Delilah, prospero, and D-Pax2 constitute a gene regulatory network essential for the development of functional proprioceptors
Figures
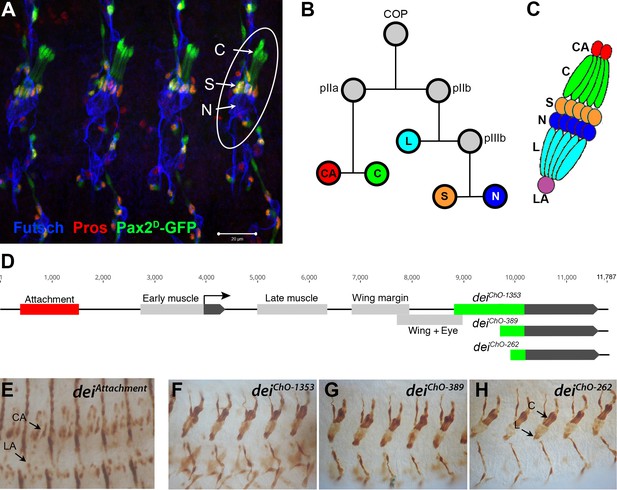
The LCh5 organ and the dei gene.
(A) Four abdominal segments of a stage 16 embryo carrying the sv/Pax2D1-GFP reporter (green) which labels the cap and scolopale cells, stained with anti-Pros (red) which labels the nuclei of scolopale cells, and anti-Futsch (blue), which labels the neurons. One LCh5 organ is circled and the cap cells (C), scolopale cells (S) and neurons (N) are indicated. The cap-attachment, ligament-attachment and ligament cells are not stained. Scale bar = 20 μm. (B–C) the ChO lineage (B) and schematic illustration of an LCh5 organ (C). The neurons are depicted in blue, the scolopale cells in orange, the ligament cells in cerulean, the cap cells in green, the cap-attachment cells in red and the ligament-attachment cell in purple. (D) Schematic representation of the dei locus showing the two exons (black boxes) and the cis regulatory modules (CRM) that drive gene expression within the ChO lineage: the deiattachment CRM which drives expression in the attachment cells (red box) and the deiChO CRM which drives expression in the cap and ligament cells (green box). The deiChO CRM was originally mapped to a 1353 bp fragment (deiChO-1353) and was then delimited to a smaller 389 bp fragment located immediately upstream to the second exon (deiChO-389). As part of this work, the ChO-specific CRM was further delimited to a 262 bp fragment (deiChO-262). (E–H) The embryonic expression patterns driven by the deiattachment (E), deiChO-1353 (F), deiChO-389 (G), and deiChO-262 (H) enhancers are shown. C, cap cell; L, ligament cell; CA, cap-attachment cell, LA, ligament-attachment cell.
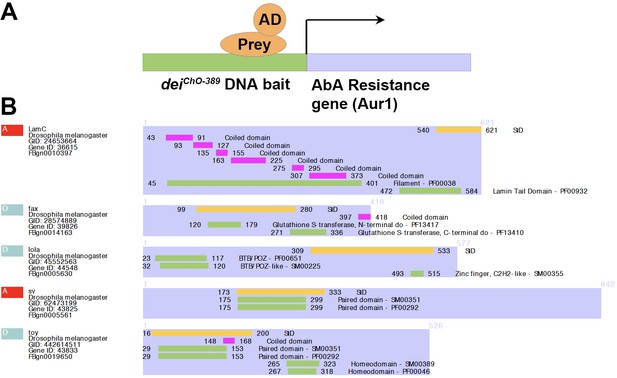
Identifying potential direct regulators of dei in the ChO using a Y1H screen.
(A) Schematic representation of the screening system that used the deiChO-389 enhancer as a bait and the Aurobasidin A selection system to screen an embryonic cDNA library for preys that bind to the bait (Hybergenics Services). (B) A DomSight graph showing the top candidates identified in the screen in very high confidence in the interaction (red) or moderate confidence (green). This score represents the probability of an interaction to be non-specific based on the comparison between the number of independent prey fragments found for an interaction and the chance of finding them at random (background noise). For each interaction, a Predicted Biological Score (Global PBS) is computed to assess the interaction reliability and the results are ranked in four categories from A (the highest confidence rank) to D (Formstecher et al., 2005). The orange bars mark the SID fragment (selected interaction domain), which is the amino acid sequences shared by all prey.
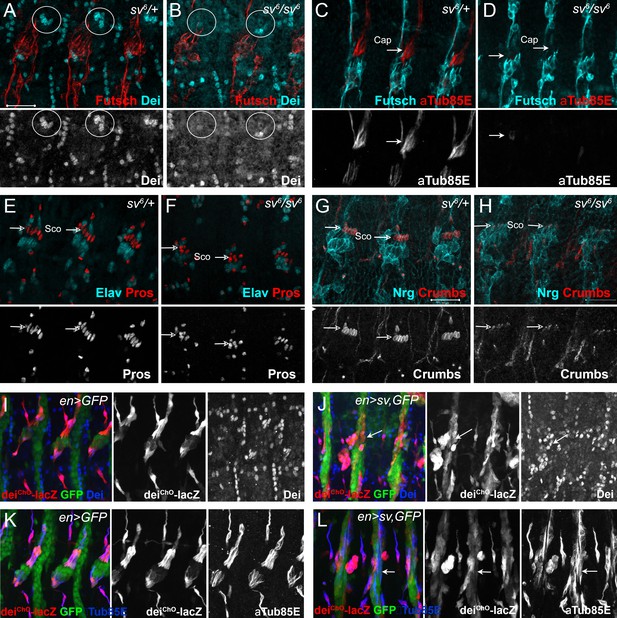
Sv/D-Pax2 activates dei expression in the cap cell.
