Spatial alanine metabolism determines local growth dynamics of Escherichia coli colonies
Figures
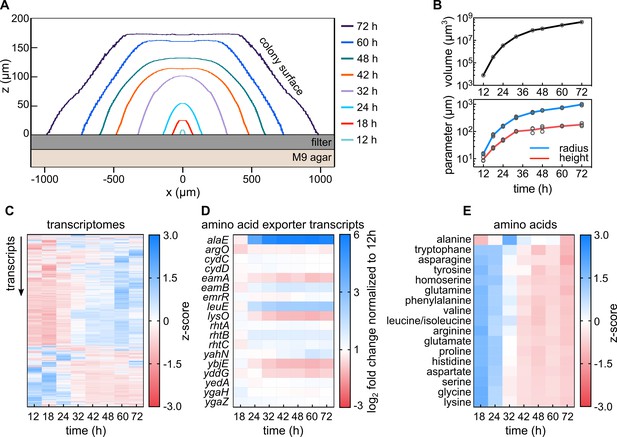
Transcriptomes and metabolomes during E. coli colony biofilm growth reveal metabolic transition and a unique role of alanine.
(A) Cross-sectional profiles of representative E. coli colonies grown on membrane filters placed on solid M9 agar at different time points. (B) Volume (top panel), radius and height (bottom panel) of colonies as a function of time indicate a growth rate transition around 24–32 hr. All replicates are shown as individual data points, with a line connecting the mean values, n = 3 biological replicates. (C) Dynamics of expression profiles of 4231 genes during colony growth reveal physiological transition between 24 and 32 hr, n = 3 biological replicates. (D) Mean expression fold-changes of known amino acid exporters during colony growth, calculated in comparison to 12 hr colonies, n = 3 biological replicates. (E) Comparison of amino acid profiles in whole colonies, measured with mass spectrometry during colony growth; n = 3 biological replicates. Levels of amino acids are shown in Figure 1—figure supplement 5.
-
Figure 1—source data 1
Source data for Figure 1.
- https://cdn.elifesciences.org/articles/70794/elife-70794-fig1-data1-v1.xlsx
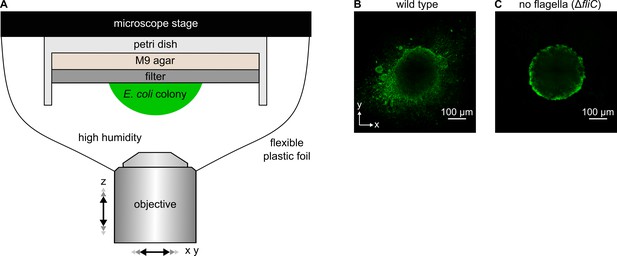
In contrast to wild-type colonies, flagella-deficient (ΔfliC) colonies are highly symmetric in shape.
(A) Experimental arrangement for investigating colony growth using an inverted confocal microscope. Dimensions are not drawn to scale. (B, C) Confocal microscopy images of xy-planes of colonies on filter membranes on M9 agar, made by the wild type strain (panel B) or a strain lacking the flagellin gene fliC (panel C) grown for 24 hr. The ΔfliC strain produces no flagella and is incapable of swimming motility.
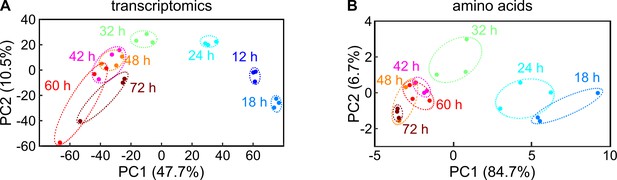
Principal component analysis of transcriptomic and metabolomic data.
(A) Principal component analysis of the transcriptomic data presented in Figure 1C. The axes represent the 1st principal component (PC1) and the 2nd principal component (PC2). (B) Principal component analysis of amino acid data presented in Figure 1E. Each circle represents a different independent transcriptome or metabolome biological replicate. Replicates for the same colony growth timepoint have the same color and are enclosed by an ellipse with a dashed line.
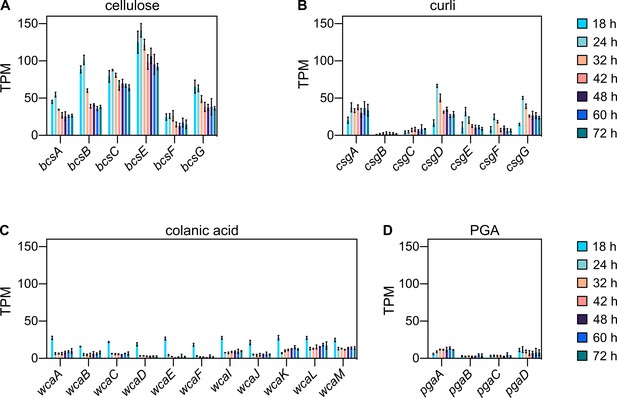
E. coli biofilm colonies express extracellular matrix genes.
E. coli extracellular matrix is composed of cellulose, curli, colanic acid, β–1,6-N-acetyl-D-glucosamine (PGA) and flagella (Hobley et al., 2015). Here, transcripts per million (TPM) of genes involved in the production of cellulose (A), curli (B), colonic acid (C) and PGA (D) are shown for colonies of the parental strain (KDE722) grown for 18, 24, 32, 42, 48, 60, or 72 hr, measured using RNA-seq. Flagellar genes are not shown here, because the flagellin gene fliC is deleted in the parental strain. Data are mean ± standard deviation (s.d.), n = 3 biological replicates.
