtained from three individual experiments. Line is set at median. Ns stands for not significant (Student’s t test). (G) Confocal immunofluorescence images (63×) presenting the organization of total alpha tubulin (red) or MACF1 (green) in mature primary myofibers following treatment with either scramble shRNA (left panel) or a pool of four distinct shRNAs (right panel) targeting Macf1. Scale bar = 10 µm. (H–J) Mean values for fluorescent intensity of total alpha tubulin (H), myonuclei ferret (I) and distance between adjacent myonuclei (J) measured on mature primary myofibers following treatment with either scramble shRNA or a pool of four distinct shRNAs targeting Macf1. All panels were generated from data obtained from three individual experiments. Line is set at median. *p<0.05 and ***p<0.001 (Student’s t test).
-
Figure 1—source data 1
Table showing data from experiments plotted in Figure 1F,H,I and J.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Whole immunoblotting membrane for MACF1 and GAPDH plotted in Figure 1C.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig1-data2-v2.pdf
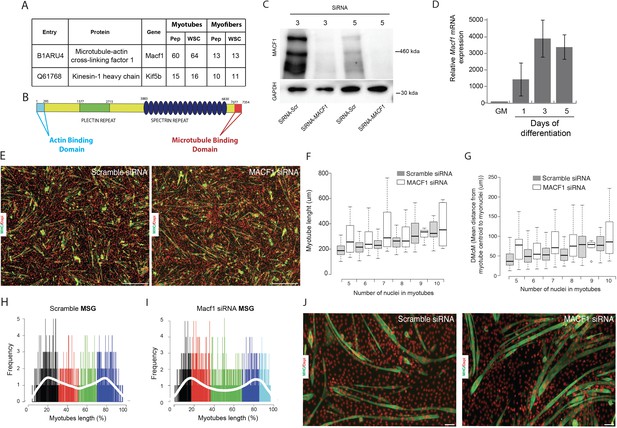
Reduction of MACF1 does not affects myotubes formation or myonuclei spreading within myotubes.
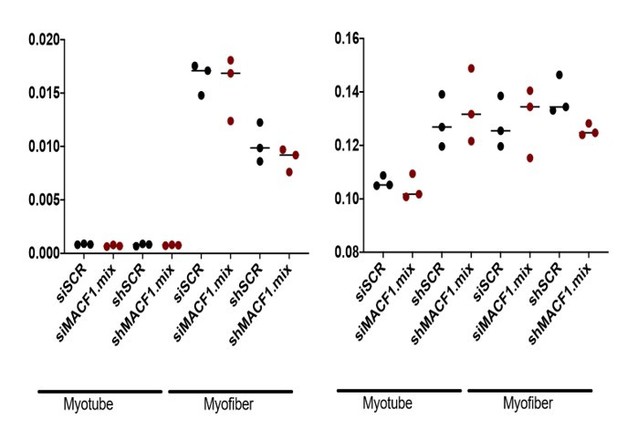
(A) Table showing mass spectrometry identification of MACF1 and Kif4b in 3- and 13-day-old primary myotubes and myofibers, respectively. (B) Schematic representation of MACF1 identified domains. ABD is the ‘Actin Binding Domain’ and MTBD is the ‘Microtubule Binding Domain’. (C) Western blot analysis of MACF1 protein expression (main two isoforms at 830 and 630 kD) in total protein extracts of primary cells in myotubes and myofibers at 3- and 5 days post-differentiation, respectively. Cells were treated with a scramble siRNA or a pool of three individual siRNAs targeting Macf1. GAPDH was used as loading control. (D) QRT-PCR analysis of Macf1 gene expression level relative to housekeeping genes in proliferating C2C12 cells (GM) and after 1, 3, or 5 days of differentiation. Data are from three individual experiments. Mean (with SEM) values are presented. (E) Immunofluorescence staining of Myosin Heavy Chain (green) and myonuclei (red) in primary myotubes treated with scramble siRNA (left panel) or a pool of three distinct siRNAs targeting Macf1 (right panel) after 3 days of differentiation. Scale Bar = 150 µm. (F–G) Values for myotube’s length (F) and mean distance between myonuclei and myotube’s centroid (G) ranked by myonuclei content per myotubes were quantified after 3 days of differentiation on cells treated with a scramble siRNA or a pool of three individual Macf1 siRNAs. At least three individual experiments per condition were quantified. Line is set at median. (H–I) Myonuclei Spreading Graph (MSG) representing the statistical spatial distribution of myonuclei along myotubes in cells treated with scramble siRNA (H) or with a pool of three individual siRNAs (I) after 3 days of differentiation. (J) Immunofluorescence staining of Myosin Heavy Chain (green) and myonuclei (red) in 5-day-differentiated C2C12 myotubes treated with scramble siRNA (left panel) or a pool of three individual siRNAs targeting Macf1 (right panel). Scale Bar = 50 µm.
-
Figure 1—figure supplement 1—source data 1
Table showing data from experiments plotted in Figure 1—figure supplement 1D,F and G.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig1-figsupp1-data1-v2.xlsx
-
Figure 1—figure supplement 1—source data 2
Whole immunoblotting membrane for MACF1 and GAPDH plotted in Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig1-figsupp1-data2-v2.pdf
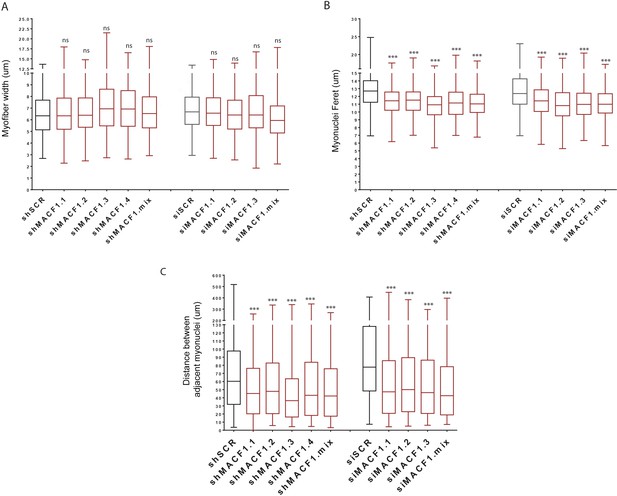
MACF1 is essential for myonuclei positioning and shape in mature myofibers.