(A–B) Representative abdominal segments of stage 16 embryos stained for Dei (cyan) and the neuronal marker Futcsh (red). The anti-Dei staining is shown separately in the lower panel. (A) A sv6 heterozygous embryo demonstrating the expression of Dei in the cap cell nuclei (circled). (B) The expression of Dei is lost in homozygous sv6 embryos. (C–D) Heterozygous (C) and homozygous (D) sv6 embryos stained for αTub85E (red) and Futsch (cyan). The arrows mark the cap cells. The anti-αTub85E staining is shown separately in the lower panel. (E–F) Heterozygous (E) and homozygous (F) sv6 embryos stained for Elav (cyan) and Pros (red). The arrows mark the nuclei of scolopale cells. The anti-Pros staining is shown separately in the lower panel. (G–H) Heterozygous (G) and homozygous (G) sv6 embryos stained for Nrg (cyan) and Crumbs (red). The arrows mark the nuclei of scolopale (Sco) cells. The anti-Crumbs staining is shown separately in the lower panel. (I–L) Representative abdominal segments of stage 16 embryos that express GFP (I, K) or GFP and sv (J, L) under the regulation of en-Gal4. The embryos carry the deiChO-262-lacZ marker (anti bGal staining is shown in red) and are stained with anti-Dei (blue in I-J) or anti-αTub85E (blue in K-L). The staining patterns of deiChO-262-lacZ, Dei and αTub85E are shown separately on the right. Note the ectopic expression of Dei, deiChO-262-lacZ and αTub85E in epidermal cells within the en domain (arrows in J, L).
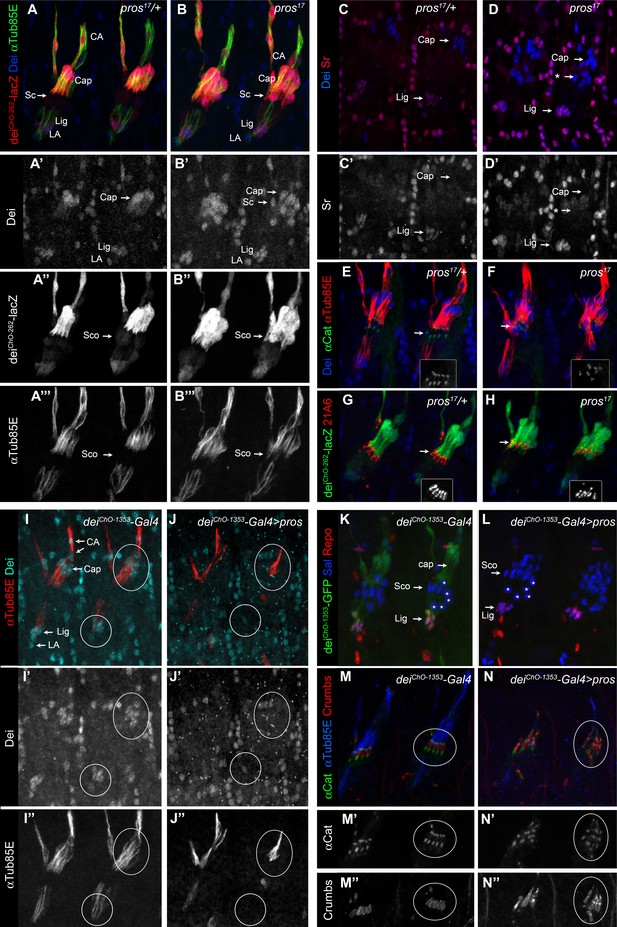
Pros represses dei in the scolopale cell.
(A–B) Representative abdominal segments of stage 16 pros17 heterozygous (A) and homozygous (B) embryos carrying the deiChO-262-lacZ marker (shown in red) and stained for Dei (blue) and αTub85E (green). Each of the three channels is shown separately below. Note the expansion of Dei and the deiChO-262-lacZ marker into the scolopale cells in pros17 mutant embryos. The arrows point to the scolopale cells. (C–D) pros17 heterozygous (C) and homozygous (D) embryos stained for Dei (blue) and Sr (red). Note that the ectopic expression of Dei in the scolopale cells of pros mutant embryo is not accompanied by ectopic expression of Sr (the arrow labeled with asterisks in D). (E–F) pros17 heterozygous (E) and homozygous (F) embryos carrying an α-Catenin-GFP reporter and stained for Dei (blue, shown separately in the inset) and αTub85E (red). (G–H) pros17 heterozygous (G) and homozygous (H) embryos carrying the deiChO-262-lacZ reporter (shown in green) and stained with the scolopale marker anti-Eyes Shut (MAb21A6, red, shown separately in the inset). Note that the expression of both Eyes Shut/21A6 and α—Catenin is maintained in the pros-deficient scolopale cells. (I–J) deiChO-1353-Gal4 (I) and deiChO-1353> UAS pros (J) embryos stained for Dei (cyan, shown separately in I’-J’) and αTub85E (red, shown separately in I’’-J’’). The cap and ligament cells are circled. Note the loss of Dei and αTub85E expression upon Pros expression. (K–L) deiChO-1353 (K) and deiChO-1353> UAS pros (L) embryos carrying the deiChO-1353-lacZ reporter (shown in green) stained for Sal (blue) and Repo (red). The cap, scolopale (Sco) and ligament (Lig) cells are indicated; the asterisks mark oenocyte cell nuclei. (M–N) deiChO-1353-Gal4 (M) and deiChO-1353> UAS pros (N) embryos carrying an α—Catenin-GFP reporter (green, shown separately in M’-N’) and stained for Crumbs (red, shown separately in M’’-N’’) and αTub85E (blue). Note the duplication of scolopale-specific structures in the cap cell expressing Pros ectopically (circled).
