Control of spinal motor neuron terminal differentiation through sustained Hoxc8 gene activity
Figures
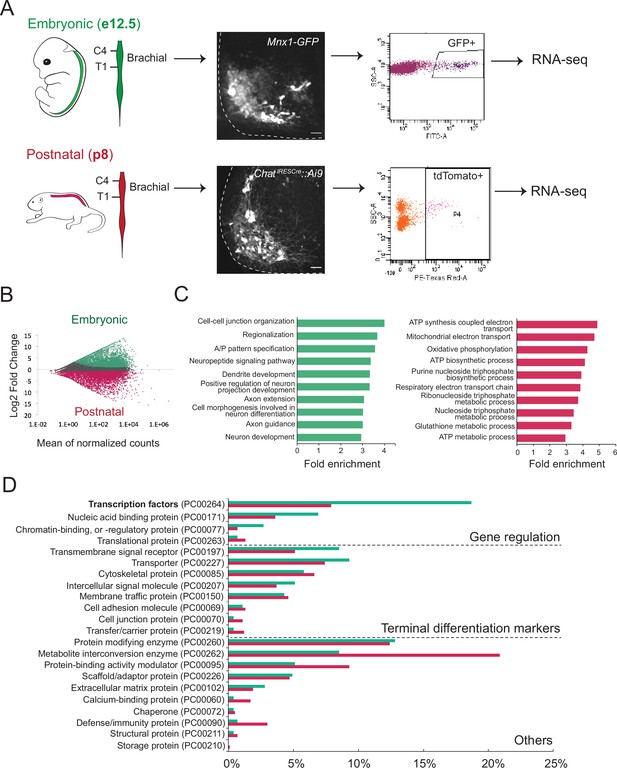
Molecular profiling of mouse brachial motor neurons (MNs) at embryonic and postnatal stages.
(A) Schematic representation of the workflow used in the comparison of embryonic and postnatal transcriptomes. The brachial domain (C4–T1) of Mnx1-GFP (in green) and ChatIRESCre::Ai9 (in red) mice was microdissected. Brachial GFP+ (at e12.5, scale bar: 20 μm) and tdTomato+ (at p8, scale bar: 100 μm) MNs were fluorescence-activated cell sorted and processed for RNA-sequencing. Spinal cord is outlined with white dashed line. (B) MA plot of differentially expressed genes. Green and red dots represent individual genes that are significantly (p<0.05) expressed (fourfold and/or higher) in embryonic and postnatal MNs, respectively. (C) Graphs showing fold enrichment for genes involved in specific biological processes. (D) Gene onthology analysis comparing protein class categories of highly expressed genes in embryonic (e12.5) and postnatal (p8) MNs. Green and red bars represent embryonic and postnatal genes, respectively.
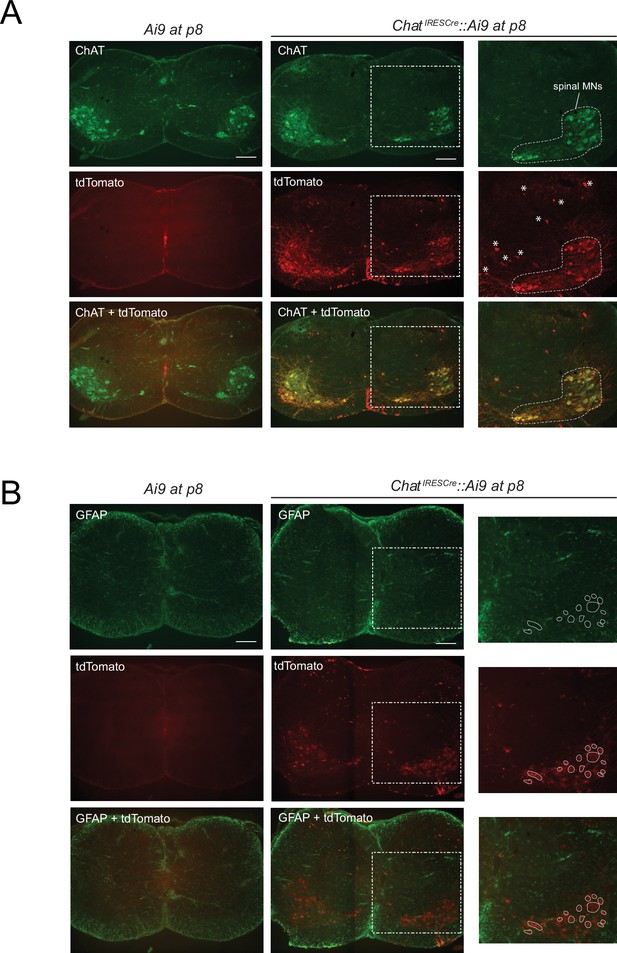
Testing the specificity of genetic labeling of brachial motor neurons (MNs) at p8.