(A–C) Myofibers width (A), myonuclei ferret (B) and distance between adjacent myonuclei (C) measured on mature primary myofibers treated with either scramble siRNA, three individual siRNAs targeting Macf1 or a pool of the three siRNAs targeting Macf1 in addition to cells treated with scramble shRNA, four individual shRNAs targeting Macf1 or a pool of the four shRNAs targeting Macf1. All panels were generated from data obtained from three individual experiments. Line is at median. Ns stands for not significant and ***p<0.001 (Student’s t test).
-
Figure 1—figure supplement 2—source data 1
Table showing data from experiments plotted in Figure 1—figure supplement 2A–C.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig1-figsupp2-data1-v2.xlsx
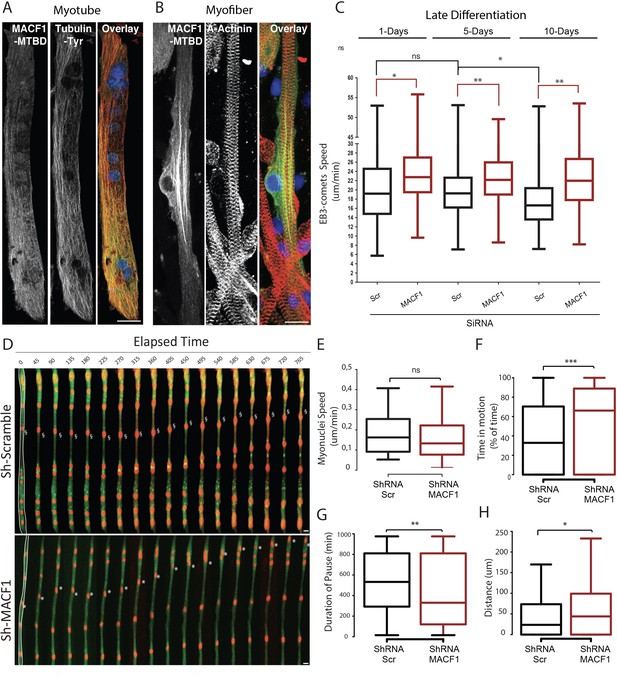
MACF1 regulates myonuclei dynamics through the control of microtubules.
(A–B) representative confocal immunofluorescence images (63×) presenting MACF1-MTBD (green) and tyrosinated alpha tubulin (red) in primary myotubes (A) or alpha actinin (red) in primary myofibers (B). Scale bar = 10 µm. (C) Quantification of EB3 comets speed in primary cells treated with either scramble siRNA or a pool of 3 individual siRNAs targeting Macf1 in primary myotubes at three different time points during maturation. At least 200 comets from three individual experiments were mapped per condition. Line is set at median. Ns stands for not significant, *p<0.05 and **p<0.01 (Student’s t test). (D) Frames from a 14 hr time-lapse movie of two channels (shRNA in green and lamin-chromobody in red) taken 5 days post differentiation from primary myotubes treated with either a scramble shRNA (upper panel) or a mix of shRNAs targeting Macf1 (lower level). In the first frame (on the left), myofibers are selected in white, which corresponds to the region used to create the adjacent kymograph. Scale bar = 10 µm. (E–H) Speed (E), Time in motion (F), Duration of pauses (G), and Distance travelled by myonuclei (H) were quantified using SkyPad analysis (Cadot et al., 2014). Quantifications were done on myonuclei within different myofibers from three individual experiments. Line is set at median. Ns stands for not significant, *p<0.05, **p<0.01, and ***p<0.001 (Student’s t test).
-
Figure 2—source data 1
Table showing data from experiments plotted in Figure 2C and E–H.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig2-data1-v2.xlsx
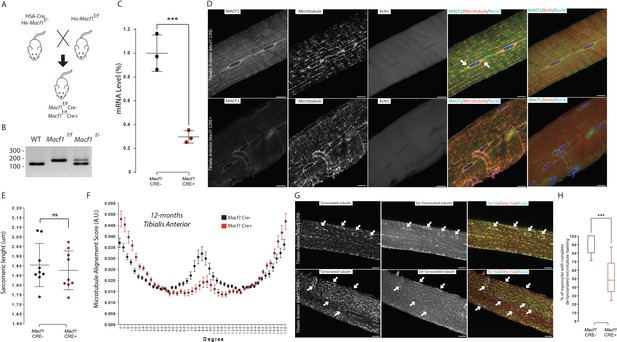
MACF1 is necessary for global and perinuclear microtubules organization within skeletal muscles in vivo.