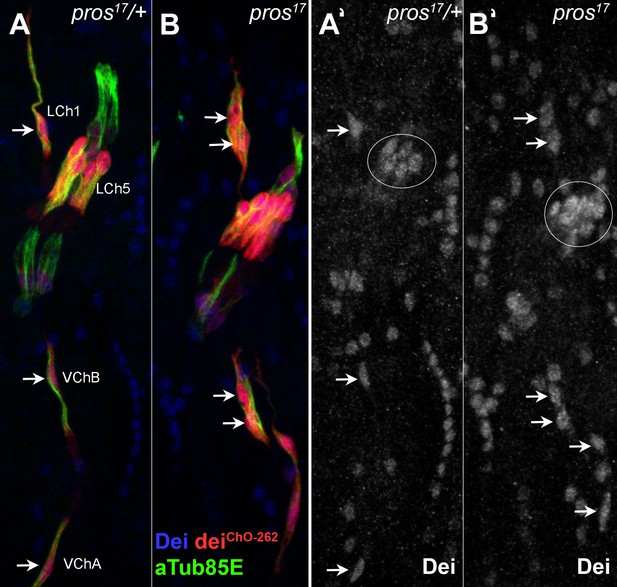
The loss of pros affects similarly the various types of larval ChOs.
(A–B) Representative segments of stage 16 heterozygous (A) and homozygous (B) pros17 mutant embryos that carry the deiChO-262-lacZ reporter (red) and stained for Dei (blue) and αTub85E (green). The staining of Dei is shown separately in A’ and B’. Note the ectopic expression of Dei and the deiChO262-lacZ reporter in the scolopale cells of pros-deficient LCh5, LCh1, and VChB organs (arrows in B and B’).
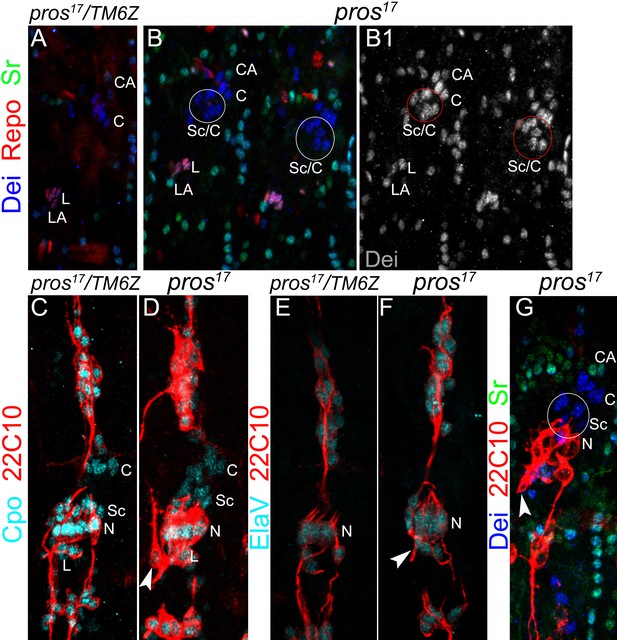
The number of neurons and ligament cells remain normal in pros mutant embryos.
(A–B) Representative segments of stage 16 wt (A) and pros17 mutant (B) embryos stained for Dei (blue), Repo (red), and Sr (green). Repo staining shows the normal number of ligament cells in the mutant. (C–D) Representative LCh5 organs of wt (C) and pros17 mutant (D) embryos stained for CPO (cyan) and 22C10 (anti Futsch, red). CPO is expressed in all of the LCh5 cells. (E–F) Representative LCh5 organs of wt (E) and pros17 mutant (F) embryos stained for the neuronal markers Elav (cyan) and 22C10 (red). The data shown in C-F demonstrate that the number of neurons remains unaltered in pros mutant embryos. (G) An LCh5 of pros mutant embryo stained for the neuronal marker 22C10 (anti-Futsch), Dei and Sr. The arrowhead points to the typical axonal pathfinding defect of pros mutant embryos.
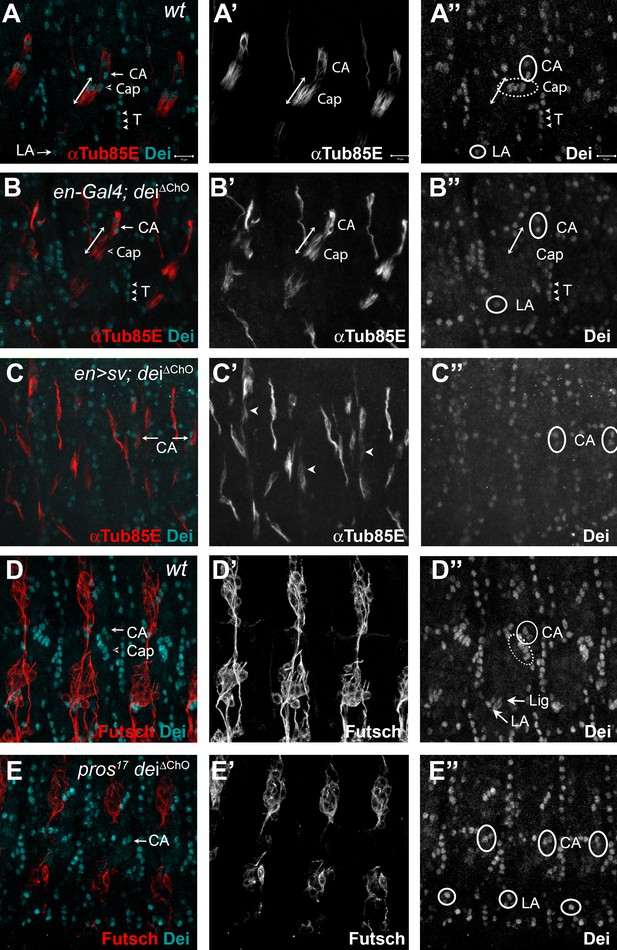
D-Pax2/Sv and Pros regulate the transcription of dei via the deiChO-262 regulatory module.