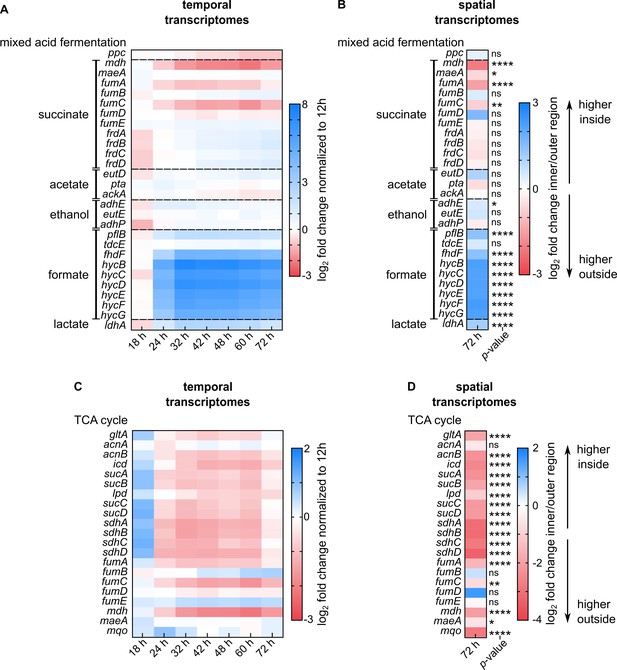
Spatial and temporal expression of genes involved in mixed acid fermentation and TCA cycle in colonies on M9 agar.
(A, B) Heatmaps show genes involved in mixed acid fermentation. (A) Fold-changes of genes from whole-colony measurements at different growth stages, normalized to transcriptomes of 12 hr colonies. (B) Fold-changes of genes from spatially-resolved transcriptomes, measured for 72 hr colonies. The inner region corresponds to non-fluorescent cells, the outer region corresponds to those cells with mRuby2 fluorescence. (C, D) Heatmaps show genes involved in the TCA cycle. (C) Fold-changes of genes from whole-colony measurements at different growth stages, normalized to transcriptomes of 12 hr colonies. (D) Fold-changes of genes from spatially-resolved transcriptomes, measured for 72 hr colonies. Non-significant differences are labeled ‘ns’. p-Values correspond to FDR-adjusted p-values: * p < 0.05, ** p< 0.01, **** p < 0.0001. For panels A and C, n = 3 biological replicates; for B and D, n = 4 biological replicates. The genes mdh, fumABCDE, and maeA are involved in the mixed acid fermentation and the TCA cycle pathways, and are therefore displayed in all heatmaps.
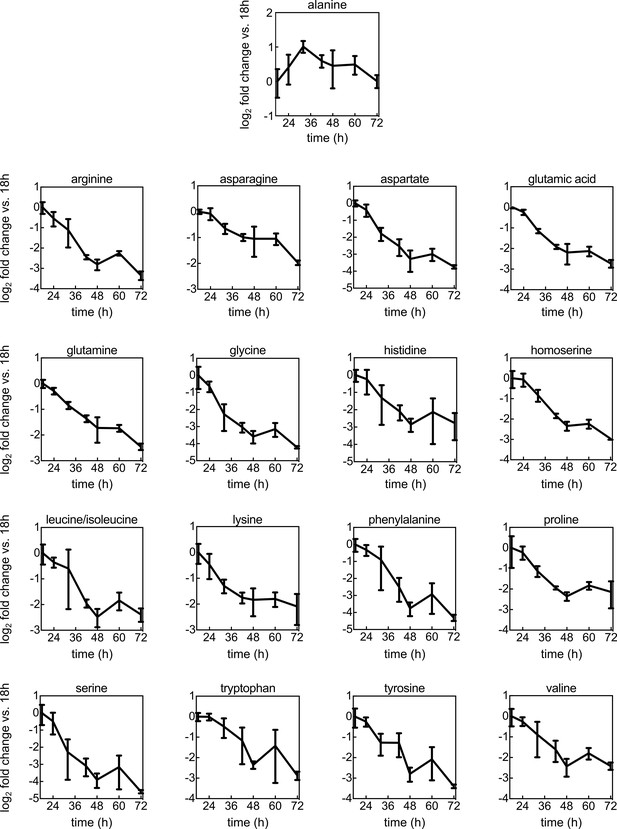
Amino acid levels during colony growth.
Fold-changes of amino acid levels as a function of time during colony development on M9 agar, normalized to amino acid levels from 18 hr colonies. Measurements were performed using mass spectrometry. Data are mean values ± s.d., n = 3 biological replicates.
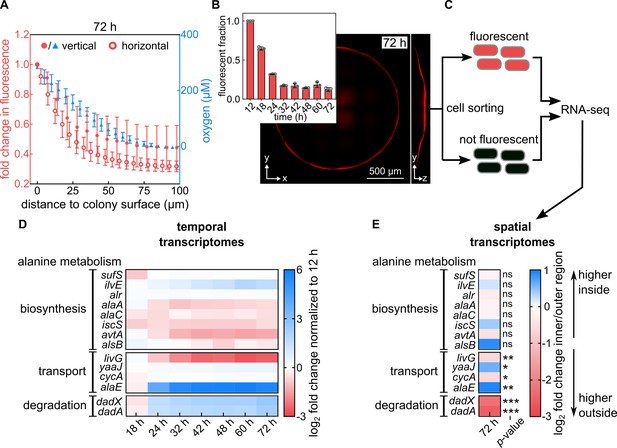
Alanine transport and degradation are spatially regulated within colony biofilms.
