A MET-PTPRK kinase-phosphatase rheostat controls ZNRF3 and Wnt signaling
Figures
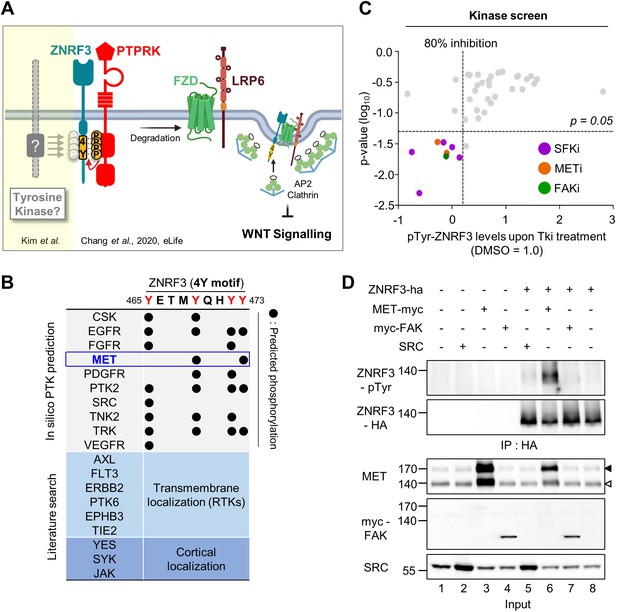
Identification of SRC and MET as ZNRF3 kinases.
(A) Model for tyrosine phosphorylation regulating ZNRF3 and Wnt signaling (Chang et al., 2020). ZNRF3 E3 ubiquitin ligase reaching the plasma membrane is co-internalized with Wnt receptors and targets them for lysosomal degradation to reduce Wnt signaling. An unphosphorylated 4-tyrosine (4Y) motif serves as ZNRF3 internalization signal. The phosphatase PTPRK dephosphorylates and unmasks the 4Y motif, promoting internalization and lysosomal targeting of ZNRF3 and Wnt receptors, and reducing Wnt signaling. PTPRK is counteracted by an unknown tyrosine kinase(s) that phosphorylates the 4Y motif, impairs ZNRF3/Wnt receptor internalization, and increases Wnt signaling. Created with Biorender.com. (B) Scheme of 4Y PTK candidate selection. Eighteen tyrosine kinases were selected by combined in silico prediction and reported cellular localization. Kinase inhibitor screening (C) was then conducted for validation. (C) Inhibitor screen for ZNRF3 4Y kinases. TetOn ZNRF3-HA H1703 cells were treated with 40 inhibitors targeting 18 PTKs selected as shown in (B). pTyr-ZNRF3 was analyzed by immunoblot for pan-phosphotyrosine and normalized to total ZNRF3. Results from two independent screens were pooled to generate the dot graph. X-axis indicates the relative tyrosine phosphorylation of ZNRF3 upon inhibitor (Tki) treatment relative to DMSO control, y-axis indicates p-value. Three kinases (SFK, MET, FAK) with phosphorylation inhibition ≥80% and p-value ≤ 0.05 were selected for downstream analysis and are highlighted. (D) Analysis of pTyr-ZNRF3 in HEK293T cells co-transfected with ZNRF3-HA and MET-myc, myc-FAK or SRC. Cell lysates were pulled down with anti-HA antibody and subjected to immunoblot analysis using the indicated antibodies. Data show a representative result from three independent experiments with similar outcome. Black arrowhead: Immature MET precursor. White arrowhead: Mature, processed MET (β-chain). Source files of all blots used in this figure are available in Figure 1—source data 1.
-
Figure 1—source data 1
Uncropped immunoblot images for Figure 1D.
- https://cdn.elifesciences.org/articles/70885/elife-70885-fig1-data1-v2.zip
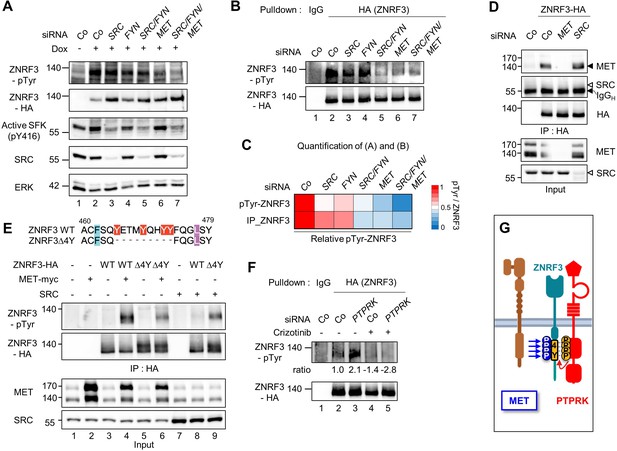
MET is a ZNRF3 4Y kinase.