(A–B) Stage 16 wt (A) and dei∆ChO (B) embryos stained for Dei (cyan) and αTub85E (red). Note that in (B) Dei is still evident in the cap-attachment and ligament-attachment cells (CA and LA, arrows in A, circled in A’’), as well as in tendon cells (T; arrowheads), but is lost from the cap and ligament cells. The double edge arrows in A and B demarcate the length of the cap cells. (C) An embryo in which sv was expressed under the regulation of en-Gal4 in a dei∆ChO background. Sv was unable to induce Dei expression in the cap and ligament cells in the absence of the deiChO-262 enhancer. Ectopic expression of αTub85E is evident within the en domain (arrowheads in C’). (D–E) wt (D) and A pros17 dei∆ChO homozygous embryo (E) stained for Dei (cyan) and anti-Futsch (red). The pros17 dei∆ChO embryo (E) presents a dei∆ChO-like and not pros17-like expression pattern of the Dei protein, indicating that the dei∆ChO deletion is epistatic to pros loss-of-function. The neurons present the typical pros axonal pathfinding defects.
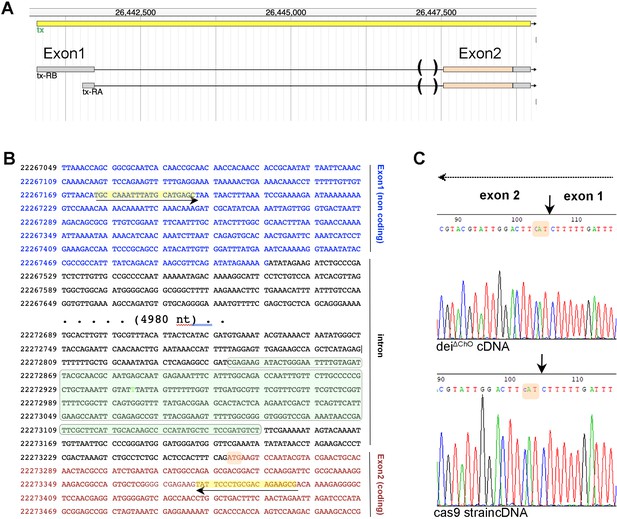
Deletion of the deiChO262 enhancer does not affect splicing.
(A) Structure of the dei (tx) locus and the two expected transcripts. The gene contains one non-coding and one coding exon that are separated by a ~ 5.3 kb intron. The parentheses mark the sequence deleted in the deiΔChO allele. (B) A partial sequence of the dei locus showing the first exon (blue), the intron (black) and part of the coding exon (red). The sequence deleted in the deiΔChO allele is highlighted in green; the initiation codon is highlighted in orange, and the primers used for the amplification of a 460 bp cDNA fragment for sequencing are highlighted in yellow. (C) Electropherograms of sequences read in the reverse orientation on cDNA prepared from flies homozygous for the deiΔChO allele and control flies from the Cas9 strain to which injection was done. The splicing site is marked with arrows and the initiation codon is highlighted in orange in both sequences. The arrow at the top marks the direction of transcription.
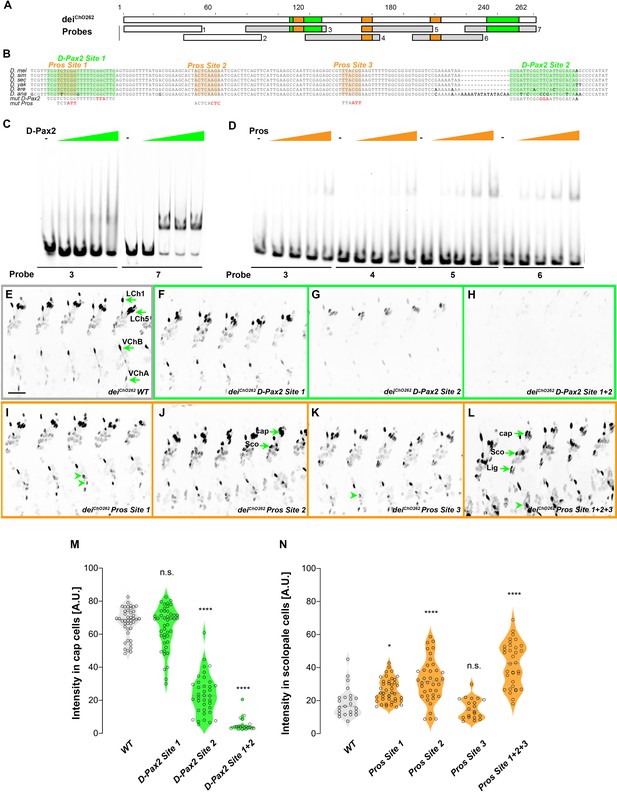
deiChO-262 contains two binding sites for the activator D-Pax2/Sv and three binding sites for the repressor Pros.
(A) Schematic representation of the deiChO-262 enhancer. Green and orange boxes represent the location of the D-Pax2/Sv and Pros binding sites identified by systematic EMSAs using oligos corresponding to the regions represented by Boxes 1–7. (B) Sequence alignment between six Drosophila species for the region of the deiChO-262 enhancer containing the three D-Pax2/Sv and Pros sites (labeled and highlighted in green and orange, respectively). Dashes indicate gaps in the aligned sequence. Mutations of the D-Pax2/Sv and Pros sites used for the in vivo assays are shown at the bottom. (C) D-Pax2/Sv binds to two sites in deiChO-262, one low affinity site in fragment 3 (D-Pax2 site 1) and one high affinity site in fragment 7 (D-Pax2 site 2), as demonstrated with EMSA. The full screen is shown in Figure 5—figure supplement 1. (D) Pros binds to three sites in deiChO-262: Pros site 1 in fragment 3, Pros site 2 in an overlapping sequence in fragments 4 and 5, Pros site 3 in fragment 6, as demonstrated with EMSA. The full screen is shown in Figure 5—figure supplement 2. (E–L) Expression of wild-type (E) and mutated (F–L) deiChO-262-lacZ reporter constructs in abdominal segments A2-A6 of representative stage 16 embryos. The name of the construct is indicated in the bottom of each panel. The green arrows in E point to the cap cells of the various ChO of one abdominal segment: LCh1-lateral ChO1, LCh5-pentascolopidial organ, VChB and VChA are two ventral ChOs. The green arrows in J and L point to cap, scolopale (Sco) and ligament (Lig) cells of LCh5 organs where elevated level of reporter expression is evident. The arrowheads in I and K point to ligament cells of the ventral ChOs. in (M–N) Quantification of reporter activity in nuclei of cap (M) and scolopale cells (N) from LCh5 of segment A2 in embryos carrying the indicated constructs (n = 10 embryos for each genotype). In violin plots, each point represents an individual nucleus, median is shown as dark gray dashed line. Asterisks denote significant difference from wild-type activities, (*) - p < 0.05, (****) – p < 0.0001, n.s. – not significant (Dunnett’s multiple comparison test).