(A) Measurement of oxygen penetration into colonies grown for 72 hr on filter membranes on M9 agar using two different methods (for strain KDE722). Left axis (red): Intensity of mRuby2 fluorescence within a vertical cylinder of radius 3.3 µm in the center of the colony (filled circles), and intensity of mRuby2 fluorescence within a horizontal plane at the base of the colony (open circles, mean ± s.d., n = 3 biological replicates). Right axis (blue): Direct measurement of oxygen levels acquired by vertical scanning of an oxygen microsensor at the center of the colony (blue triangles, mean ± s.d., n = 10 replicates). (B) Confocal image of a representative 72 hr colony of a strain that constitutively expresses mRuby2 (KDE722). Insert: Fraction of fluorescent biovolume inside colonies grown for different times. Data are mean ± s.d., n = 3 biological replicates. (C) Scheme of the sorting procedure: Cells from a 72 hr colony are separated, fixed using formaldehyde, and sorted according to their mRuby2 fluorescence, followed by RNA-seq, as described in the methods section. (D) Heatmaps showing the mean fold-change (n = 3 biological replicates) of expression levels of genes involved in alanine metabolism, from whole-colony measurements. Fold-changes are computed relative to the 12 hr timepoint. (E) Spatial transcriptome results, quantified as fold-changes between the inner region (no mRuby2 fluorescence) and outer region (high mRuby2 fluorescence) regions of colonies grown for 72 hr. Blue color in the heatmap indicates genes with higher transcript levels in the inner region of the colony, red color indicates higher transcripts in the outer region of the colony. Data are means, n = 4 biological replicates. Non-significant differences between spatial regions are labeled ‘ns’. The p-values correspond to false discovery rate (FDR)-adjusted p-values: * p < 0.05, ** p < 0.01, *** p < 0.001.
-
Figure 2—source data 1
Source data for Figure 2.
- https://cdn.elifesciences.org/articles/70794/elife-70794-fig2-data1-v1.xlsx
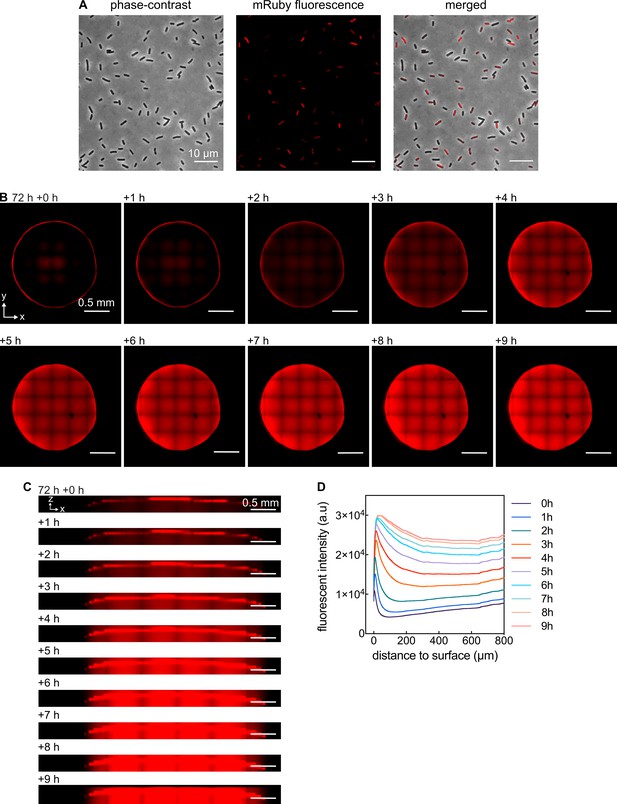
Fluorescent gradients of mRuby2 in growing colonies are not due to imaging artefacts.
(A) Images of cells from a 72 hr colony that was resuspended in PBS. Some cells are fluorescent, others display less or no fluorescence. (B) Confocal images of a xy-plane of a 72 hr colony acquired at z = 60 µm above the filter membrane on M9 agar. At 72 hr + 0 hr the filter membrane carrying the colony was moved from M9 agar to an agar plate with the same medium, but lacking glucose. As a consequence, cells stop growth and strongly reduce the consumption of molecular oxygen, allowing oxygen to penetrate into the colony. Penetration of oxygen allows the mRuby2 protein to fold into the fluorescent conformation. The square pattern in the images results from the fact that colony images were stitched together from 7 × 7 image tiles. (C) Confocal images of a xz-plane of the same 72 hr colony as in panel B, following the same experimental procedure. (D) Quantification of panels B, C: mRuby2 fluorescent intensity profiles at different times after transferring the colony to M9 agar without glucose (different colored lines), as a function of distance to the outer surface of the colony.