(A) Double immunofluorescence staining for ChAT (green) and tdTomato (red) shows overlap in spinal MNs (white dotted area in inset) of ChatIRESCre::Ai9 (Rosa26-CAGpromoter-loxP-STOP-loxP-tdTomato) mice at p8. Asterisks indicate tdTomato expression in a small fraction of cells that are not MNs (based on their position). As a negative control, no tdTomato is detected in the spinal cord of Ai9 mice (not crossed with ChatIRESCre). Scale bar: 200 μm. (B) Double immunofluorescence staining for the astrocyte marker GFAP (glial fibrillary acidic protein, ingreen) and tdTomato (red) shows no overlap in the spinal cord of ChatIRESCre::Ai9 (Rosa26-CAGpromoter-loxP-STOP-loxP-tdTomato) mice at p8. White circles indicate tdTomato expression in brachial MNs (based on their position). As a negative control, no tdTomato is detected in the spinal cord of Ai9 mice (not crossed with ChatIRESCre). Scale bar: 200 μm.
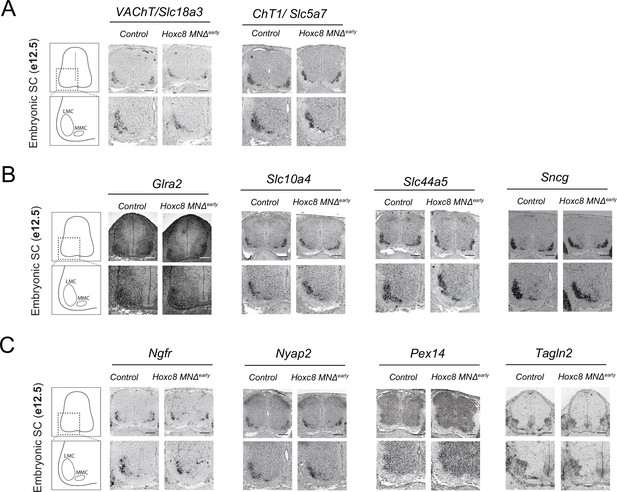
RNA ISH analysis of terminal differentiation markers in Hoxc8 MNΔ early mice.
. Expression of Slc18a3, Slc5a7, Glra2, Slc10a4, Slc44a5, Sncg, Ngfr, Nyap2, Pex14, and Tagln2 was detected in the ventrolateral region of embryonic (e12.5) spinal cords (SC) in control (Hoxc8 fl/fl) and Hoxc8 MNΔ early embryos (N = 4). Zoomed images of the ventrolateral SC are provided below each image. Scale bar: 100 μm.
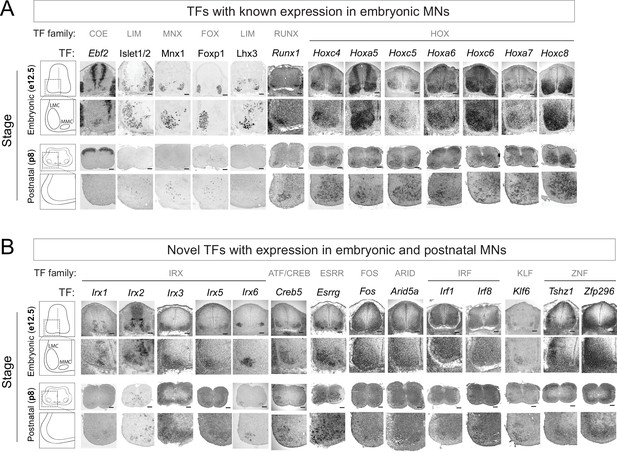
Known and novel transcription factors (TFs) are continuously expressed in brachial motor neurons (MNs) during embryonic and postnatal stages.
(A) The expression of TFs with previously published roles in MN development was assessed in embryonic (e12.5) and postnatal (p8) spinal cords (N = 4) with RNA ISH (Ebf2, Runx1, Hoxc4, Hoxa5, Hoxc5, Hoxa6, Hoxc6, Hoxa7, Hoxc8) and immunohistochemistry (Islet1/2, Mnx1 [Hb9], Lhx3, Foxp1). Zoomed area of one side of the ventral spinal cord is shown below each image. (B) The expression of novel TFs was assessed in embryonic (e12.5) and postnatal (p8) spinal cords with RNA ISH (N = 4). Scale bar for e12.5 images: 50 μm; scale bar for p8 images: 250 μm.
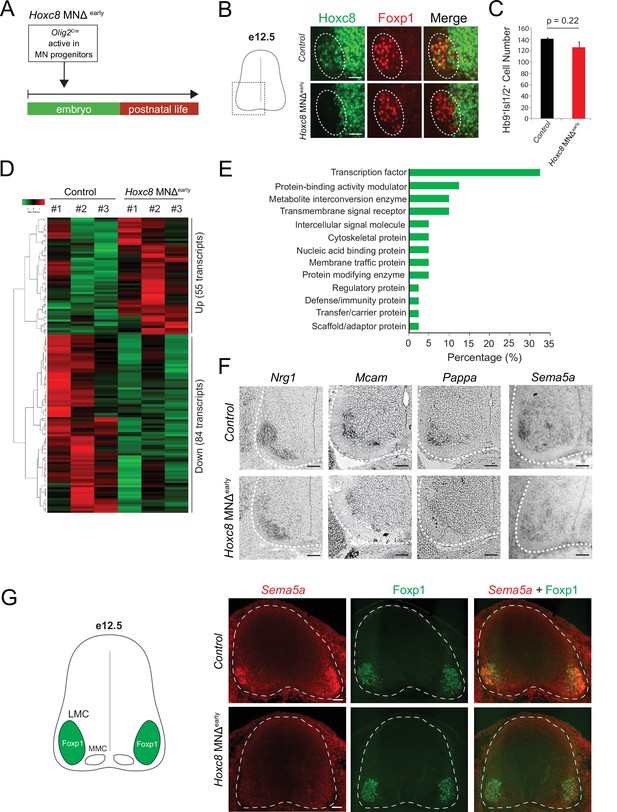
Early Hoxc8 gene inactivation in brachial motor neurons (MNs) affects the expression of terminal differentiation genes.