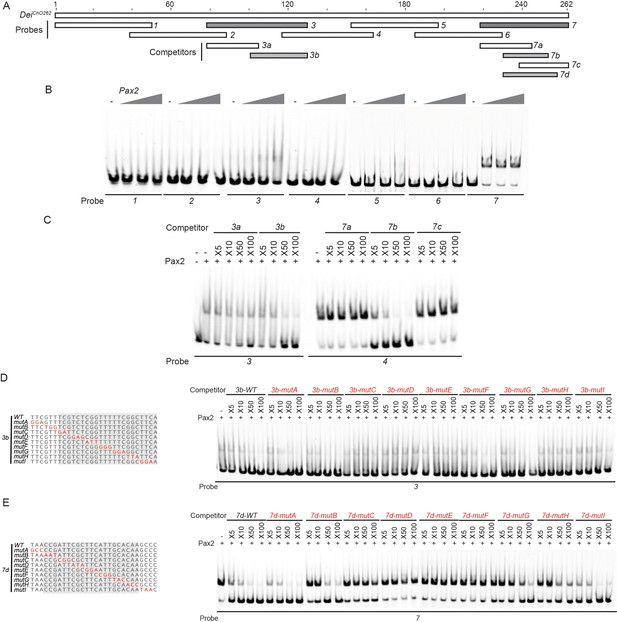
Identification of the regions in the deiChO-262 enhancer that bind D-Pax2/Sv in vitro.
(A) A schematic representation of the regions tested for their ability to bind D-Pax2/Sv in EMSAs. In gray are fragments that show specific binding. (B) EMSA scan for D-Pax2/Sv binding sites in deiChO-262 using the oligos marked in (A). Two sub-regions of deiChO-262 (fragments 3 and 7) bind to D-Pax2/Sv. (C–E) Dissection of the D-Pax2/Sv binding sites through competition assays. EMSAs were performed in the presence of increasing concentrations of unlabeled oligos (competitors). (C) Sub-fragments 3b and 7b unlabeled probes completely compete the binding of fragments 3 and 7, respectively, to D-Pax2/Sv. (D–E) On the left, alignment of the mutated competitors used in the competition EMSAs shown on the right. The nucleotides that comprise the D-Pax2/Sv binding sites are marked in gray.
-
Figure 5—figure supplement 1—source data 1
I dentification of the regions in the deiChO-262 enhancer that bind D-Pax2 in vitro - the full raw unedited gels.
- https://cdn.elifesciences.org/articles/70833/elife-70833-fig5-figsupp1-data1-v1.zip
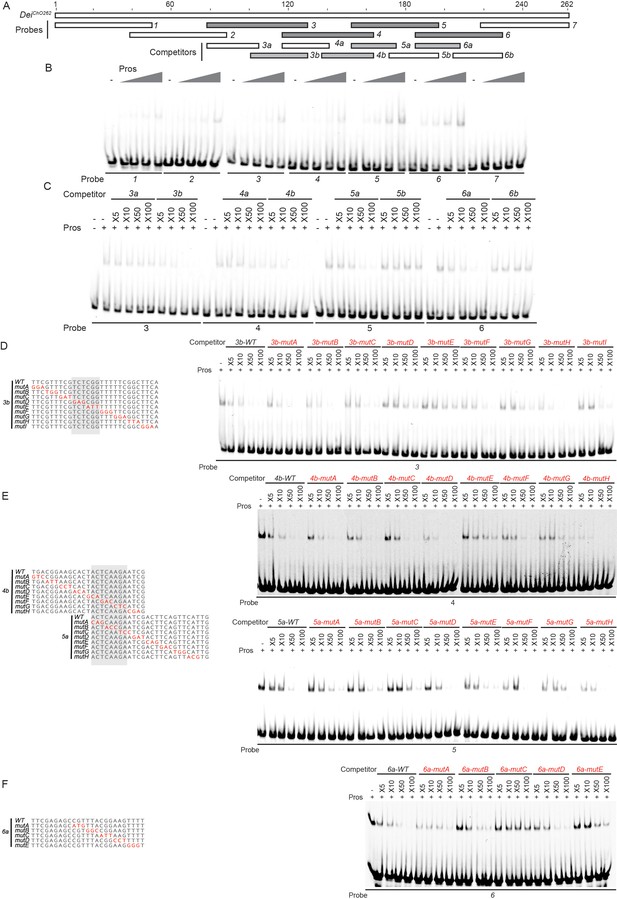
Identification of the regions in the deiChO262 enhancer that bind Pros in vitro.
(A) A schematic representation of the regions tested for their ability to bind Pros in EMSAs. In gray are fragments that show specific binding (stable upon increase of non-specific and specific competitors). (B) EMSA scan for Pros binding sites in deiChO-262 using the oligos marked in (A). Four sub-regions of deiChO-262 (fragments 3, 4, 5, and 6) bind specifically to Pros. (C–F) Dissection of the Pros binding sites through competition assays. EMSAs were performed in the presence of increasing concentrations of unlabeled oligos (competitors). (C) Sub-fragments 3b, 4b, 5a, and 6a unlabeled probes completely compete the binding of fragments 3, 4, 5, and 6, respectively, to Pros. (D–F) On the left, alignment of the mutated competitors used in the competition EMSAs shown on the right. The nucleotides that comprise the Pros binding sites are marked in gray.