(A) Tyrosine phosphorylation of ZNRF3 requires endogenous MET and SRC. pTyr-ZNRF3 was monitored in TetOn ZNRF3-HA H1703 cells upon siRNA knockdown of SRC, FYN, MET or combinations as indicated. Cells were induced with Dox for 48 hr before harvest. Cells without Dox treatment were used as control for background pTyr signal. Data show a representative result from two independent experiments with similar outcome. (B) Analysis of pTyr-ZNRF3 in TetOn ZNRF3-HA H1703 cells upon siRNA knockdown of SRC, FYN, MET or combination as indicated. Lysates were pulled down with anti-HA antibody or control IgG and subjected to immunoblot analysis with the indicated antibodies. (C) Visualization of normalized pTyr-ZNRF3 from immunoblots (A) and ZNRF3 pulldown (B) as a heat map. Background signal (lane 1 from (A, B)) was subtracted from pTyr-ZNRF3 before normalization to total ZNRF3. Color code is represented on the right. (D) ZNRF3 binds endogenous MET but not SRC. Co-immunoprecipitation (Co-IP) analysis in TetOn ZNRF3-HA H1703 cells upon siRNA knockdown of MET or SRC. Data show a representative result from three independent experiments with similar outcome. Black arrow: IgG heavy chain. Black arrowhead: MET. White arrowhead: expected position of SRC. (E) MET but not SRC is a 4Y-specific kinase. Tyrosine phosphorylation of ZNRF3-HA or ZNRF3(Δ4Y)-HA in HEK293T cells co-transfected with the indicated kinases. Lysates were pulled down with anti-HA antibody and subjected to immunoblot analysis using the indicated antibodies. Data show a representative result from three independent experiments with similar outcome. (F) MET and PTPRK oppose each other on 4Y phosphorylation. Analysis of pTyr-ZNRF3 in TetOn ZNRF3-HA H1703 cells upon siRNA knockdown of PTPRK and/or overnight crizotinib treatment as indicated. Lysates were pulled down with anti-HA antibody or control IgG and subjected to immunoblot analysis with the indicated antibodies. Ratio, relative pTyr-ZNRF3 normalized to total ZNRF3. Data show a representative result from two independent experiments with similar outcome. (G) Model: MET and PTPRK phosphorylate and dephosphorylate the 4Y motif on ZNRF3, respectively. Created with Biorender.com. Source files of all blots used in this figure are available in Figure 2—source data 1. Source files of densitometric analysis for heat map in (C) are available in Figure 2—source data 2. A source file of densitometric analysis for (F) is available in Figure 2—source data 3.
-
Figure 2—source data 1
Uncropped immunoblot images for Figure 2A–B, D–F.
- https://cdn.elifesciences.org/articles/70885/elife-70885-fig2-data1-v2.zip
-
Figure 2—source data 2
Densitometric analysis of immunoblots for Figure 2A–B to generate heap map for Figure 2C.
- https://cdn.elifesciences.org/articles/70885/elife-70885-fig2-data2-v2.zip
-
Figure 2—source data 3
Densitometric analysis of immunoblots for Figure 2F.