(A) Scheme of the strategy used to generate muscle specific Macf1 knockout mouse model. (B) Representative PCR gel confirming the floxed Macf1 gene in mutant mice carrying Cre transgene. (C) Quantification of efficacy of the Cre-lox system in our muscle specific mouse model by analyzing the Macf1 gene expression level. qRT-PCR was carried out on total mRNA extracted from Gastrocnemius of 3 WT and 3 Macf1 KO mice of 12-month-old. Following the calculation of relative gene expression level on housekeeping genes for each mouse, the mean expression level of Macf1 was calculated for the WT mice. To generate the graph, the Macf1 expression level of each WT and KO mouse was then normalized on the previously calculated mean value. Line is set at mean (with SEM). ***p<0.001 (Student’s t test). (D) Representative confocal images (63×) of isolated myofibers from Tibialis Anterior of Macf1 Cre- (upper panel) and Cre+ (lower panel) mice, showing MACF1 (green), myonuclei (blue) and microtubules or F-actin (red). Scale Bar = 15 µm. (E) Sarcomere length quantified from the alpha actinin staining from 3 Macf1 Cre- and 3 Macf1 Cre+ 12-month-old mice. Following myofibers isolation from Tibialis Anterior, the distance between alpha actinin transverse bands of 3 myofibers per mouse was measured. Line is set at mean (with SD). Ns stands for not significant (Mann-Whitney test). (F) Microtubules network organization analysis of at least three myofibers from each of the three WT and three conditional Macf1-KO mice using TeDT software. The final generated graph presents a global score for each given degree of microtubules orientation, with 0 and 180 degrees corresponding to the longitudinal microtubules and 90 degrees corresponding to the transverse microtubules. Mean values (with SEM) is presented. (G) Representative confocal images (63×) from myofibers isolated from Tibialis Anterior issued from 12-month-old Macf Cre- (upper panel) or Cre+ (lower panel) mice, presenting tyrosinated tubulin (green), de-tyrosinated tubulin (red) and myonuclei (blue). White arrows point to myonuclei. Scale Bar = 15 µm. (H) Quantification of the percentage of myonuclei presenting the total de-tyrosinated tubulin ring. Quantifications were done on confocal images obtained from several isolated myofibers from Tibialis Anterior of three WT and three conditional Macf1-KO mice at the age of 12 months. Line is set at median. ***p<0.001 (Student’s t test).
-
Figure 3—source data 1
Table showing data from experiments plotted in Figure 3C,E,F and H.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig3-data1-v2.xlsx
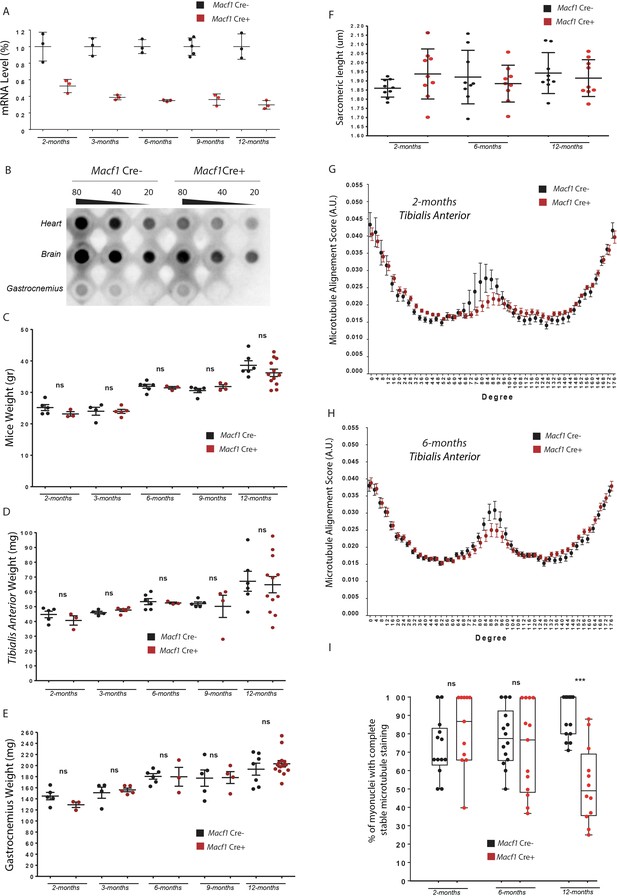
MACF1 conditional KO mice present progressive microtubules disorganization.
(A) Quantification of efficacy of Cre-lox system in our muscle specific mouse model by analyzing the Macf1 gene expression level. QRT-PCR was carried out on total mRNA extracted from Gastrocnemius of three individual Macf1 Cre- and Macf1 Cre+ mice at different ages. Following the calculation of relative gene expression level to housekeeping genes for each mouse, the mean expression level of Macf1 was calculated for the WT mice. To generate the graph, the Macf1 expression level of each WT and KO mouse was then normalized on the previously calculated mean value. Line is set at mean (with SEM). (B) Dot blot assay for MACF1 expression in Heart, Brain and Gastrocnemius lysates from 12-month-old Macf1 Cre- and Macf1 Cre+ mice. (C–E) Evaluation of total body weight (C), weight of Tibialis Anterior (D) and weigh of Gastrocnemius (E) of Macf1 Cre- and Macf1 Cre+ mice at different ages. Line is set at mean (with SEM). Ns stands for not significant (Student’s t test). (F) Measurements of sarcomere length quantified on alpha actinin staining in three individual Macf1 Cre- and Macf1 Cre+ mice at each given age. Following myofibers isolation from Tibialis Anterior, the distance between alpha actinin transverse bands of 3 myofibers per mouse was measured. Line is set at mean (with SD). (G–H) Microtubule network organization analysis from at least three myofibers from three individual Macf1 Cre- or conditional KO Macf1 Cre+ mice at 2 months (G) or 6 months (H) of age, using TeDT software. The final graph presents a global score for each given degree of microtubules orientation, with 0 and 180 degrees corresponding to the longitudinal microtubules and 90 degrees to the transverse microtubules. Mean values (with SEM) is presented. (I) Quantification of the percentage of myonuclei presenting the total de-tyrosinated tubulin ring. Quantifications were done on confocal images obtained from several myofibers isolated from Tibialis Anterior muscles of three Macf1 Cre- and three conditional KO Macf1 Cre+ mice at different ages. Line is set at median. Ns stands for not significant and ***p<0.001 (Student’s t test).