(A) Diagram illustrating genetic approach for Hoxc8 gene inactivation during early MN development (Hoxc8 MNΔ early mice). (B) Immunohistochemistry showing that Hoxc8 protein (green) is not detected in Foxp1+ MNs (red, indicated with dashed ellipse) of Hoxc8 MNΔearly spinal cords at e12.5. Images of one side of the spinal cord are shown (boxed region in schematic at left). Scale bar: 50 μm. (C) Quantification of Mnx1+(Hb9+) Isl1/2+ MNs in e12.5 brachial spinal cords of Hoxc8 MNΔearly and control (Hoxc8fl/fl) embryos (N = 4). (D) Heatmap showing upregulated and downregulated genes detected by RNA-Seq in control (Hoxc8 fl/fl) and Hoxc8 MNΔearly e12.5 MNs. Green and red colors, respectively, represent lower and higher gene expression levels. (E) Graphical percentage (%) representation of protein classes of the downregulated genes in Hoxc8 MNΔearly spinal cords. (F) RNA ISH showing downregulation of Nrg1, Mcam, Pappa, and Sema5a mRNAs in brachial MNs of e12.5 Hoxc8 MNΔearly spinal cords (N = 4). Spinal cord is outlined with a white dotted line. Scale bar: 50 μm. (G) RNA FISH for Sema5a coupled with antibody staining against Foxp1 (LMC marker) shows reduced Sema5a mRNA expression in Foxp1 +MNs of e12.5 Hoxc8 MNΔearly spinal cords (N = 4). Images of a cross-section of the entire e12.5 spinal cord are shown. Scale bar: 40 μm.
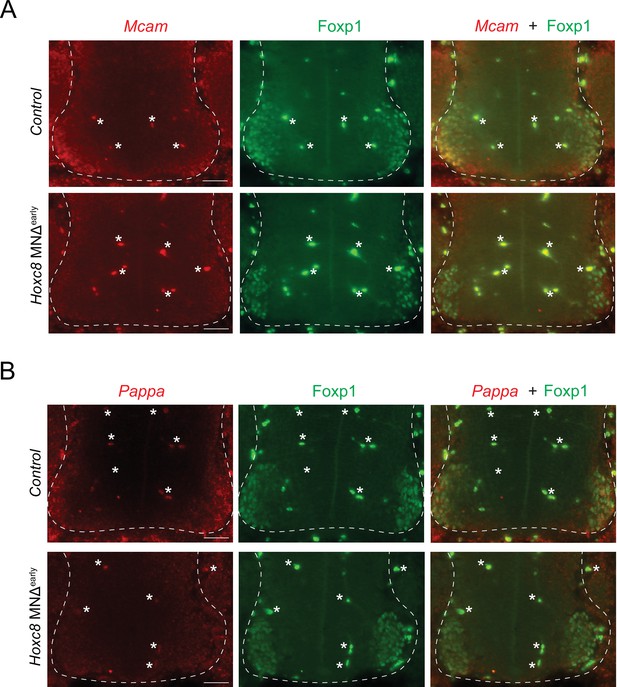
RNA FISH analysis of terminal differentiation markers in Hoxc8 MNΔ early mice.
(A) RNA FISH for Mcam coupled with antibody staining against Foxp1 (LMC marker) shows reduced Mcam mRNA expression in Foxp1 +MNs of e11.5 Hoxc8 MNΔearly spinal cords (N = 4). Note that Mcam partially overlaps with Foxp1-expressing motor neurons (MNs) in the control (Hoxc8 fl/fl) spinal cord. (B) RNA FISH for Pappa coupled with antibody staining against Foxp1 (LMC marker) shows reduced Pappa mRNA expression in Foxp1 +MNs of e11.5 Hoxc8 MNΔearly spinal cords (N = 4). Scale bar: 50 μm. Spinal cord is outlined with a white dashed line. White asterisks (*) indicate autofluorescence from blood cells.
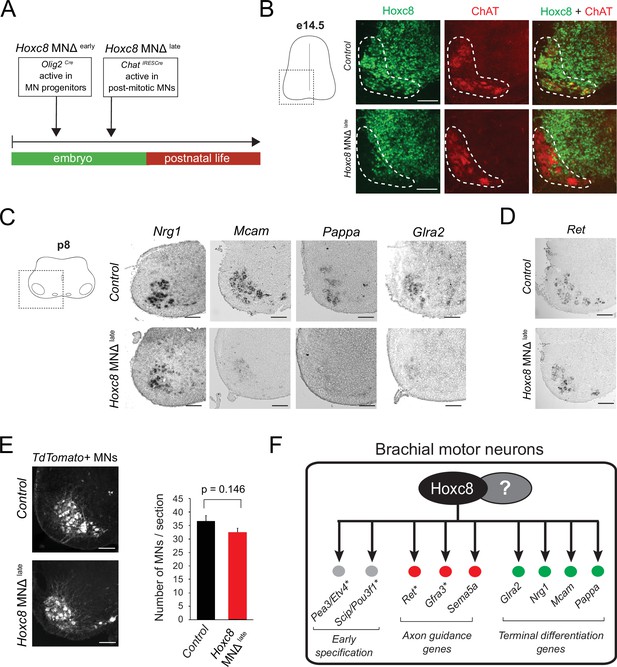
Late Hoxc8 gene inactivation in brachial motor neurons (MNs) affects expression of terminal differentiation genes.