- https://cdn.elifesciences.org/articles/70885/elife-70885-fig2-data3-v2.xlsx
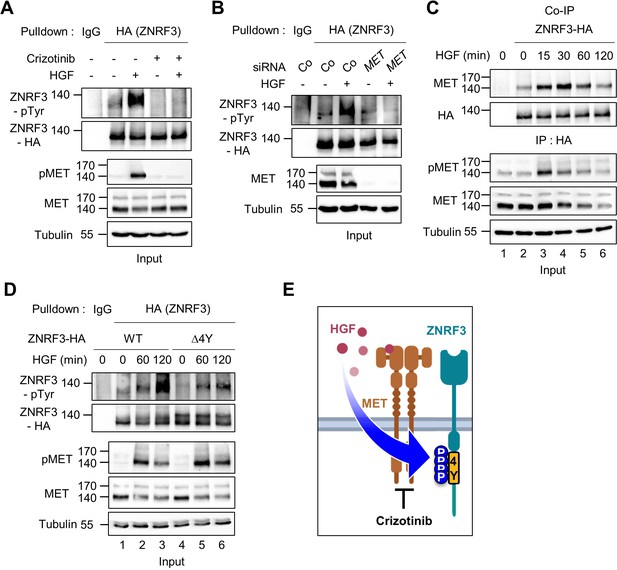
HGF-MET signaling triggers 4Y phosphorylation of ZNRF3.
(A) HGF induces tyrosine phosphorylation of ZNRF3 and requires MET kinase activity. Analysis of pTyr-ZNRF3 in TetOn ZNRF3-HA H1703 cells upon overnight Crizotinib treatment. HGF was added for 2 hr before harvest. Lysates were pulled down with anti-HA antibody or control IgG and subjected to immunoblot analysis. Data show a representative result from two independent experiments with similar outcome. (B) HGF-induced tyrosine phosphorylation of ZNRF3 requires MET. pTyr-ZNRF3 in TetOn ZNRF3-HA H1703 cells was analyzed upon siRNA knockdown of MET. Cells were treated with HGF for 2 hr before harvest. Lysates were pulled down with anti-HA antibody or control IgG and subjected to immunoblot analysis using the indicated antibodies. Data show a representative result from two independent experiments with similar outcome. (C) HGF promotes the interaction of endogenous MET with ZNRF3. Co-IP analysis in TetOn ZNRF3-HA H1703 cells upon HGF treatment. Lysates were pulled down anti-HA antibody or control IgG and subjected to immunoblot analysis. Data show a representative result from two independent experiments with similar outcome. (D) HGF promotes tyrosine phosphorylation of the 4Y motif. Analysis of tyrosine phosphorylation of ZNRF3-HA (WT) or ZNRF3(Δ4Y)-HA in H1703 cells upon HGF treatment. Lysates were pulled down with anti-HA antibody or control IgG and subjected to immunoblot analysis. Data show a representative result from two independent experiments with similar outcome. (E) Model: HGF-MET phosphorylates 4Y motif on ZNRF3. Crizotinib blocks the 4Y phosphorylation by HGF. Created with Biorender.com. Source files of all blots used in this figure are available in Figure 3—source data 1.
-
Figure 3—source data 1
Uncropped immunoblot images for Figure 3A–D.
- https://cdn.elifesciences.org/articles/70885/elife-70885-fig3-data1-v2.zip
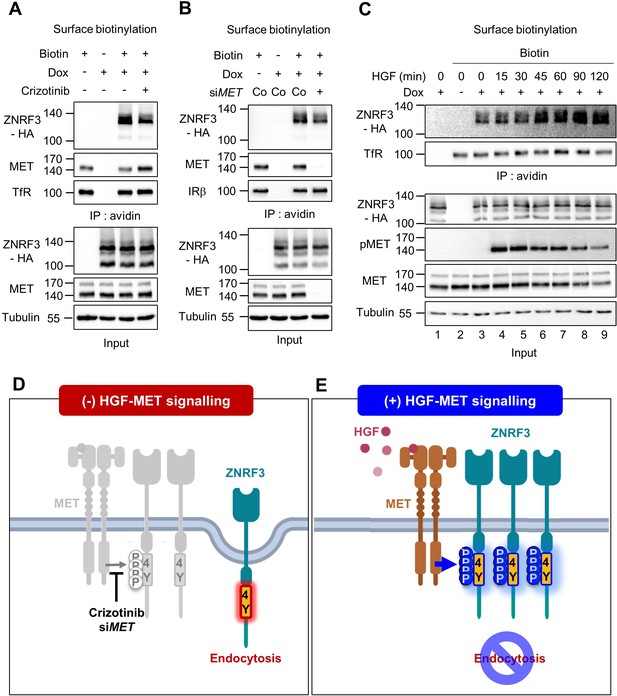
HGF-MET signaling stabilizes ZNRF3 at the cell surface.