-
Figure 3—figure supplement 1—source data 1
Table showing data from experiments plotted in Figure 3—figure supplement 1A and C–I.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig3-figsupp1-data1-v2.xlsx
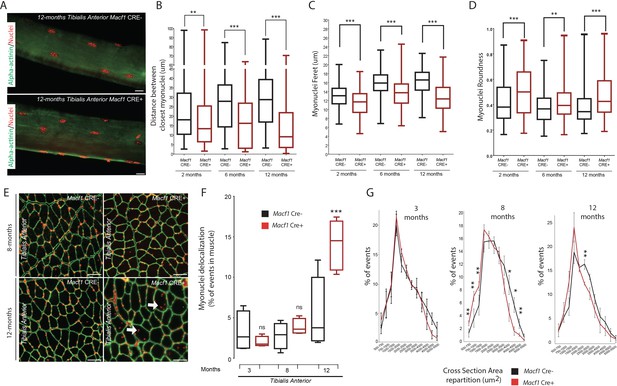
Loss of MACF1 alters myonuclei shape and positioning progressively with age.
(A) Representative confocal images (63×) of myofibers isolated from Tibialis Anterior of Macf1 Cre- (upper panel) and Cre+ (lower panel) mice at the age of 12 months presenting alpha-actinin (green) and myonuclei (red). Scale Bar = 15 µm. (B–D) Quantification of distance between the nearest myonuclei (B), myonuclei feret (C) and myonuclei roundness (D) measured on isolated myofibers from Tibialis Anterior of Macf1 Cre- and Cre+ mice (as presented in panel A) at 2-, 6-, and 12 months of age. Several myofibers were subjected to analysis per mouse. Each group gathers data obtained from myofibers of 3 or more mice. Line is set at median. **p<0.01 and ***p<0.001 (Student’s t test). (E) Representative images of Tibialis Anterior muscle cross-section from Macf1 Cre- (left panels) and Macf1 Cre+ (right panels) mice at the age of 8 and 12 months, stained for myonuclei (red) and Laminin (green). Examples of myofibers with mis-localized myonuclei are indicated by white arrows. Scale Bar = 150 µm. (F) Quantification of the percentage of myofibers with mis-localized myonuclei in Tibialis Anterior of 3, 8, and 12 months-old Macf1 Cre- and Macf1 Cre+ mice. Line is set at median. Ns stands for not significant and ***p<0.001 (Student’s t test). (G) Distribution of myofibers cross-section area in the Tibialis Anterior from 3-, 8-, and 12-months-old Macf1 Cre- and Macf1 Cre+ mice. Line is set at mean (with SEM). Ns stands for not significant, *p<0.05 and **p<0.01 (Student’s t test).
-
Figure 4—source data 1
Table showing data from experiments plotted in Figure 4B–D and F–G.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig4-data1-v2.xlsx
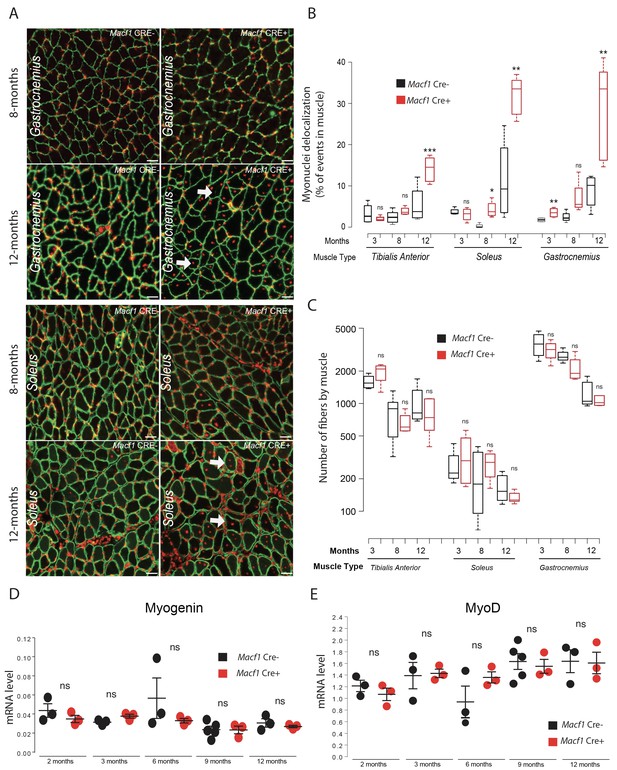
Mis-localization of myonuclei in MACF1 conditional KO mice is independent from muscular regeneration.
(A) Representative images of Gastrocnemius and Soleus muscles cross-sections from Macf1 Cre- (left panels) and Macf1 Cre+ (right panels) mice at the age of 8 and 12 months, stained for myonuclei (red) and Laminin (green). Example of myofibers with mis-localized myonuclei are indicated by white arrows. Scale Bar = 100 µm. (B) Quantification of the percentage of myofibers with mis-localized myonuclei in Tibialis Anterior, Soleus and Gastrocnemius of 3-, 8-, and 12-month-old Macf1 Cre- and Macf1 Cre+ mice. Line is set at median. Ns stands for not significant, *p<0.05, **p<0.01, and ***p<0.001 (Student’s t test). (C) Quantification of the number of muscle fibers per cross-sections of Tibialis Anterior, Soleus and Gastrocnemius of 3-, 8-, and 12-month-old Macf1 Cre- and Macf1 Cre+ mice. Line is set at median. Ns stands for not significant (Student’s t test). (D–E) QRT-PCR results presenting relative gene expression levels for Myogenin (D) and MyoD (E) in total mRNA extracts from Gastrocnemius of Macf1 Cre- and Cre+ at each given age. Target genes relative expression level to housekeeping genes is presented on each panel. Line is set at mean (with SEM). Ns stands for not significant (Student’s t test).