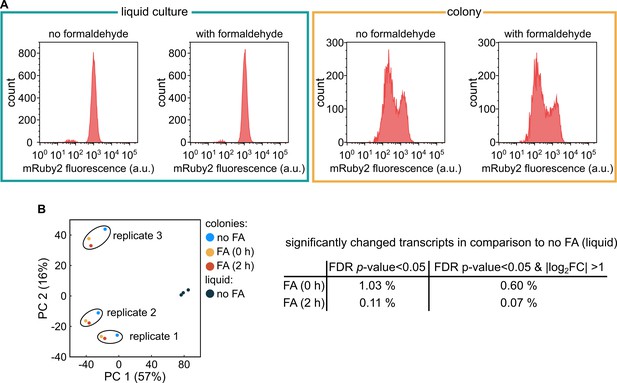
Formaldehyde fixation does not affect mRuby2 fluorescence or the colony transcriptome.
(A) Flow cytometry results for fluorescence of E. coli constitutively expressing mRuby2 (strain KDE722) with or without 2 hr of fixation using 4 % formaldehyde. For the two panels on the left (enclosed by green box), the cells were grown in liquid shaking M9 medium. For the two panels on the right (enclosed by orange box), the cells were grown as a colony on M9 agar. (B) Principal component analysis of the transcriptomes of colonies grown for 72 hr, which were either fixed with 4 % formaldehyde (FA) for 0 hr or 2 hr, or without fixation. As a control, the transcriptome of cells from an exponentially growing M9 liquid shaking culture without fixation was also analyzed. Each data point corresponds to an independent biological replicate (n = 3). The table on the right indicates the number of differentially expressed genes in colonies, in comparison to the colonies without fixation, using as threshold for differentially expressed genes either a FDR-adjusted p-value < 0.05, or a FDR-adjusted p-value < 0.05 and an absolute log2 fold-change (log2 FC) > 1.
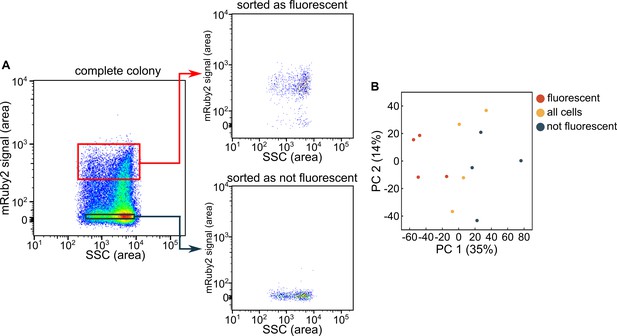
Application of flow cytometry with fluorescence-activated cell sorting to separate cells from the inner and outer regions of colonies.
(A) Representative sorting results of a colony grown for 72 hr, using the mRuby2 fluorescence signal and the side scatter (SSC) signal. Cells were sorted into two separate bins (red, black). (B) Principal component analysis of RNA-seq results of the two separated bins, or the entire population, from colonies that were grown for 72 hr, showing the 1st (PC1) and 2nd (PC2) principal component axes. Each data point corresponds to an independent replicate (n = 4 biological replicates).

Key pathways in alanine metabolism.
(A) Alanine biosynthesis pathways. (B) Alanine degradation and export pathways. Cells are drawn with a schematic rod shape.
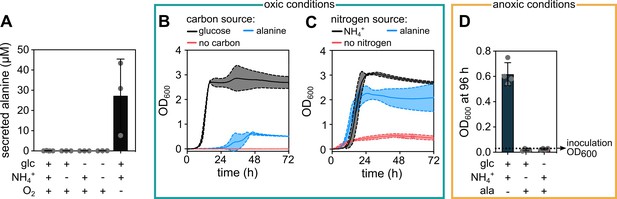
Alanine can be secreted in anoxic conditions and consumed in oxic conditions by E. coli.
(A) Extracellular alanine concentration in the supernatant of liquid cultures grown in presence or absence (indicated by+ or -) of glucose (glc), ammonium (NH4+), and molecular oxygen (O2). Data are mean ± s.d., n = 3 biological replicates. (B) Growth curves using M9 minimal salts medium with ammonium as nitrogen source and different carbon sources: Either glucose (5 g/L, as in our standard M9 medium), alanine (10 mM), or no carbon source. (C) Growth curves using M9 minimal salts medium with glucose as a carbon source and different nitrogen sources: Either ammonium (22.6 mM, as in our standard M9 medium), alanine (5 mM) or no nitrogen source. For panels B and C, continuous middle lines correspond to the mean and the dotted lines to the standard deviation, n = 3 biological replicates. The growth curves shown for alanine in panels B and C correspond to the alanine concentrations that resulted in the highest final optical density at 600 nm (OD600) in each condition. (D) E. coli cultures starting with OD600 = 0.03 were incubated for 96 hr in different anoxic media. The medium contained combinations of glucose, NH4+, and alanine as indicated. Alanine was provided either as the sole nitrogen source (middle bar) or sole carbon source (right bar) with a concentration of 5 mM and 10 mM, respectively. The final OD600 after 96 hr of incubation is shown, corresponding to a time point when the culture without alanine reached stationary phase. Continuous measurements of OD600 in anoxic conditions was not possible in our laboratory due to technical limitations. Data are mean ± s.d., n = 4 biological replicates.
-
Figure 3—source data 1
Source data for Figure 3.