(A–B) Cell surface biotinylation assay of ZNRF3 in TetOn ZNRF3-HA H1703 cells upon Crizotinib treatment (A) or siRNA knockdown of MET (B). Cells were treated with Crizotinib (A) overnight before harvest. After labeling surface proteins with biotin, lysates were pulled down with streptavidin beads and subjected to immunoblot analysis. Transferrin receptor (TfR) or Insulin receptor beta (IRβ) was used as a loading control for avidin pulldown. Tubulin was used as a loading control for total cell lysate (Input). Data show a representative result from two independent experiments with similar outcome. (C) HGF stabilizes cell surface ZNRF3 within 45 min. Cell surface biotinylation assay in TetOn ZNRF3-HA H1703 cells upon HGF treatment for the indicated time. After labeling surface protein with biotin, lysates were pulled down with streptavidin beads and subjected to immunoblot analysis. Transferrin receptor (TfR) was used as loading control for avidin pulldown. Tubulin was used as loading control for total cell lysate (Input). Data show a representative result from two independent experiments with similar outcome. (D–E) Model: In the absence of HGF-MET signaling (D), unphosphorylated 4Y motif promotes endocytosis of ZNRF3. In the presence of HGF-MET signaling (E), activated MET phosphorylates and masks the endocytic 4Y motif, stabilizing ZNRF3 at the cell surface. Created with Biorender.com. Source files of all blots used in this figure are available in Figure 4—source data 1.
-
Figure 4—source data 1
Uncropped immunoblot images for Figure 4A–C.
- https://cdn.elifesciences.org/articles/70885/elife-70885-fig4-data1-v2.zip
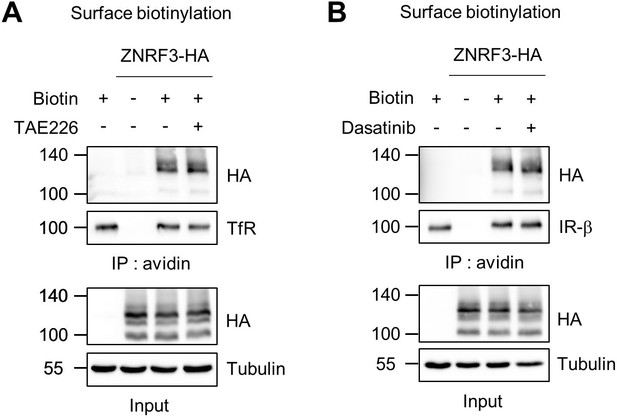
Surface ZNRF3 is not changed by FAK or SFKs inhibitor.
(A–B) FAK or SFK inhibitor does not affect ZNRF3 cell surface levels. Cell surface biotinylation assay in TetOn ZNRF3-HA H1703 cells upon TAE226 (FAK inhibitor) (A) or Dasatinib (SFK inhibitor) (B) treatment overnight. After labeling surface proteins with biotin, lysates were pulled down with streptavidin beads and subjected to immunoblot analysis. Transferrin receptor (TfR) or Insulin receptor beta (IRβ) was used as loading control for avidin pulldown. Tubulin was used as loading control for total cell lysate (Input). Data show a representative result from two independent experiments with similar outcome. Source files of all blots used in this figure are available in Figure 4—figure supplement 1—source data 1.
-
Figure 4—figure supplement 1—source data 1
Uncropped immunoblot images for Figure 4—figure supplement 1A–B.