(A) Diagram illustrating genetic approach for Hoxc8 gene inactivation during late MN development. Hoxc8 conditional mice were crossed with the ChatIRESCre mouse line (Hoxc8 MNΔlate). (B) Immunohistochemistry showing that Hoxc8 protein (green) is not detected in ChAT-exprressing MNs (red) of Hoxc8 MNΔ late spinal cords at e14.5 (N = 4). MN location is indicated with white dashed line. Hoxc8 is also expressed in other cell types outside the MN territory. Images of one side of the spinal cord are shown (boxed region in schematic at left). Scale bar: 100 μm. (C) RNA ISH showing reduced expression of Pappa, Mcam, Glra2, and Nrg1 in Hoxc8 MNΔlate spinal cords at p8 (N = 4). Scale bar: 200 μm. (D) Ret expression is comparable between control and Hoxc8 MNΔlate spinal cords at p8 (N = 4). Scale bar: 200 μm. (E) Representative images and quantification of TdTomato-labeled MNs in p8 control (Hoxc8fl/fl::Ai9) and Hoxc8 MNΔ late (Hoxc8fl/fl::ChatIRESCre::Ai9) spinal cords (N = 4). Scale bar: 200 μm. (F) Schematic summarizing Hoxc8 target genes in brachial MNs. Asterisks indicate previously known Hoxc8 target genes.
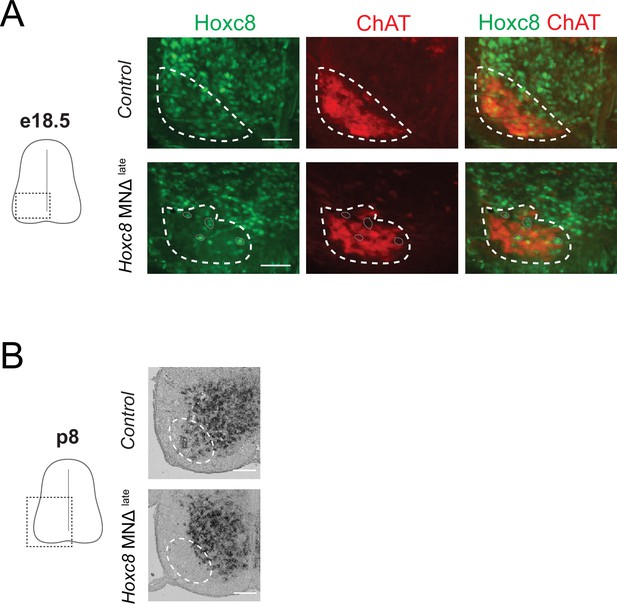
Depletion of Hoxc8 in brachial motor neurons (MNs) of Hoxc8 MNΔ late mice.
(A) Immunohistochemistry showing that Hoxc8 protein (green) is not detected in ChAT-expressing MNs (red) of Hoxc8 MNΔlate spinal cords at e18. (N = 3). MN location is indicated with white dashed line. Cells still expressing Hoxc8 (gray circles) are not MNs because they do not express ChAT. Scale bar: 100 μm. (B) RNA ISH shows no Hoxc8 expression in the expected location of brachial MNs of Hoxc8 MNΔlate mice at p8. Expression in other cells outside the MN territory is unaffected in Hoxc8 MNΔlate spinal cord. Scale bar: 200 μm.
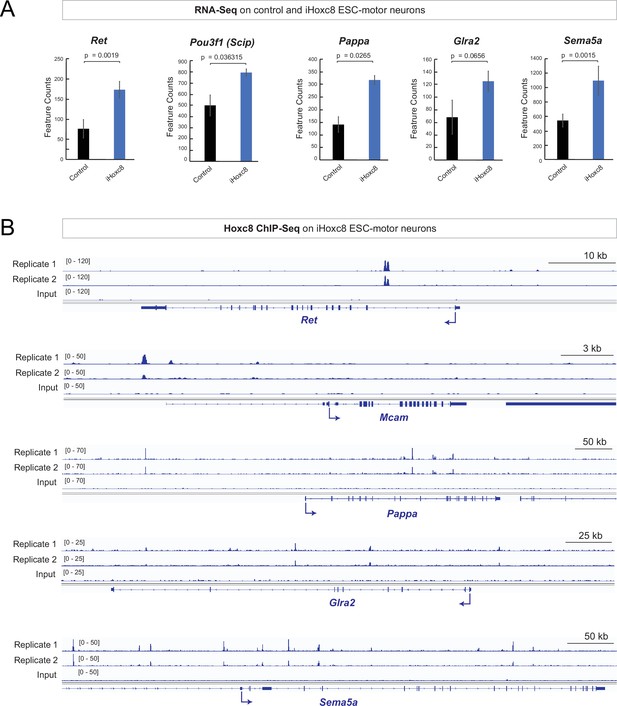
Hoxc8 sufficiency and direct mode of action.