-
Figure 5—figure supplement 2—source data 1
I dentification of the regions in the deiChO-262 enhancer that bind Pros in vitro - the full raw unedited gels.
- https://cdn.elifesciences.org/articles/70833/elife-70833-fig5-figsupp2-data1-v1.zip

Sequence of the deiChO-262 enhancer and tested mutations.
Sequence alignment of the wild-type deiChO-262 enhancer and the mutated constructs used in Figure 5. Verified D-Pax2/Sv and Pros binding sites are marked with green and orange boxes, respectively. Red letters denote mutated sites.
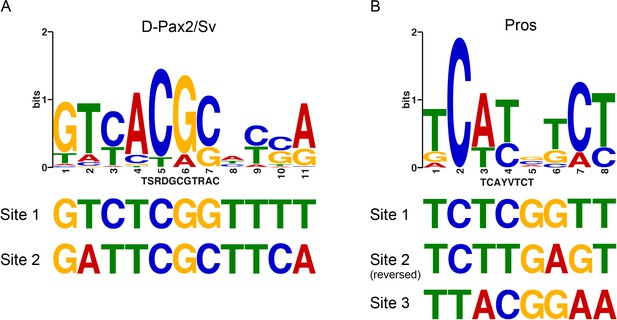
The deiChO-262 enhancer contains non-canonical binding sites for Pros and D-Pax2.
Consensus positional weight metrics (PWM) motifs for D-Pax2/Sv (A) and Pros (B) aligned to binding sites discovered in this study.
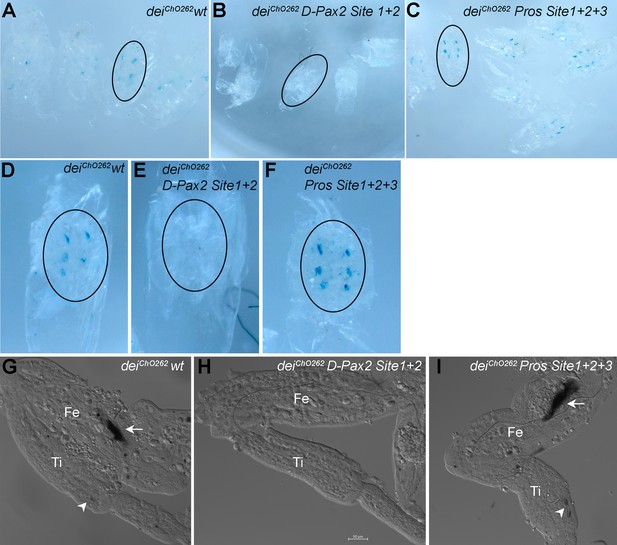
The D-Pax2 and Pros binding sites regulate the activity of the deiChO-262 enhancer in femoral ChOs.
b-galactosidase expression pattern driven by the wild-type and mutated deiChO-262 enhancers in pupal femoral ChOs. Pupae were stained with X-gal 40 hr after pupa formation. (A–C) An overview of pupae carrying the deiChO-262 wt (A), the deiChO-262 D-Pax site 1 + 2 (B) and the deiChO-262 Pros Site 1 + 2 + 3 (C) enhancers. A representative pupa of each genotype is circled and shown in higher magnification in (D–F). (G–I) A closeup view of a femur (Fe) and tibia (Ti) of legs dissected from the pupae shown in (D–F). The arrows point to femoral ChOs and arrowhead pont to the tibial ChOs (in genotypes where staining is evident).
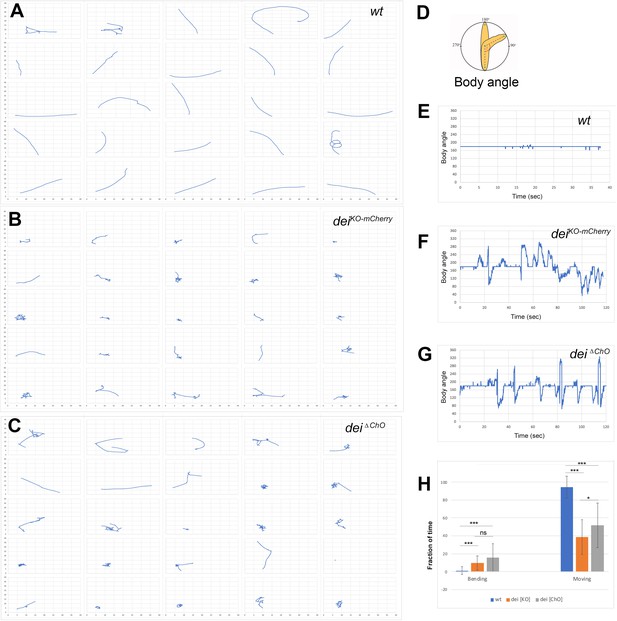
The deiChO-262 enhancer is essential for normal larval locomotion.
(A–C) Crawling trajectories of 25 wild-type (A) deiKO-mCherry (B) and deiΔChO (C) larvae. Each trajectory is shown in a square that represents 40 × 40 mm area. (D) Schematic representation of the body angle, γ, defined as the angle between the head and the body axis. (E–G) Representative time evolutions of the body angle of a wild-type (E), deiKO-mCherry (F) and deiΔChO (G) larvae. The wild-type larva walks persistently, and the body angle stays 180 degrees most of the time (a 40 s interval is shown, after which the larva exited the filmed arena). The deiKO-mCherry and deiΔChO mutant larvae display frequent changes in the direction of motion and long pauses accompanied by extensive head swiping (120 s intervals are shown). (H) A graph showing the average fraction of the time the larvae were crawling (GoPhase) and the fraction of time in which the body was bended more than 70 degrees (the measured angle γ was higher than 250 degrees or lower than 110 degrees). n = 24–27 for all genotypes; error bars represent the standard deviation. *** p < 0.0001, * p < 0.05, ns = non-significant. p Values were calculated using the unpaired two-tailed Mann-Whitney test.