- https://cdn.elifesciences.org/articles/70885/elife-70885-fig4-figsupp1-data1-v2.zip
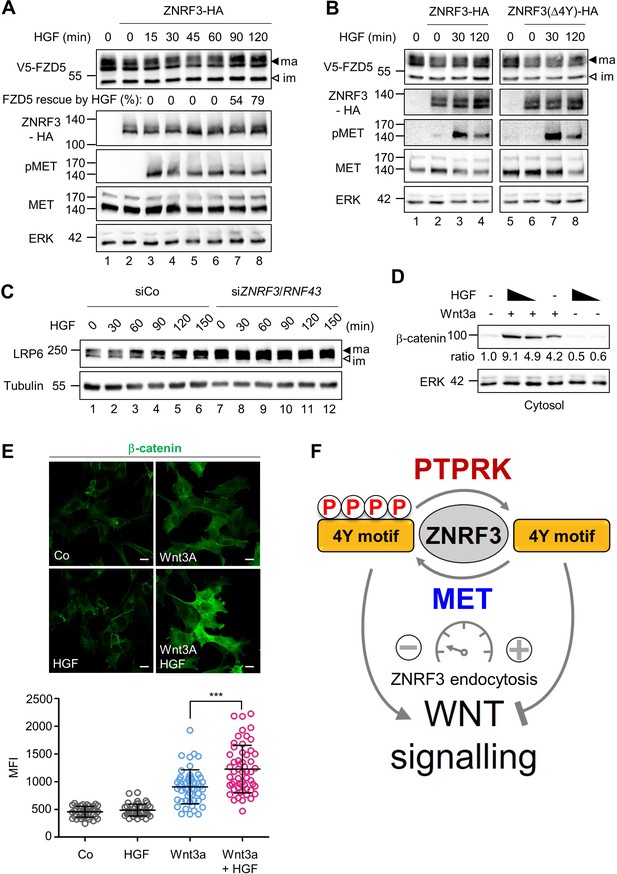
HGF increases Wnt receptor levels by counteracting ZNRF3 function.
(A) HGF protects mature FZD5 against ZNRF3. Immunoblot analysis of V5-FZD5 in H1703 cells with ZNRF3-HA transfection upon HGF treatment as indicated. ma, im: Mature and immature forms of V5-FZD5, respectively. Rescue of ZNRF3-reduced FZD5 levels by HGF was quantified by normalizing V5-FZD5 to ERK, setting V5-FZD5 in lanes 1 and 2 to 100- and 0%, respectively, and calculating HGF-rescued levels of V5-FZD5 relative to this scale. Data show a representative result from two independent experiments with similar outcome. (B) HGF stabilizes mature FZD5 in presence of ZNRF3 but not ZNRF3(Δ4Y). Immunoblot analysis of V5-FZD5 in H1703 cells with ZNRF3-HA or ZNRF3(Δ4Y)-HA transfection upon HGF treatment as indicated. ma, im: Mature and immature forms of V5-FZD5, respectively. Data show a representative result from two independent experiments with similar outcome. (C) HGF stabilizes LRP6 levels in a ZNRF3/RNF43-dependent manner. Immunoblot of total LRP6 protein in H1703 cells upon HGF treatment without or with siZNRF3/RNF43 knockdown as indicated. ma, im: Mature and immature form of LRP6, respectively. (D) HGF enhances β-catenin levels upon Wnt3a stimulation. Immunoblot analysis of cytosolic (saponin-extracted) β-catenin in H1703 cells treated overnight as indicated. Ratio, relative levels of β-catenin normalized to ERK1/2. Data show a representative result from three independent experiments with similar outcome. (E) HGF enhances β-catenin levels upon Wnt3a stimulation. Immunofluorescence microscopy (IF) showing nuclear and cytosolic β-catenin in H1703 cells. Cells were treated overnight as indicated. Top, representative IF images. Bottom, quantification of β-catenin (Mean ± SD, ***p˂0.001, student t-test, MFI: Mean fluorescence intensity). (F) Model for how the phospho-regulated 4Y motif acts as a molecular rheostat for ZNRF3 endocytosis in Wnt signaling. MET phosphorylates and PTPRK dephosphorylates the 4Y motif, which attenuates or promotes ZNRF3 endocytosis to activate or inhibit Wnt signaling, respectively. Source files of all blots used in this figure are available in Figure 5—source data 1. A source file of densitometric analysis for Figure 5 (A and D) is available in Figure 5—source data 2.
-
Figure 5—source data 1
Uncropped immunoblot images for Figure 5A–D.
- https://cdn.elifesciences.org/articles/70885/elife-70885-fig5-data1-v2.zip
-
Figure 5—source data 2
Densitometric analysis of immunoblots for Figure 5A and D.