(A) Analysis of RNA-sequencing (RNA-Seq) data from control and iHoxc8 motor neurons (MNs) shows Hoxc8 is sufficient to induce the expression of previously known (Ret, Pou3f1[Scip]) and new (Pappa, Glra2, Sema5a) Hoxc8 target genes. GEO accession numbers: Control (GSM4226469, GSM4226470, GSM4226471) and iHoxc8 (GSM4226475, GSM4226476, GSM4226477). (B) Analysis of chromatin immunoprecipitation-sequencing (ChIP-Seq) data from iHoxc8 MNs shows Hoxc8 directly binds to the cis-regulatory region of its target genes (Ret, Mcam, Pappa, Glra2, Sema5a). GEO accession numbers: Input (GSM4226461) and iHoxc8 replicates (GSM4226436, GSM4226437). Snapshots of each gene locus were generated with Integrative Genomics Viewer (IGV, Broad Institute).
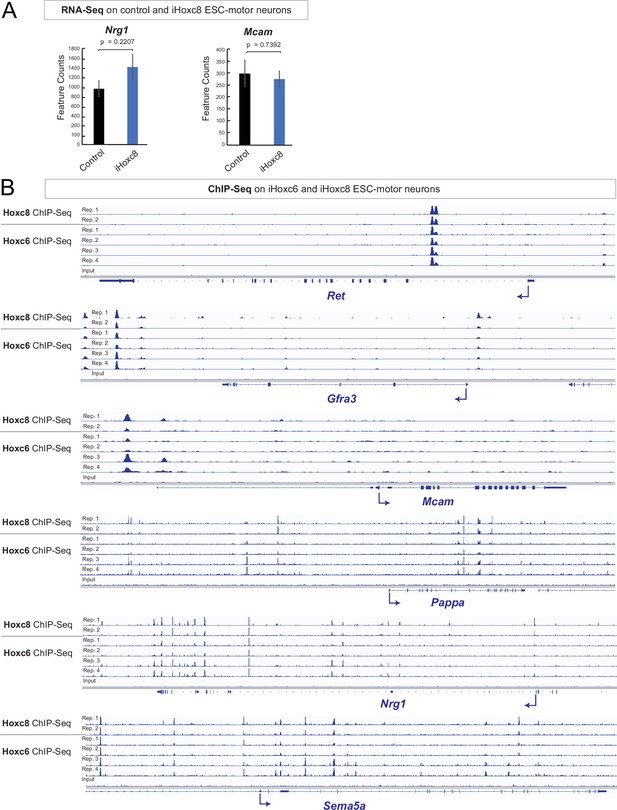
Hoxc6 and Hoxc8 bind to the same cis-regulatory regions of Hoxc8 target genes.
(A) Analysis of RNA-sequencing (RNA-Seq) data from control and iHoxc8 motor neurons (MNs) shows Hoxc8 is not sufficient to induce expression of Nrg1 and Mcam. GEO accession numbers: Control (GSM4226469, GSM4226470, GSM4226471) and iHoxc8 (GSM4226475, GSM4226476, GSM4226477). (B) Analysis of chromatin immunoprecipitation-sequencing (ChIP-Seq) data from iHoxc8 and iHoxc6 MNs shows both Hox proteins directly bind to the cis-regulatory region of several Hoxc8 target genes (Ret, Mcam, Gfra3, Pappa, Nrg1, Sema5a). GEO accession numbers: Input (GSM4226461), iHoxc8 replicates (GSM4226436, GSM4226437) and iHoxc6 replicates (GSM4226434, GSM4226435, GSM4226450, GSM4226451). Snapshots of each gene locus were generated with Integrative Genomics Viewer (IGV, Broad Institute).
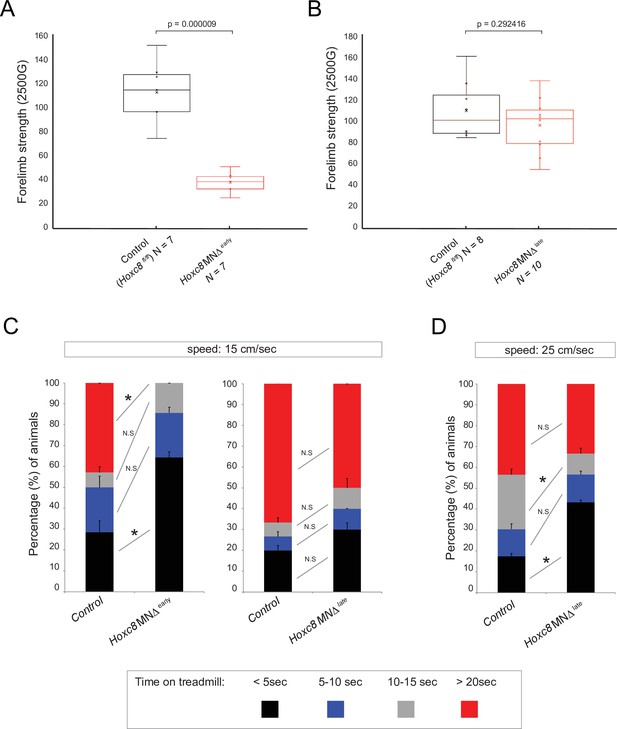
Brachial motor neuron (MN) function is impaired upon Hoxc8 depletion.