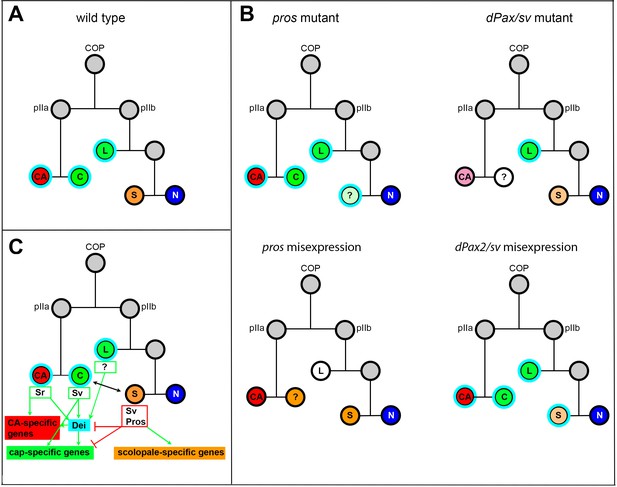
Summary of the relations between Sv, Pros and Dei in the ChO lineage and their effect on ChO development.
(A) A wt ChO lineage. The cap (C) and ligament (L) cells are depicted in green, the scolopale cell (S) is depicted in orange and the neuron (N) is depicted in blue. Cells that expressed Dei are circled in light blue. (B) The loss of pros leads to upregulation of Dei in the scolopale cell and to failure of scolopale cell differentiation. In contrast, the loss of Sv leads to loss of Dei expression from the cap cell and failure of cap cell differentiation. The Sv-deficient scolopale cells are also abnormal. The CA cells which depend on the cap cell for their development/maintenence also appear abnormal is sv mutants. Misexpression of Pros leads to repression of Dei in the cap and ligament cells, preventing their normal differentiation. The Pros-expressing cap cells adopt some scolopale-specific features. In contrast, over-expression of Sv leads to ectopic expression of Dei. Due to the presence of Pros, the level of expression of dei in the scolopale cell is restricted. (C) A schematic summary showing the relations between Pros, Sv and Dei and their relations to cell-type-specific differentiation programs. In the CA cells, dei is activated by Sr via the deiattachment enhancer. Both Sr and Dei are required there for the activation of CA-specific genes. In the cap cell dei is activated by Sv via the deiChO-262 enhancer. Sv is required for activating cap-specific genes in both Dei-dependent and independent ways. In the scolopale cells, dei is repressed by Pros via the deiChO-262 enhancer. Pros is required in addition for activating scolopale-specific genes. Dei is also expressed in the ligament cells and is required for their correct differentiation. The regulators of dei in the ligament cell are yet to be identified.
Videos
A video showing the locomotion of a wt (Canton-S) larva.
A video showing the locomotion of a wt (Canton-S) larva.
A video showing the locomotion of a deiKO-mCherry larva.
A video showing the locomotion of a deiKO-mCherry larva.
A video showing the locomotion of a deiΔChO larva.
A video showing the locomotion of a deiΔChO larva.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | dei/tx | FlyBase | CG5441, FBgn0263118 | |
| Gene (Drosophila melanogaster) | pros | FlyBase | CG17228, FBgn0004595 | |
| Gene (D. melanogaster) | sv/D-Pax2 | FlyBase | CG11049, FBgn0005561 | |
| Genetic reagent (D. melanogaster) | sv6/act-GFP | Kavaler et al., 1999 | N/A | |
| Genetic reagent (D. melanogaster) | Dpax2D1-GFP | Johnson et al., 2011 | N/A | |
| Genetic reagent (D. melanogaster) | pros17/TM6B, Tb1 | Bloomington Drosophila Stock Center | BDSC:5458 | |
| Genetic reagent (D. melanogaster) | deiChO-1353-GFP,deiattachment-RFP;en-Gal4 | Halachmi et al., 2016 | N/A | |
| Genetic reagent (D. melanogaster) | deiKO-mCherry | Hassan et al., 2018 | N/A | |
| Genetic reagent (D. melanogaster) | en-Gal4 | Brand and Perrimon, 1993 | ||
| Genetic reagent (D. melanogaster) | P{UAS-3xFLAG-pros.S}14 c, y1 w*; Pin1/CyO | Bloomington Drosophila Stock Center | BDSC:32245 | |
| Genetic reagent (D. melanogaster) | UAS-sv-RNAi | Vienna Drosophila Resource Center | VDRC:107343 | |
| Genetic reagent (D. melanogaster) | UAS-sv | Kavaler et al., 1999 | N/A | |
| Genetic reagent (D. melanogaster) | UAS-D-α-Catenin-GFP | Oda and Tsukita, 1999 | N/A | |
| Genetic reagent (D. melanogaster) | deiattachment-lacZ | Nachman et al., 2015 | N/A | |
| Genetic reagent (D. melanogaster) | deiChO-1353-lacZ | Nachman et al., 2015 | N/A | |
| Genetic reagent (D. melanogaster) | deiChO-1353 -GFP | Halachmi et al., 2016 | N/A | |
| Genetic reagent (D. melanogaster) | deiChO-389 -lacZ | Nachman et al., 2015 | N/A | |
| Genetic reagent (D. melanogaster) | deiChO-Gal4 | This study | N/A | |
| Genetic reagent (D. melanogaster) | deiChO-262- lacZ(in pH-Pelican) | This study | N/A | P element transgenesis. Available on 1st, 2nd, and 3rd chromosomes |
| Genetic reagent (D. melanogaster) | deiChO-262-placZ-wildtype(in placZattB) | This study | N/A | Inserted at attP2 |
| Genetic reagent (D. melanogaster) | deiChO-262-placZ-Pros-site 1(in placZattB) | This study | N/A | Inserted at attP2 |
| Genetic reagent (D. melanogaster) | deiChO-262-placZ-Pros-site 2(in placZattB) | This study | N/A | Inserted at attP2 |
| Genetic reagent (D. melanogaster) | deiChO-262-placZ-Pros-site 3(in placZattB) | This study | N/A | Inserted at attP2 |