-
Figure 4—figure supplement 1—source data 1
Table showing data from experiments plotted in Figure 4—figure supplement 1B–E.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig4-figsupp1-data1-v2.xlsx
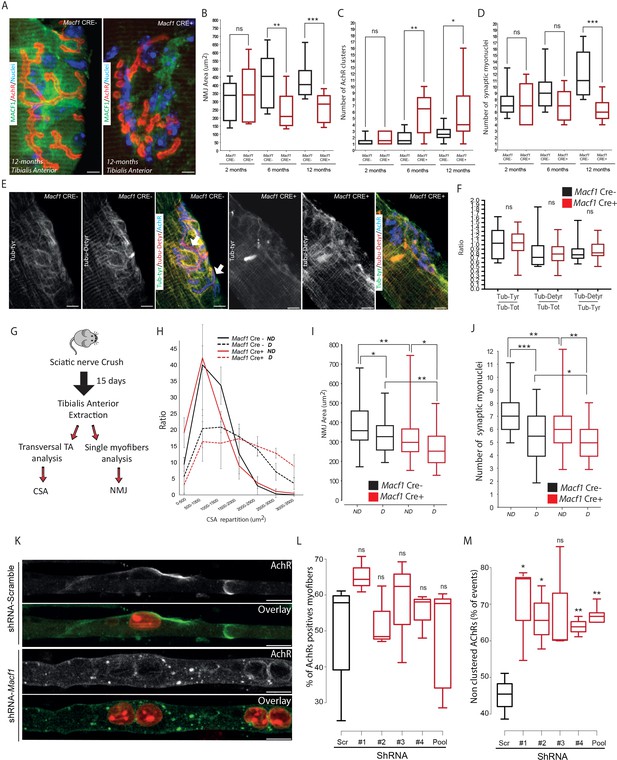
MACF1 controls AChRs clustering but has no impact on synaptic myonuclei at the NMJ.
(A) Representative confocal images (63×) of post-synaptic NMJs within isolated myofibers from Tibialis Anterior of Macf1 Cre- (left panel) and Cre+ (right panel) mice at the age of 12 months presenting MACF1 (green), AChRs clusters (red) and myonuclei (blue). Scale Bar = 10 µm. (B–D) Quantification of post-synaptic NMJs area (B), number of AChRs clusters (C) and number of synaptic myonuclei (D) measured on myofibers isolated from Tibialis Anterior of Macf1 Cre- and Cre+ mice (as presented in panel A) at 2-, 6-, and 12 months of age. Several myofibers were subjected to analysis per mouse and each group is formed from at least three mice. Line is set at median. Ns stands for not significant, *p<0.05, **p<0.01 and ***p<0.001 (Student’s t test). (E) Representative confocal images (63×) presenting tyrosinated tubulin (green), de-tyrosinated tubulin (red), and AChRs clusters (blue) in myofibers isolated from Tibialis Anterior of Macf1 Cre- (three left panels) and Cre+ (three right panels) mice at the age of 12 months. Scale Bar = 10 µm. (F) Quantification of the ratios of fluorescence intensity from tyrosinated tubulin to total tubulin, de-tyrosinated tubulin to total tubulin and de-tyrosinated tubulin to tyrosinated tubulin. The mean fluorescence intensity of each given staining was measured specifically for the post-synaptic NMJs area (as presented in panel E) on NMJs of myofibers isolated from Tibialis Anterior of Macf1 Cre- and Cre+ mice at the age of 12 months. Line is set at median. Ns stands for not significant (Student’s t test). (G) Scheme of the strategy used to induce sciatic nerve crush and subsequent analysis of muscle fibers from Macf1 Cre- and Cre+ mice. (H) Distribution of myofibers cross-section areas from non-denervated (referred as ND) and denervated (referred as D) Tibialis Anterior muscles of Macf1 Cre- and Cre+ mice at the age of 8 months. Line is set at mean (with SEM). (I–J) Quantification of post-synaptic NMJs area (I) and number of synaptic myonuclei (J) measured on isolated myofibers from non-denervated (referred as ND) and denervated (referred as D) Tibialis Anterior muscles of Macf1 Cre- and Cre+ mice at the age of 8 months. Line is set at median. *p<0.05, **p<0.01, and ***p<0.001 (Student’s t test). (K) Representative fluorescence images (63×) of AChRs clusters (green) and myonuclei (red) in myofibers treated with scramble shRNA (two upper panels) or with a pool of the four distinct shRNAs targeting Macf1 (two lower panels) after 10 days of differentiation in presence of Agrin in culture medium. Scale Bar = 10 µm. (L) Percentage of myofibers expressing immunofluorescence staining for AChRs in primary myofibers treated with scramble shRNA, with each of the four distinct shRNAs or with a pool of these four distinct shRNAs targeting Macf1 after 10 days of differentiation in the presence of Agrin. Data are from three individual experiments. Line is set at median. Ns stands for not significant (Student’s t test). (M) AChRs clustering distribution in cells treated with scramble shRNA, with each of the four individual shRNAs or with a pool of the four individual shRNAs targeting Macf1 after 10 days of differentiation in the presence of Agrin. Data was collected from three individual experiments for each condition. Line is set at median. Ns stands for not significant, *p<0.05 and **p<0.01 (Student’s t test).