(A) Forelimb grip strength analysis on control (Hoxc8 fl/fl, N = 7) and Hoxc8 MNΔ early (N = 8) adult mice. See Methods for details. (B) Forelimb grip strength analysis on control (Hoxc8 fl/fl, N = 7) and Hoxc8 MNΔ late (N = 8) adult mice. (C). Treadmill analysis (at 15 cm/s speed) on control (Hoxc8 fl/fl, N = 7) and Hoxc8 MNΔ early (N = 8) adult mice, as well as on control (Hoxc8 fl/fl, N = 8) and Hoxc8 MNΔ late (N = 10) adult mice. See Methods for details. Asterisk (*) indicates p=0.0108. Experiment repeated twice. (D). Treadmill analysis (at 25 cm/s speed) on control (Hoxc8 fl/fl, N = 8) and Hoxc8 MNΔ late (N = 10) adult mice. Treadmill speed at 25 cm/s. Asterisk (*) indicates p=0.0461. Experiment repeated three times. The 30-s long videos were analyzed and data were binned into four categories based on the duration of each mouse’s stay on the treadmill (category 1 [black]: <5 s; category 2 [blue]: 5–10 s; category 3 [gray]: 10–15 s; category 4 [red]: >20 s).
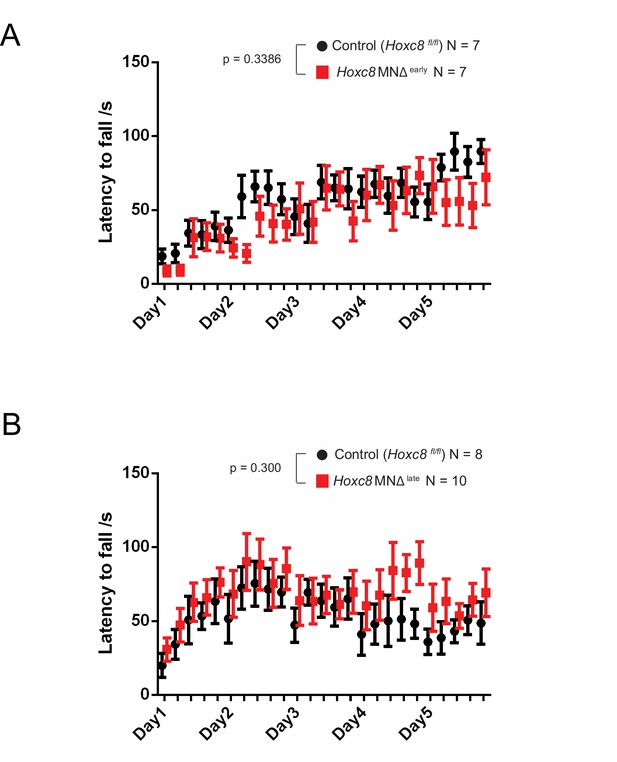
Rotarod performance test on Hoxc8 MNΔ early and Hoxc8 MNΔ late mice.
(A) Rotarod performance test conducted on 4–5 month-old Hoxc8 MNΔ early female mice (N = 7) and control littermates (Hoxc8 fl/fl) (N = 7). Two-way ANOVA (Prism Software), Genotype main effect, F (1, 12) = 0.9932; p=0.3386. (B) Rotarod performance test conducted on 2–5 month-old Hoxc8 MNΔ late female mice (N = 8) and control littermates (Hoxc8 fl/fl) (N = 10). Two-way ANOVA (Prism Software), Genotype main effect, F (1, 16) = 1.147; p=0.300.
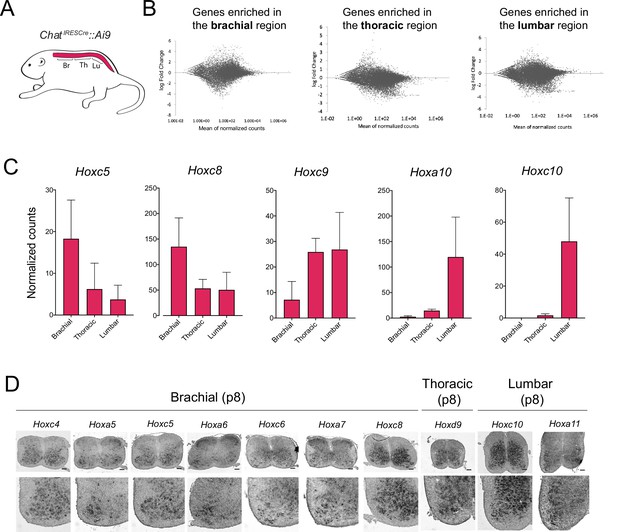
Different Hox genes are expressed in brachial, thoracic, and lumbar motor neurons (MNs) at postnatal day 8.
(A) Schematic showing the different regions of the p8 spinal cord (brachial, thoracic, lumbar) used for RNA-sequencing (RNA-Seq) analysis. The ChatIRESCre::Ai9 mouse line was used to fluorescently label MNs (see Materials and methods). (B) MA plots showing the differential expression of genes (each dot is an individual transcript) in brachial, thoracic, and lumbar regions. (C) Plots showing the normalized counts of Hoxc5, Hoxc8, Hoxc9, Hoxa10, and Hoxc10 transcripts in the brachial, thoracic, and lumbar regions of the spinal cord. (D) RNA ISH was used to independently validate the RNA-Seq data shown in panels B-C. Expression of Hoxc4, Hoxa5, Hoxc5, Hoxa6, Hoxc6, Hoxa7, and Hoxc8 was assessed in p8 brachial MNs, and is also shown in Figure 2 (panel A). Expression of Hoxd9 was assessed in p8 thoracic MNs, whereas Hoxc10 and Hoxa11 were evaluated in p8 lumbar MNs (N = 4). Scale bar: 200 μm.