- https://cdn.elifesciences.org/articles/70794/elife-70794-fig3-data1-v1.xlsx
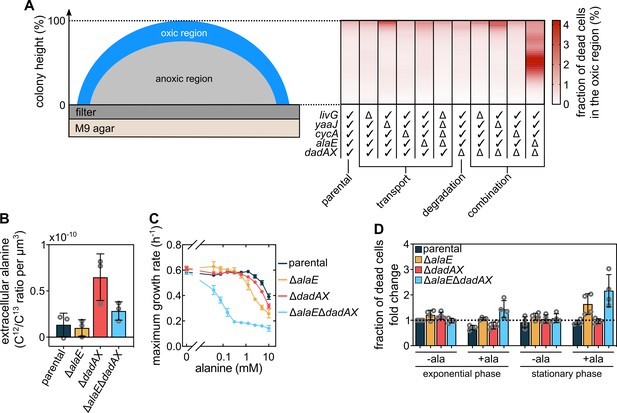
Alanine influences cell viability and growth.
(A) Measurement of the fraction of dead cells as a function of height in colonies grown for 72 hr on M9 agar. These measurements were performed only for cells located within 30 µm from the outer colony surface, which is a conservative measurement of the oxic region (Figure 2A). The table designates the genotype of strains that were investigated: ‘✓’ indicates that the gene is intact and ‘Δ’ that the gene was deleted. Data in the heatmaps shows means, n = 3 biological replicates. Errors are shown in Figure 4—figure supplement 2. (B) Extracellular alanine levels measured from colonies (mean ± s.d., n = 3 biological replicates). (C) Maximum liquid culture growth rate (in presence of glucose and ammonium) for different strains, as a function of the concentration of exogenously added alanine. Data are mean ± s.d., n = 3 biological replicates. (D) Fraction of dead cells, measured using SYTOX Green fluorescence normalized by OD600, in cultures grown with glucose and ammonium in presence or absence of 5 mM alanine. Fold-changes are calculated relative to the parental strain in exponential phase without alanine. Measurements were performed when cultures reached half of the maximum OD600 (for exponential phase measurements) or when they reached their maximum OD600 (for stationary phase measurements). Bars indicate mean ± s.d., n = 4 biological replicates.
-
Figure 4—source data 1
Source data for Figure 4.
- https://cdn.elifesciences.org/articles/70794/elife-70794-fig4-data1-v1.xlsx
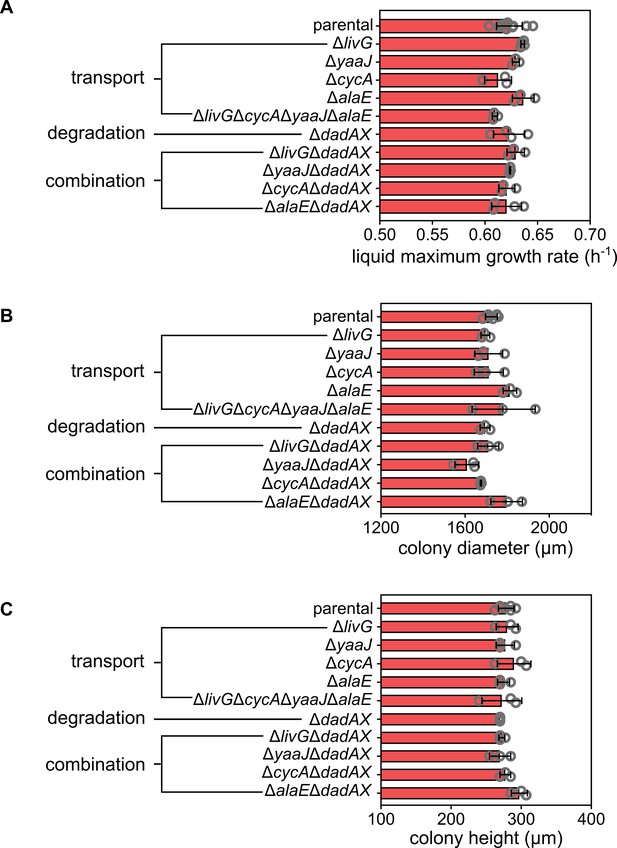
Growth rates and colony sizes for alanine metabolism mutants.
(A) Maximum growth rate in liquid shaking cultures in M9 medium. Colony diameter (B) and height (C) after 72 hr of growth on M9 agar for different mutants in alanine transport and degradation, compared to the parental E. coli strain. The parental and ΔdadAXΔalaE colonies do not show significant differences in colony diameter and height. Data are mean values ± s.d., each biological replicate is shown as a gray circle.
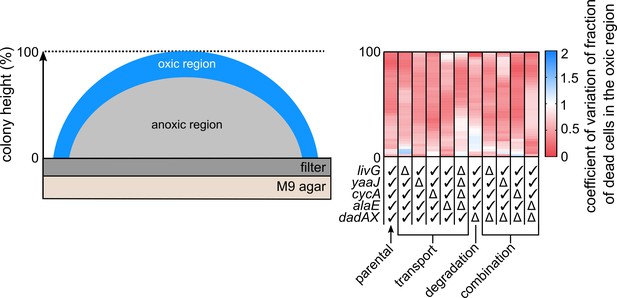
Coefficient of variation of the fraction of dead cells in the oxic region of the colony.
Heatmap shows the coefficient of variation for the data shown in Figure 4A.