- https://cdn.elifesciences.org/articles/70885/elife-70885-fig5-data2-v2.xlsx
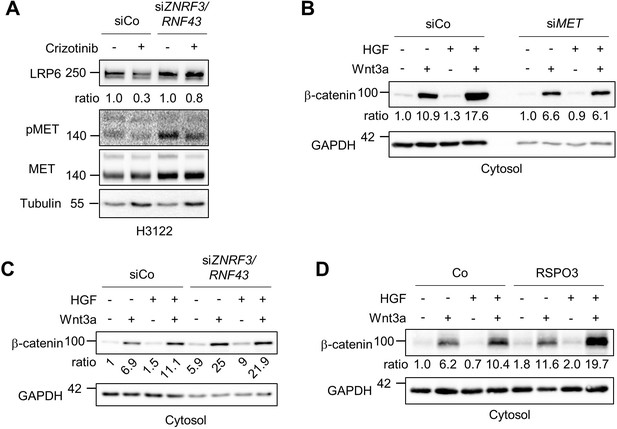
RSPO and HGF synergize to increase Wnt signaling.
(A) HGF-MET stabilizes LRP6 levels in a ZNRF3/RNF43-dependent manner in H3122 cells. Immunoblot of total LRP6 protein in H3122 cells upon Crizotinib treatment without or with siZNRF3/RNF43 knockdown as indicated. Ratio, relative levels of LRP6 normalized to Tubulin. Data show a representative result from two independent experiments with similar outcome. (B–C) HGF-enhanced β-catenin levels upon Wnt3a stimulation requires MET (B) and ZNRF3/RNF43 (C). Immunoblot analysis of cytosolic β-catenin in H1703 cells upon treatment overnight with or without siMET or siZNRF3/RNF43 knockdown as indicated. Ratio, relative levels of β-catenin normalized to GAPDH. Data show a representative result from two independent experiments with similar outcome. (D) HGF synergizes with RSPO on β-catenin accumulation upon Wnt3a stimulation. Immunoblot analysis of cytosolic β-catenin in H1703 cells upon treatment of Wnt3a, HGF and/or RSPO3 for 2 hr as indicated. Ratio, relative levels of β-catenin normalized to GAPDH. Data show a representative result from three independent experiments with similar outcome. Source files of all blots used in this figure are available in Figure 5—figure supplement 1—source data 1. A source file of all densitometric analysis in this figure is available in Figure 5—figure supplement 1—source data 2.
-
Figure 5—figure supplement 1—source data 1
Uncropped immunoblot images for Figure 5—figure supplement 1A–D.
- https://cdn.elifesciences.org/articles/70885/elife-70885-fig5-figsupp1-data1-v2.zip
-
Figure 5—figure supplement 1—source data 2
Densitometric analysis of immunoblots for Figure 5—figure supplement 1A–D.
- https://cdn.elifesciences.org/articles/70885/elife-70885-fig5-figsupp1-data2-v2.xlsx

Bafilomycin treatment enhances pTyr-ZNRF3 levels.
Tyrosine phosphorylation of ZNRF3 in TetOn ZNRF3-HA H1703 cells upon siRNA transfection with or without bafilomycin treatment overnight. Cells were treated with Dox for 48 hr before harvest. As a positive control, cells were treated with Na-pervanadate (PV, phosphatase inhibitor) for 30 min before harvest. Lysates were pulled down with anti-HA antibody or control IgG and subjected to Western blot analysis. Ratio, tyrosine phosphorylation of ZNRF3 normalized to total ZNRF3. Note that bafilomycin is enhancing pTyr-ZNRF3 levels but does not qualitatively affect the siPTPRK outcome.
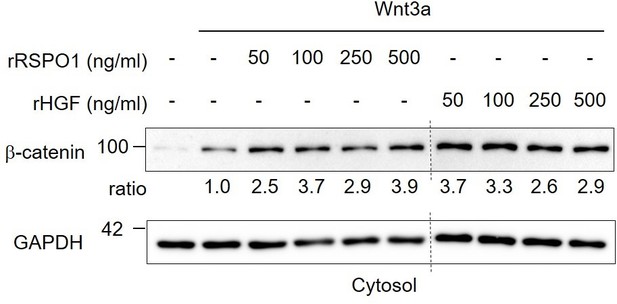
Comparison of recombinant human RSPO1 and HGF in Wnt activation.