-
Figure 5—source data 1
Table showing data from experiments plotted in Figures 5B–D,F,H–J and L–M.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig5-data1-v2.xlsx
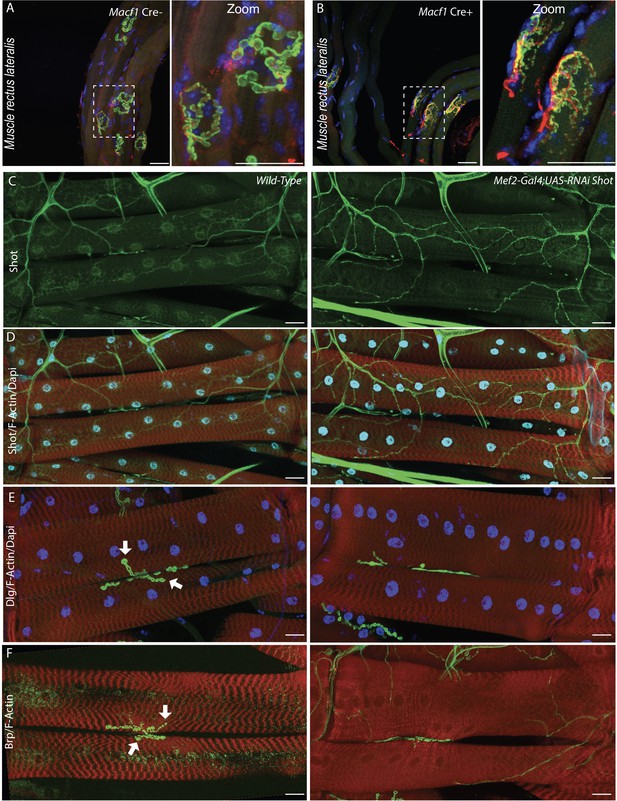
MACF1 orthologue, Shot, is essential for NMJs organization in Drosophila.
(A–B) Representative images of myofibers extracted from Rectus Lateralis muscle stained for myonuclei (blue), presynaptic markers (Neurofilament and Synaptic vesicles 2) (red) and AChRs (green) from 4-month-old Macf1 Cre- (A) and Macf1 Cre+ (B) mice. Scale Bar = 150 µm. (C–D) Larval muscles labeled for Shot (Green), myonuclei (blue) and F-actin (red) in WT (left panels) or RNAi treated muscle-targeting Shot (right panels). (E) Larval muscles labeled for Discs-Large, Dlg (Green), myonuclei (blue) and F-actin (red) in WT (left panel) or RNAi treated muscle targeting Shot (right panel). (F) Larval muscles labeled for Bruchpilot, BRP (Green), myonuclei (blue) and F-actin (red) in WT (left panel) or RNAi-treated muscle targeting Shot (right panel). Scale Bar = 50 µm.
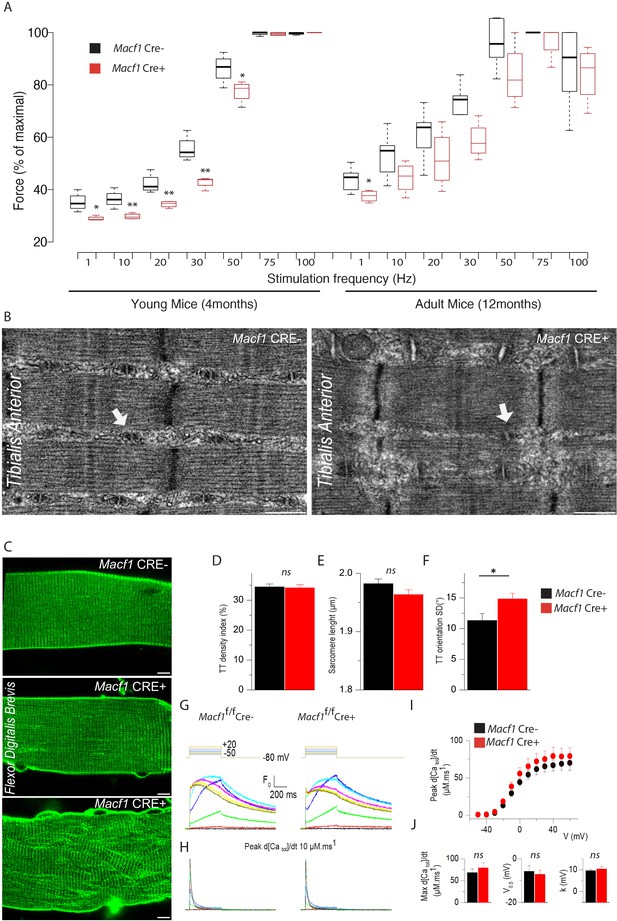
Muscle-specific Macf1 knockout affects muscle force production and T-tubule organization but leaves SR Ca2+ release unchanged.
(A) Force production from the Hindlimb muscles in young (4 months) and adult (12 months) Macf1 Cre- and Macf1 Cre+ mice in response to various incremental stimulation frequencies (from 1 to 100 Hz). Line is set at median. *p<0.05 and **p<0.01 (Student’s t test). (B) Representative electron microscopy images of myofibrils and T-tubule organization within Tibialis Anterior muscles from 12-month-old Macf1 Cre- and Macf1 Cre+ mice. Scale Bar = 0.5 µm. (C) Representative fluorescence microscopy images of di-8-anepps staining of the T-tubule network in Flexor Digitalis Brevis muscle fibers of 12-month-old Macf1 Cre- and Macf1 Cre+ mice. Scale Bar = 15 µm. (D–F) Quantification of T-tubules density index (D), Sarcomere length (E) and T-tubules orientation (F) in Flexor Digitalis Brevis muscle fibers from 12-month-old Macf1 Cre- and Macf1 Cre+ mice. Mean with SD is showing. Ns stands for not significant and *p<0.05 (Student’s t test). (G) Representative rhod-2 Ca2+ transients in a Macf1 Cre- and in a Macf1 Cre+ fiber in response to 0.5 s-long depolarizing pulses from −80 mV to the range of indicated values, with intervals of 10 mV. (H) Corresponding Ca2+ release flux (d[CaTot]/dt) traces calculated as described in the Materials and methods. (I) Mean voltage-dependence of the peak rate of SR Ca2+ release in Macf1 Cre- and in a Macf1 Cre+ fibers. (J) Inset shows the mean values for maximal rate, half-activation voltage and steepness factor (k) for SR Ca2+ release in the two groups of fibers, as assessed from Boltzmann fits to data from each fiber.