| Genetic reagent (D. melanogaster) | deiChO-262-placZ-Pros-site 1 + 2 + 3(in placZattB) | This study | N/A | Inserted at attP2 |
| Genetic reagent (D. melanogaster) | deiChO-262-placZ-Pax2-site 1(in placZattB) | This study | N/A | Inserted at attP2 |
| Genetic reagent (D. melanogaster) | deiChO-262-placZattB Pax2-site 2 | This study | N/A | Inserted at attP2 |
| Genetic reagent (D. melanogaster) | deiChO-262-placZ-Pax2-site 1 + 2(in placZattB) | This study | N/A | Inserted at attP2 |
| Genetic reagent (D. melanogaster) | deiΔChO/(TM6) | This study | N/A | |
| Genetic reagent (D. melanogaster) | M{nos-Cas9.P}ZH-2A | Bloomington Drosophila Stock Center | BDSC:54591 | |
| Antibody | Anti Sv/D-Pax2 (rabbit polyclonal) | Johnson et al., 2011 | N/A | (1:10,000) |
| Antibody | Anti-α85E-Tubulin (rabbit polyclonal) | Klein et al., 2010 | N/A | (1:200) |
| Antibody | Anti α85E-Tubulin (mouse monoclonal) | Nachman et al., 2015 | N/A | (1:20) |
| Antibody | Anti-Dei (rabbit polyclonal) | Egoz-Matia et al., 2011 | N/A | (1:50) |
| Antibody | Anti-Spalt (rabbit polyclonal) | Halachmi et al., 2007 | N/A | (1:500) |
| Antibody | Anti-NRG (rat polyclonal) | Banerjee et al., 2006 | N/A | (1:1000) |
| Antibody | Anti-βGal (mouse monoclonal) | Promega | Z3781 | (1:1000) |
| Antibody | Anti-Pros (mouse monoclonal) | The Developmental Studies Hybridoma Bank | MR1A | (1:20) |
| Antibody | Anti Futsch (mouse monoclonal) | The Developmental Studies Hybridoma Bank | 22C10 | (1:20) |
| Antibody | Anti-ELAV (rat monoclonal) | The Developmental Studies Hybridoma Bank | 7E8A10 | (1:50) |
| Antibody | Anti-Eys (mouse monoclonal) | The Developmental Studies Hybridoma Bank | 21A6 | (1:20) |
| Antibody | Anti-Repo (mouse monoclonal) | The Developmental Studies Hybridoma Bank | 8D12 | (1:10) |
| Antibody | Anti Crb (mouse monoclonal) | The Developmental Studies Hybridoma Bank | Cq4 | (1:10) |
| Antibody | Anti-Cpo (rabbit polyclonal) | Bellen et al., 1992 | N/A | (1:5000) |
| Antibody | Anti-Sr (Chicken polyclonal) | This study | N/A | (1:20) |
| Antibody | Cy3-conjugated goat anti-mouse | Jackson Laboratories, Bar-Harbor, Maine, USA | 115-165-166 | (1:100) |
| Antibody | Cy3-conjugated goat anti-rabbit | Jackson Laboratories, Bar-Harbor, Maine, USA | 115-165-144 | (1:100) |
| Antibody | Cy2-conjugated goat anti-rabbit | Jackson Laboratories, Bar-Harbor, Maine, USA | 111-225-144 | (1:100) |
| Antibody | Alexa Fluor-647-conjugated goat anti-mouse | Jackson Laboratories, Bar-Harbor, Maine, USA | 115-605-166 | (1:100) |
| Antibody | Alexa Fluor-647-conjugated goat anti-rabbit | Jackson Laboratories, Bar-Harbor, Maine, USA | 115-605-144 | (1:100) |
| Antibody | Cy3-conjugated donkey anti-chicken IgY | Jackson Laboratories, Bar-Harbor, Maine, USA | 703-165-155 | (1:100) |
| Antibody | Alexa Fluor-647-conjugated Donkey anti-Rat | Jackson Laboratories, Bar-Harbor, Maine, USA | 712-605-153 | (1:100) |
| Commercial assay or kit | qScript cDNA Synthesis kit | Quanta BIOSCIENCES | 95047–100 | |
| Commercial assay or kit | PrimeSTAR Max DNA polymerase | TAKARA | R045A | |
| Chemical compound, drug | DAKO fluorescence mounting medium | Agilent Technologies, Santa Clara, CA, USA | S3023 | |
| Chemical compound, drug | TRI Reagent | Sigma | T9424 | |
| Software, algorithm | ImajeJ | National Institute of Health | SCR_003070 | |
| Software, algorithm | FIMTrack tracking software | Risse et al., 2017 | https://www.uni-muenster.de/FRIA/en/FIM/ | |
| Software, algorithm | MATLAB | Mathworks | SCR_001622 | |
| Software, algorithm | Imaris | Bitplane | SCR_007370 |
Additional files
-
Supplementary file 1
Results of the 1YH screen.
This table summarizes the sequencing data of 146 positive clones identified in the 1YH screen.
- https://cdn.elifesciences.org/articles/70833/elife-70833-supp1-v1.xlsx
-
Supplementary file 2
Transgenic constructs used in this study.
This table lists all the dei-related transgenes used in this study, including the vectors used and landing sites. The sequences of the wildtype and mutated deiChO-262 fragments are shown. The associated Figures are indicated.
- https://cdn.elifesciences.org/articles/70833/elife-70833-supp2-v1.xlsx
-
Supplementary file 3
Oligos and proteins used in the Electro Mobility Shift Assays.
The Oligos and Protein Concentrations Used in the described Electro Mobility Shift Assays performed in this study. The mutated nucleotides in the competitor oligos are underlined.
- https://cdn.elifesciences.org/articles/70833/elife-70833-supp3-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70833/elife-70833-transrepform1-v1.pdf