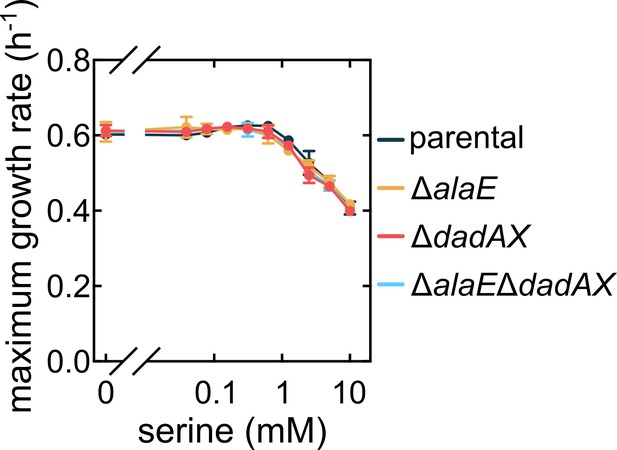
Effects of exogenously added alanine on maximum growth rate for alanine metabolism mutants are specific to alanine, and are not triggered by exogenously added serine.
The maximum liquid culture growth rate as function of the concentration of exogenically added serine in M9 medium is the same for all strains. Data are mean values ± s.d., n = 3 biological replicates.
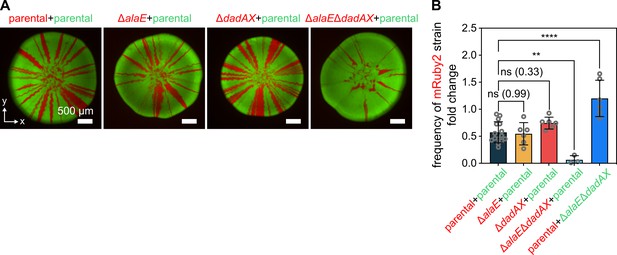
Cells capable of secreting and consuming alanine have a fitness advantage during colony growth.
(A) Confocal images of a xy-plane of representative 72-hr-old colonies grown by inoculating a 1:1 mixture of two strains onto a membrane filter placed on solid M9 agar. The strains in red and green express the fluorescent proteins mRuby2 and sfGFP respectively, from the same chromosomal locus, using the same promoter. Left image: Outcome of a control experiment in which the red and green strains are isogenic except for the fluorescent protein, showing that the mRuby2 expression results in a slight fitness cost compared to the sfGFP expression. Center left, center right, and right images: Outcome of two-strain competition experiments, which should be compared to the outcome shown in the image on the far left. The color and genotype of each strain is indicated above each image. (B) Quantification of the fold change between the frequency of the red strain at 350 µm from the edge of the inoculation region and the frequency of the red strain in the inoculation suspension measured by flow cytometry. Data are mean values ± s.d., n = 3–15 biological replicates. Statistical significances were calculated using one-way ANOVA with Dunnett’s correction. Non-significant differences are labeled ‘ns’ and the p-value is shown in brackets. **p < 0.01, ****p < 0.0001.
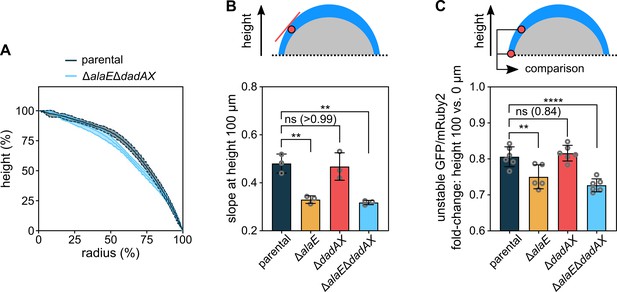
Colony morphology is influenced by alanine cross-feeding.
(A) Colony height as a function of the colony radius, for colonies grown for 72 hr. Central lines correspond to the mean and the shaded area to the s.d., n = 3 biological replicates. (B) Slope of curves in panel A at height 100 µm. Data are mean ± s.d., n = 3 biological replicates. (C) Fluorescence ratio of constitutively expressed unstable GFP and stable mRuby2, which is a measure for the cellular growth rate, as shown in Figure 5—figure supplement 1. The unstable GFP was constructed by adding the ASV-tag to superfolder GFP. The fold-change of this fluorescent protein ratio was calculated for the oxic colony region between the heights 100 µm and 0 µm. The fluorescent protein ratio was only measured for cells located within 30 µm from the outer colony surface, as a conservative measure of the oxic region. Data are mean ± s.d., n = 5–6 biological replicates. Statistical significances were calculated using one-way ANOVA with Dunnett’s correction. Non-significant differences are labeled ‘ns’ and the p-value is shown in brackets; **p < 0.01, ****p < 0.0001.
-
Figure 5—source data 1
Source data for Figure 5.
- https://cdn.elifesciences.org/articles/70794/elife-70794-fig5-data1-v1.xlsx
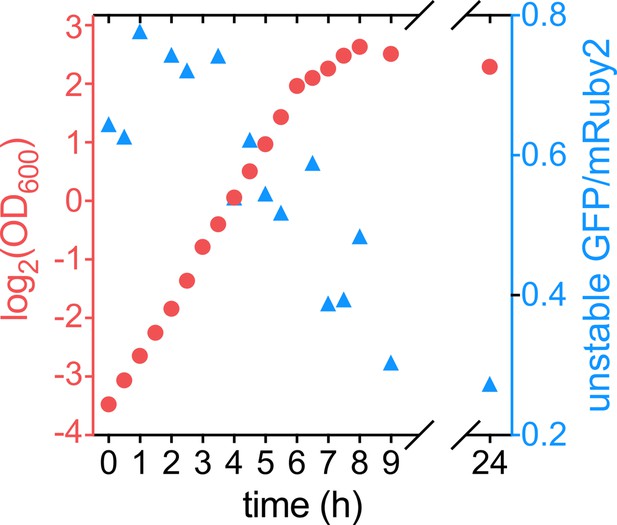
Ratio between unstable GFP and stable mRuby2 correlates with bacterial growth rate.