-
Figure 6—source data 1
Table showing data from experiments plotted in Figure 6A,D–F and I.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig6-data1-v2.xlsx
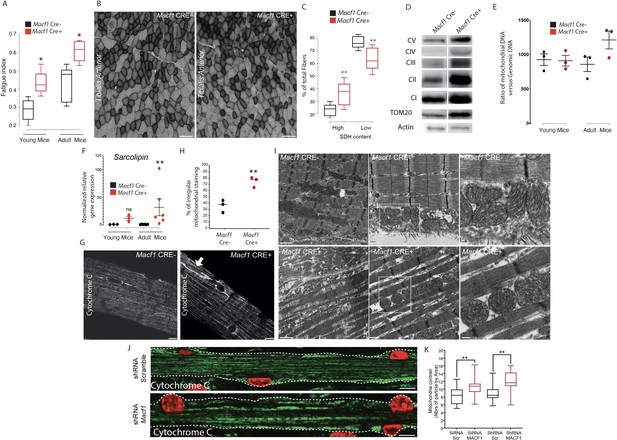
Muscle specific MACF1 knockout affects mitochondrial content and organization.
(A) Quantification of the fatigue index from the Hindlimb muscles in young (4 months) and adult (12 months) Macf1 Cre- and Macf1 Cre+. Line is set at median. *p<0.05 (Student’s t test). (B) Representative images of transversal cross-section of Tibialis Anterior muscle from 12-month-old Macf1 Cre- (left panel) and Macf1 Cre+ (right panel) mice stained for Succinate DeHydrogenase activity. Scale Bar = 150 µm. (C) Quantification of the Succinate DeHydrogenase activity relative to myofibers distribution in conditional Macf1-KO mice compared to control mice. Line is set at median. Ns stands for not significant, **p<0.01 (Student’s t test). (D) Western blot analysis of OXPHOS mitochondrial electron transport chain, TOM20 and actin proteins expression in total extracts from Gastrocnemius muscles of 12-month-old Macf1 Cre- and Macf1 Cre+ mice. (E) QPCR quantification of the ratio between mitochondrial and genomic DNA in total DNA extracts obtained from Gastrocnemius muscles of three individual Macf1 Cre- and Macf1 Cre+ mice at each given age. Line is set at mean (with SEM). (F) QRT-PCR results presenting relative gene expression level for SLN in Macf1 Cre- and Cre+ at each given age. Following the calculation of relative gene expression level with reference to housekeeping genes for each mouse, the mean expression level of SLN was calculated for the WT mice. Next, the expression level of each WT and KO mouse was normalized to the previously calculated mean value. Line is set at mean (with SEM). Ns stands for not significant and **p<0.01 (Mann-Whitney test). (G) Representative confocal images (63×) from isolated myofibers of Tibialis Anterior stained for Cytochrome C in Macf1 Cre- (left panel) and Cre+ (right panel) mice of 12-month-old mice. Scale Bar = 15 µm. (H) Quantification of the percentage of myofibers with irregular (broken/oriented/accumulated) mitochondrial staining in isolated myofibers of Tibialis Anterior muscles from three individual Macf1 Cre- and Cre+ mice at the age of 12 months. Line is set at median. **p<0.01 (Student’s t test). (I) Representative electron microscopy images of myofibrils and mitochondria organization within Tibialis Anterior muscles from 12-month-old Macf1 Cre- (upper panels) and Macf1 Cre+ (lower panels) mice. Scale Bar = 1, 0.5, 0.2 µm. (J) Confocal immunofluorescence images (63×) presenting the organization of Cytochrome C (green) in primary myofibers treated either with scramble shRNA (upper panel) or with a pool of four distinct shRNAs targeting Macf1 (lower panel) after 13 days of differentiation. Myonuclei are presented in red. Scale Bar = 10 µm. (K) Quantification of mitochondrial content per area in primary myofibers treated either with scramble shRNA or with a pool of four distinct shRNAs targeting Macf1 after 13 days of differentiation. Line is set at median. **p<0.01 (Student’s t test).
-
Figure 7—source data 1
Table showing data from experiments plotted in Figure 7A,C,E–F,H and K.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig7-data1-v2.xlsx
-
Figure 7—source data 2
Whole immunoblotting membrane for OXPHOS, TOM20, and actin plotted in Figure 7D.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig7-data2-v2.pdf
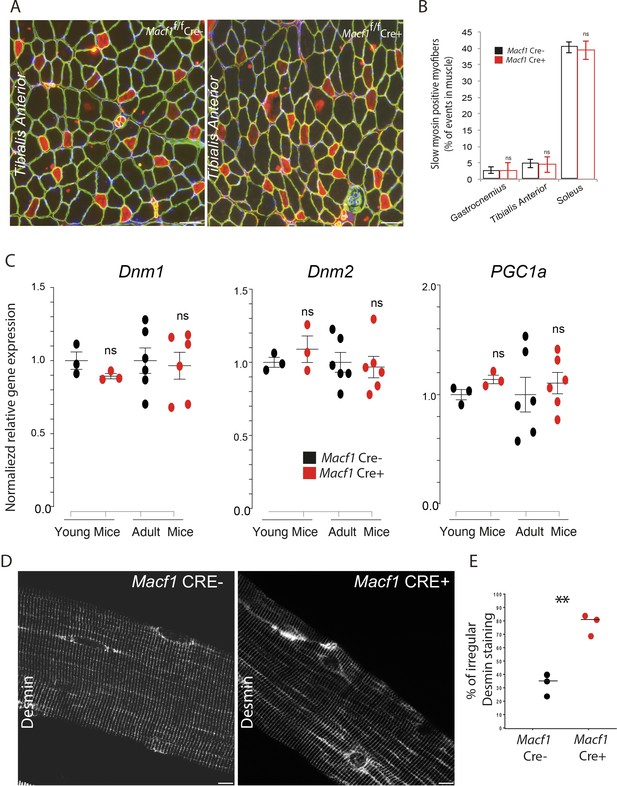
MACF1 conditional KO mice present normal distribution of different skeletal muscles types but disorganized Desmin network.