Immunoblot analysis of cytosolic β-catenin in H1703 cells upon 2h treatment of Wnt3a with different concentrations of recombinant human HGF or RSPO1. Ratio, relative levels of β-catenin normalized to GAPDH. Data show a representative result from two independent experiments with similar outcome.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene(Homo sapiens) | MET | NCBI | NM_000245 | |
| Gene(Homo sapiens) | SRC | Gary DavidsonPMID:25391905 | ||
| Gene(Homo sapiens) | PTK2 | GenBank | AAH35404.1 | |
| Cell line (Homo-sapiens) | H1703 | ATCC | CRL-5889RRID:CVCL_1490 | |
| Cell line (Homo-sapiens) | H3122 | Cellosaurus | RRID:CVCL_5160 | Prof. Dr. Rocio Sotillo |
| Cell line (Homo-sapiens) | 293T | ATCC | CRL-3216RRID:CVCL_0063 | |
| Cell line (Homo-sapiens) | H1703 TetOn ZNRF3-HA | Chang et al., 2020 | Generated from H1703 | |
| Antibody | Anti-MET (Rabbit monoclonal) | Cell signaling | 8198T RRID:AB_10858224 | WB (1:3000) |
| Antibody | Anti-phospho-MET (Y1234/1235; Rabbit monoclonal) | Cell signaling | 3077T RRID:AB_2143884 | WB (1:1000) |
| Antibody | Anti-α-Tubulin (Mouse monoclonal) | Sigma | T5168 RRID:AB_477579 | WB (1:3000) |
| Antibody | Anti-SRC (Rabbit monoclonal) | Cell signaling | 2,123 S RRID:AB_2106047 | WB (1:1000) |
| Antibody | Anti-phospho Src Family (Y416; Rabbit monoclonal) | Cell signaling | 2,101 S RRID:AB_331697 | WB (1:1000) |
| Antibody | Anti-Insulin receptor beta (Rabbit monoclonal) | Cell signaling | 3025T RRID:AB_2280448 | WB (1:1000) |
| Antibody | Anti-GAPDH (Rabbit monoclonal) | Cell signaling | 2,118 LRRID:AB_561053 | WB (1:10000) |
| Antibody | Anti-β-catenin (mouse monoclonal) | BD bioscience | 610154 RRID:AB_397555 | WB (1:5000)IF (1:500) |
| Antibody | Goat anti-mouse Alexa 488 (goat polyclonal) | Invitrogen | A11029 RRID:AB_138404 | IF (1:500) |
| Transfected construct (human) | siRNA to human MET (SMARTpool) | Horizon discovery | M-003156-02-0005 | 25 nM |
| Transfected construct (human) | siRNA to human ZNRF3 (SMARTpool) | Horizon discovery Chang et al., 2020 | ||
| Transfected construct (human) | siRNA to human RNF43 (SMARTpool) | Horizon discovery Chang et al., 2020 | ||
| Recombinant DNA reagent | pDONR223-MET | Addgene | Kit #1000000014Plasmid #23,889 | |
| Recombinant DNA reagent | pDONR223-PTK2 | Addgene | Kit #1000000014Plasmid #23,902 | |
| Recombinant DNA reagent | pDEST-Myc-C-term | DKFZ Vector repository | MYC-C | Source: Stefan Pusch, University Heidelberg |
| Recombinant DNA reagent | pDEST-Myc-N-term | DKFZ Vector repository | MYC-N | Source: Stefan Pusch, University Heidelberg |
| Recombinant DNA reagent | pDEST-hSRC | Gary Davidson PMID:25391905 | ||
| Peptide, recombinant protein | Recombinant human HGF | Peprotech | 100–39 H | 50 ng/ml |
| Chemical compound, drug | Kinase Screening Library (96well) | Cayman chemical | 10505 | 500 nM |
| Chemical compound, drug | Crizotinib | Cayman chemical | 12087 | 500 nM |
| Chemical compound, drug | Dasatinib | Cayman chemical | 11498 | 500 nM |
| Chemical compound, drug | TAE226 | Cayman chemical | 17685 | 500 nM |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70885/elife-70885-transrepform1-v2.docx
-
Supplementary file 1
Data of Inhibitor screen for ZNRF3 4Y kinases.
Table of inhibitor screen results. The table includes the name of inhibitors, relative pTyr-ZNRF3 from two screens, average, and p-value.
- https://cdn.elifesciences.org/articles/70885/elife-70885-supp1-v2.xlsx