Treadmill test on control (Hoxc8 fl/fl) littermate of Hoxc8 MNΔ early mice.
Treadmill test on a Hoxc8 MNΔ early mouse.
Treadmill test on control (Hoxc8 fl/fl) littermate of Hoxc8 MNΔ late mice.
Treadmill test on a Hoxc8 MNΔ late mouse.
Tables
Summary of candidate and unbiased approaches to reveal Hoxc8 target genes in mouse brachial MNs.
| Gene name | Expression in WT brachial MNs | Hoxc8 dependency | |||||
|---|---|---|---|---|---|---|---|
| e12 | p8 | p56 Allen Brain ISH | p60snRNA-Seq dataset | Hoxc8 MNΔ early mice | Hoxc8 MNΔ late mice | ||
| Candidate approach | Slc10a4 | + | + | + | + | No | N.D |
| Nrg1 | + | + | + | + | Yes | Yes | |
| Nyap2 | + | + | N.D | + | No | N.D | |
| Sncg | + | + | + | + | No | N.D | |
| Ngfr | + | + | + | – | No | N.D | |
| Glra2 | + | + | + | + | No | Yes | |
| Cldn1 | N.D | – | N.D | – | N.D | N.D | |
| Cacna1g | N.D | – | + | + | N.D | N.D | |
| RNA-Seq approach | Slc44a5 | + | + | + | + | No | N.D |
| Mcam | + | + | + | + | Yes | Yes | |
| Pappa | + | + | + | + | Yes | Yes | |
| Sema5a | + | + | N.D | + | Yes | N.D | |
| Pex14 | + | + | N.D | + | No | N.D | |
| Tagln2 | + | + | + | – | No | N.D | |
| Cldn19 | N.D | – | – | – | N.D | N.D | |
| Wwc2 | N.D | – | + | + | N.D | N.D | |
| Septin1 | N.D | – | N.D | N.D | N.D | N.D | |
| Irx2 | + | + | + | – | N.D | N.D | |
| Irx5 | + | + | + | – | N.D | N.D | |
| Irx6 | + | + | + | – | N.D | N.D | |
| Known Hoxc8 targets | Ret | + | + | N.D | + | Yes | No |
| Gfra3 | + | – | – | – | Yes | N.D | |
-
Expression in p56 brachial MNs was determined using the Allen Brain Map (http://portal.brain-map.org). We also interrogated the single nucleus (sn) RNA-seq datasets of p60 spinal MNs from http://spinalcordatlas.org/.
-
N.D: Not determined; + denotes expression; – denotes no expression.
-
RNA-Seq: RNA-sequencing.
Validation of transcription factor expression in brachial MNs.
| TF | Type | Novel TF with MN expression | e12 MNs | p8 MNs | p56 MNsISH Allen Brain | p60snRNA-Seq dataset | Expression in other spinal cells at e12 |
|---|---|---|---|---|---|---|---|
| Ebf2 | Ebf/COE | No | + | – | – | – | + |
| Islet1 | LIM HD | No | + | + | + | N.D | + |
| Islet2 | LIM HD | No | + | + | + | N.D | + |
| Hb9 | HD | No | + | + | + | N.D | + |
| Foxp1 | FOX | No | + | + | – | + | – |
| Lhx3 | LIM HD | No | + | + | N.D | – | + |
| Runx1 | RUNX | No | + | + | + | – | – |
| Hoxc4 | HOX | No | + | + | + | + | + |
| Hoxa5 | HOX | No | + | + | N.D | – | + |
| Hoxc5 | HOX | No | + | + | + | + | + |
| Hoxa6 | HOX | No | + | + | – | – | + |
| Hoxc6 | HOX | No | + | + | N.D | – | + |
| Hoxa7 | HOX | No | + | + | + | – | + |
| Hoxc8 | HOX | No | + | + | N.D | – | + |
| Irx1 | IRO HD | Yes | + | + | + | – | + |
| Irx2 | IRO HD | Yes | + | + | + | – | + |
| Irx3 | IRO HD | Yes | + | + | + | – | – |
| Irx5 | IRO HD | Yes | + | + | + | – | – |
| Irx6 | IRO HD | Yes | + | + | + | – | – |
| Creb5 | CRE | Yes | + | + | + | + | – |
| Esrrg | NHR | Yes | + | + | – | + | + |
| Fos | FOS | Yes | + | + | + | – | + |
| Arid5a | ARID | Yes | + | + | – | – | + |
| Irf1 | IRF | Yes | + | + | – | – | + |
| Irf8 | IRF | Yes | + | + | – | – | + |
| Klf6 | KLF | Yes | + | + | – | + | – |
| Tshz1 | C2H2 Zn | Yes | + | + | + | + | + |
| Zfp296 | ZFP | Yes | + | + | + | – | + |
| Neurod6 | bHLH | N.A | – | – | N.D | – | Dorsal interneurons |
| Arid5b | ARID | N.A | – | – | N.D | + | Dorsal interneurons |
| Pou3f3 | POU | N.A | – | – | N.D | + | Dorsal interneurons |
| Mafb | bZIP | N.A | – | – | N.D | N.D | Ventral interneurons |
| Zfhx4 | Zn HD | N.A | – | – | N.D | + | Ventral interneurons |
| Elk3 | ETS | N.A | – | – | N.D | + | Vasculature |
| Epas1 | HIF | N.A | – | – | N.D | – | Vasculature |
| Heyl | bHLH | N.A | – | – | N.D | – | Vasculature |
-
Expression in p60 brachial MNs was determined using the Allen Brain Map (http://portal.brain-map.org). We also interrogated the single nucleus (sn) RNA-seq datasets of p60 spinal MNs from http://spinalcordatlas.org/. + denotes expression; – denotes no expression; N. D: Not determined; N. A: Not applicable.