(A) Representative microscopy images of transverse cross sections from Tibialis Anterior muscle of 12-month-old Macf1 Cre- (left panel) and Macf1 Cre+ (right panel) mice stained for slow myosin (red), Laminin (green) and myonuclei (blue). Scale Bar = 150 µm. (B) Quantification of the percentage of myofibers with slow myosin expression in Tibialis Anterior, Soleus and Gastrocnemius muscles from 12-month-old Macf1 Cre- and Macf1 Cre+ mice. Mean values (with SEM) is presented. Ns stands for not significant (Student’s t test). (C) QRT-PCR results presenting relative gene expression levels for Dnm1, Dnm2, and PPARGC1A in total mRNA extracts from Gastrocnemius of Macf1 Cre- and Cre+ at each given age. Following the calculation of relative gene expression level with reference to housekeeping genes for each mouse, the mean expression level of each target gene was calculated for the WT mice. Next, the expression level of each WT and KO mouse was normalized to the previously calculated mean value. Line is set at mean (with SEM). Ns stands for not significant (Student’s t test). (D) Representative confocal images (63×) from isolated myofibers of Tibialis Anterior stained for Desmin in Macf1 Cre- (left panel) and Cre+ (right panel) mice of 12-month-old mice. Scale Bar = 15 µm. (E) Quantification of the percentage of myofibers with irregular (broken/oriented/accumulated) Desmin staining in isolated myofibers of Tibialis Anterior muscles from three individual Macf1 Cre- and Cre+ mice at the age of 12 months. Line is set at median. **p<0.01 (Student’s t test).
-
Figure 7—figure supplement 1—source data 1
Table showing data from experiments plotted in Figure 7—figure supplement 1B–C and E.
- https://cdn.elifesciences.org/articles/70490/elife-70490-fig7-figsupp1-data1-v2.xlsx
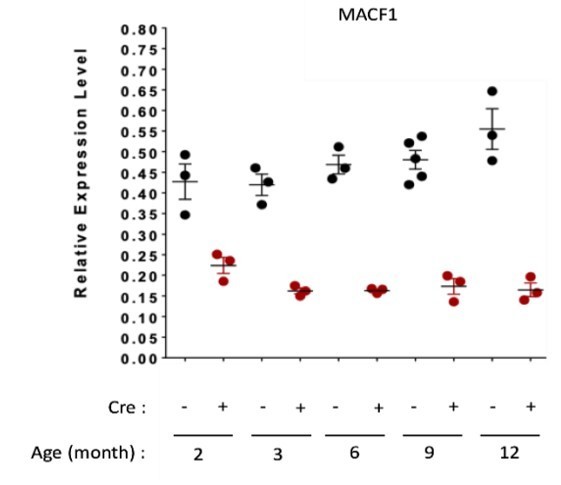
QRT-PCR results presenting relative gene expression level (to housekeeping genes) for MACF1 in Macf1cKO Cre- and Cre+ at each given age.
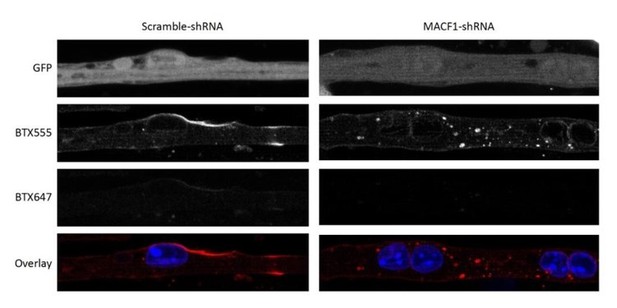
aBTX staining in primary myofibers transfected with scramble or MACF1 shRNAs.
Scale Bar 10 µm.
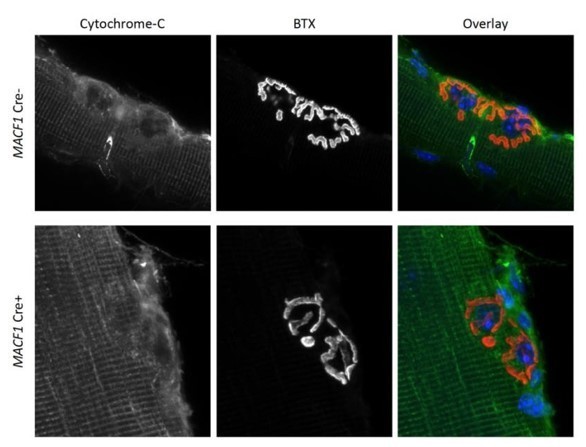
Representative confocal microscopy images (63X) from mitochondrial density (reflected by Cytochrome-C staining) at NMJs (reflected by BTX staining) of extracted Tibialis anterior of 12-month-old MACF1 Creand Cre+ mice.
In the overlay, mitochondria are presented in green, postsynaptic NMJs in red and synaptic nuclei in blue. Scale Bar 10 µm.
Videos
Time-lapse experiments of primary myoblasts co-transfected with EB3-GFP plasmid and Scramble-siRNA at 1 day, 5 days, and 10 days post ‘late differentiation’ initiation (left panels) or primary myoblasts co-transfected with EB3-GFP plasmid and Macf1- siRNA at 1 day, 5 days, and 10 days post ‘late differentiation’ initiation (right panels).
Each frames represent 500 milliseconds and was recorded for a period of time of 7 s.
Time-lapse experiments of primary myoblasts co-transfected with Lamin-Chromobody-RFP plasmids and either Scramble shRNA (left panel) or a pool of four individual Macf1 shRNA (rigth panel).
Time laps images were captured form myotubes 5 days after starting differentiation process. Primary myotubes were recorded every 15 min for a period of time of 14 hr.