Optical density at 600 nm (OD600; shown in red, left y-axis) and the ratio between unstable GFP (with the ASV-tag) and mRuby2 (shown in blue, right y-axis) as a function of time for a liquid culture of strain KDE2937.
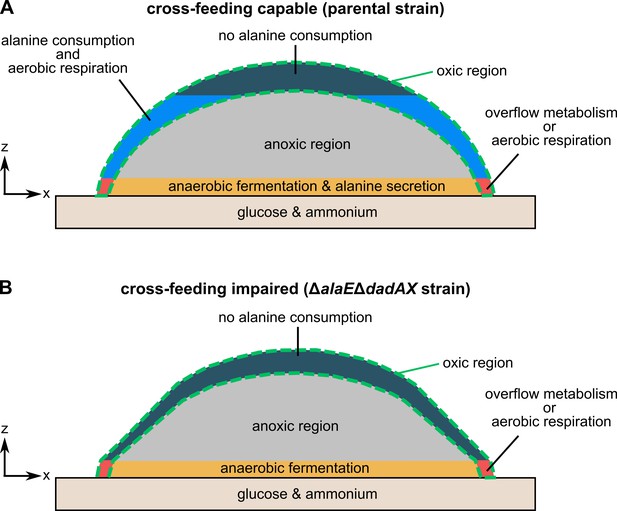
Model for alanine cross-feeding in E. coli colony biofilms.
This model applies to colonies grown for 72 hr on solid M9 agar containing glucose and ammonium. (A) In cross-feeding capable colonies, cells in the bottom layer of the biofilm have access to glucose and ammonium. Only cells in the outer periphery of the biofilm (green dashed line) have access to oxygen. Cells in the red region can use ammonium, glucose, and oxygen to perform aerobic respiration or fermentation by overflow metabolism. Cells in the orange region have access to glucose and ammonium, but no oxygen. These cells secrete alanine. The secreted alanine can be consumed by cells in the oxic region above this layer (depicted as blue), which perform aerobic respiration and convert alanine into pyruvate and ammonium that can be used for growth and to maintain cell viability. (B) Colonies of ΔalaEΔdadAX cells have a reduced ability to consume and export alanine. These colonies have a region performing aerobic respiration or overflow metabolism (red), similar to the parental strain. These colonies also have an anoxic fermentation region (orange), yet this region displays significantly less alanine secretion compared to the parental strain. Furthermore, these colonies lack an alanine consuming population in the oxic region. Due to the limited alanine secretion and alanine consumption of this strain, ΔalaEΔdadAX colonies display higher cell death and less growth in the otherwise cross-feeding-dependent oxic region, resulting in a more conical colony shape in comparison to parental colonies.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | KDE261 | Drescher lab stock | Strain carrying plasmid pCP20 | Plasmid information listed in Supplementary file 2 |
| Strain, strain background (Escherichia coli) | KDE262 | Drescher lab stock | Strain carrying plasmid pKD46 | Plasmid information listed in Supplementary file 2 |
| Strain, strain background (Escherichia coli) | KDE264 | Drescher lab stock | Strain carrying plasmid pKD3 | Plasmid information listed in Supplementary file 2 |
| Strain, strain background (Escherichia coli) | KDE265 | Drescher lab stock | Strain carrying plasmid pKD4 | Plasmid information listed in Supplementary file 2 |
| Strain, strain background (Escherichia coli) | KDE1361 | Drescher lab stock | Strain carrying plasmid pNUT1361 | Plasmid information listed in Supplementary file 2 |
| Strain, strain background (Escherichia coli) | KDE2338 | Drescher lab stock | Strain carrying plasmid pNUT2338 | Plasmid information listed in Supplementary file 2 |
| Strain, strain background (Escherichia coli) | KDE2658 | Drescher lab stock, Addgene #64,969 | Strain carrying plasmid pUC18R6KT-mini-Tn7-Km | Plasmid information listed in Supplementary file 2 |
| Strain, strain background (Escherichia coli) | KDE2659 | Drescher lab stock, Addgene #64,968 | Strain carrying plasmid pTNS2 | Plasmid information listed in Supplementary file 2 |
| Strain, strain background (Escherichia coli) | KDE2674 | This study | Strain carrying plasmid pNUT2674 | Plasmid information listed in Supplementary file 2 |
| Strain, strain background (Escherichia coli) | KDE2787 | This study | Strain carrying plasmid pNUT2787 | Plasmid information listed in Supplementary file 2 |
| Strain, strain background (Escherichia coli) | KDE2838 | This study | Strain carrying plasmid pNUT2838 | Plasmid information listed in Supplementary file 2 |
| Strain, strain background (Escherichia coli) | KDE474 | Serra et al., 2013a | E. coli AR3110 wild-type | |
| Strain, strain background (Escherichia coli) | KDE679 | Vidakovic et al., 2018 | KDE474 (E. coli AR3110), Ptac-mRuby2-mRuby2 and KanR inserted at attB site (Ptac without operator). | |
| Strain, strain background (Escherichia coli) | KDE722 | Vidakovic et al., 2018 | KDE679 with ΔfliC::scar. | |
| Strain, strain background (Escherichia coli) | KDE1899 | This study | KDE474 (E. coli AR3110) with ΔfliC::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2007 | This study | KDE679 with ΔfliC::scar, ΔalaE::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2009 | This study | KDE679 with ΔfliC::scar, ΔdadAX::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2086 | This study | KDE679 with ΔfliC::scar, ΔalaE::scar, ΔdadAX::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2183 | This study | KDE679 with ΔfliC::scar, ΔcycA::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2185 | This study | KDE679 with ΔfliC::scar, ΔlivG::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2438 | This study | KDE679 with ΔfliC::scar, ΔyaaJ::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2242 | This study | AR3110, Ptac-sfgfp-sfgfp and KanR inserted at attB site (Ptac without operator), with ΔfliC::scar, ΔalaE::scar, ΔdadAX::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2445 | This study | AR3110, Ptac-sfgfp-sfgfp and KanR inserted at attB site (Ptac without operator), with ΔfliC::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2533 | This study | KDE679 with ΔfliC::scar ΔcycA::scar, ΔlivG::scar, ΔalaE::scar, ΔyaaJ::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2564 | This study | KDE679 with ΔfliC::scar, ΔcycA::scar, ΔdadAX::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2607 | This study | KDE679 with ΔfliC::scar, ΔyaaJ::scar, ΔdadAX::scar. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2937 | This study | KDE679 with ΔfliC::scar, Ptac-sfgfp(ASV) at the Tn7 insertion site, coding for an unstable superfolder GFP with an AANDENYAASV-tag. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2938 | This study | KDE679 with ΔfliC::scar, ΔalaE::scar, Ptac-sfgfp(ASV) at the Tn7 insertion site. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2939 | This study | KDE679 with ΔfliC::scar, ΔdadAX::scar, Ptac-sfgfp(ASV) at the Tn7 insertion site. | This strain can be obtained from the Drescher lab upon request |
| Strain, strain background (Escherichia coli) | KDE2940 | This study | KDE679 with ΔfliC::scar, ΔalaE ΔdadAX, Ptac-sfgfp(ASV) at the Tn7 insertion site. | This strain can be obtained from the Drescher lab upon request |
| Sequence-based reagent | KDO834 | This study | Insertions at the attB site | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO894 | This study | fliC deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO895 | This study | fliC deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO1662 | This study | Insertions at the attB site | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO2562 | This study | alaE deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO2563 | This study | alaE deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO2566 | This study | dadAX deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO2567 | This study | dadAX deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO2845 | This study | cycA deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO2846 | This study | cycA deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO2841 | This study | livG deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO2842 | This study | livG deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO3481 | This study | yaaJ deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO3482 | This study | yaaJ deletion | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO3256 | This study | pNUT2338 construction | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO3257 | This study | pNUT2338 construction | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO3817 | This study | pNUT2838 construction | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO3818 | This study | pNUT2838 construction | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO3785 | This study | pNUT2674 construction | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO3786 | This study | pNUT2674 construction | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO4121 | This study | pNUT2787 construction | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO4121 | This study | pNUT2787 construction | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Sequence-based reagent | KDO4127 | This study | pNUT2838 construction | The sequence of this oligonucleotide can be found in Supplementary file 3 |
| Software, algorithm | Matlab | MathWorks | Version R2019b | |
| Software, algorithm | Prism | GraphPad Software | Version 9.2.0 | |
| Software, algorithm | NIS-Elements | Nikon | Version 4.5.2 | |
| Software, algorithm | Inkscape | Inkscape | Version 1.0.1 | |
| Software, algorithm | CLC Genomics Workbench | Qiagen | Version 11.0 | |
| Chemical compound, drug | Na2HPO4 | Carl Roth | P030.2 | |
| Chemical compound, drug | CoCl2 | Carl Roth | 7095.1 | |
| Chemical compound, drug | MnSO4 | Sigma-Aldrich | M8179 | |
| Chemical compound, drug | CuCl2 | Sigma-Aldrich | 307,483 | |
| Chemical compound, drug | ZnSO4 | Sigma-Aldrich | Z0251 | |
| Chemical compound, drug | thiamine-HCl | Sigma-Aldrich | T4625 | |
| Chemical compound, drug | FeCl3 | Sigma-Aldrich | 31,332 | |
| Chemical compound, drug | MgSO4, | Sigma-Aldrich | M2643 | |
| Chemical compound, drug | CaCl2 | Sigma-Aldrich | C5670 | |
| Chemical compound, drug | NaCl | Carl Roth | HN00.2 | |
| Chemical compound, drug | (NH4)2SO4 | Carl Roth | 3746.2 | |
| Chemical compound, drug | KH2PO4 | Carl Roth | 3904.1 |
Additional files
-
Supplementary file 1
Bacterial strains used in this study.
Abbreviations: Kan = kanamycin. Superscript “R” = resistance. “-” = fusion. “::” = insertion. The scar corresponds to 5’-GAAGTTCCTATACTTTCTAGAGAATAGGAACTTC-3’ sequence.
- https://cdn.elifesciences.org/articles/70794/elife-70794-supp1-v1.docx
-
Supplementary file 2
Plasmids used in this study.
Abbreviations: Kan = kanamycin, Amp = ampicillin, Chl = chloramphenicol. Superscript “R” = resistance. “-“ = fusion.
- https://cdn.elifesciences.org/articles/70794/elife-70794-supp2-v1.docx
-
Supplementary file 3
DNA oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/70794/elife-70794-supp3-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70794/elife-70794-transrepform1-v1.pdf