-
RNA-Seq: RNA-sequencing.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Mnx1-GFP | PMID:12176325 | Not available | Not available |
| Genetic reagent (M. musculus) | Ai9 | PMID:20023653 | MGI: J:155,793 | Not available |
| Genetic reagent (M. musculus) | Hoxc8 fl/fl | PMID:19621436 | Not available | Not available |
| Genetic reagent (M. musculus) | Olig2Cre | PMID:18046410 | MGI: 3774124 | Not available |
| Genetic reagent (M. musculus) | ChatIRESCre | PMID:21284986 | MGI: J:169,562 | Not available |
| Antibody | anti-ChAT(Goat polyclonal) | Millipore | Cat# AB144P, RRID:AB_2079751 | IF (1:100) |
| Antibody | anti-FoxP1 (Rabbit polyclonal) | Dasen lab | CU1025 | IF(1:32000) |
| Antibody | anti-RFP (Rabbit polyclonal) | Rockland | Cat# 600-401-379S, RRID:AB_11182807 | IF(1:500) |
| Antibody | anti-Alexa 488-Hoxc8 (mouse monoclonal) | Dasen lab | Not applicable | IF(1:1500) |
| Antibody | anti-GFAP (Chicken polyclonal) | Millipore | Cat# AB5541, RRID:AB_177521 | IF(1:200) |
| Antibody | anti-CD11b (Rat monoclonal) | Bio-Rad | Cat# MCA711, RRID:AB_321292 | IF(1:50) |
| Antibody | anti-mPea3 (Rabbit polyclonal) | Dasen lab | Not applicable | IF(1:32000) |
| Antibody | anti-Digoxigenin-POD, Fab fragments (Sheep polyclonal) | Roche Diagnostics Deutschland GmbH | Cat# 11207733910 | IF(1:3000) |
| Antibody | Cy3 AffiniPure anti-Goat IgG (Donkey polyclonal) | Jackson ImmunoResearch Labs | Cat# 705-165-147, RRID:AB_2307351 | IF(1:800) |
| Antibody | Alexa Fluor 488 anti-Rabbit IgG (Donkey) | Thermo Fisher Scientific | Cat# A-21206, RRID:AB_2535792 | IF(1:1000) |
| Antibody | Cy3 AffiniPure anti- Rabbit IgG (Donkey polyclonal) | Jackson ImmunoResearch Labs | Cat# 711-165-152, RRID:AB_2307443 | IF(1:800) |
| Antibody | Alexa Fluor 488 anti-Goat IgG (Donkey polyclonal) | Thermo Fisher Scientific | Cat# A-11055, RRID:AB_2534102 | IF(1:1000) |
| Antibody | Alexa Fluor 488 anti-mouse IgG (Donkey polyclonal) | Thermo Fisher Scientific | Cat# A-21202, RRID:AB_141607 | IF(1:1000) |
| Antibody | Alexa Fluor 488 anti-Chicken IgY (Goat polyclonal) | Thermo Fisher Scientific | Cat# A32931, RRID:AB_2762843 | IF(1:1000) |
| Antibody | Alexa Fluor 488 anti-Rat IgG (Goat polyclonal) | Thermo Fisher Scientific | Cat# A-11006, RRID:AB_2534074 | IF(1:1000) |
| Software, algorithm | ZEN | ZEISS | RRID: SCR_013672 | Version 2.3.69.1000, Blue edition |
| Software, algorithm | Fiji | Image J | RRID: SCR_003070 | Version 1.52i |
Additional files
-
Supplementary file 1
List of enriched transcripts in embryonic (e12.5) and postnatal (p8) brachial motor neurons.
- https://cdn.elifesciences.org/articles/70766/elife-70766-supp1-v1.xlsx
-
Supplementary file 2
Gene ontology (GO) analysis on enriched transcripts from embryonic (e12.5) and postnatal (p8) brachial motor neurons.
- https://cdn.elifesciences.org/articles/70766/elife-70766-supp2-v1.xlsx
-
Supplementary file 3
List of downregulated and up-regulated transcripts in brachial motor neurons of Hoxc8Δ early mice at e12.5.
- https://cdn.elifesciences.org/articles/70766/elife-70766-supp3-v1.xlsx
-
Supplementary file 4
Mendelian ratios at weaning stage for Hoxc8 MNΔ early and Hoxc8 MNΔ late animals.
- https://cdn.elifesciences.org/articles/70766/elife-70766-supp4-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70766/elife-70766-transrepform1-v1.pdf





