Genetically engineered insects with sex-selection and genetic incompatibility enable population suppression
Figures
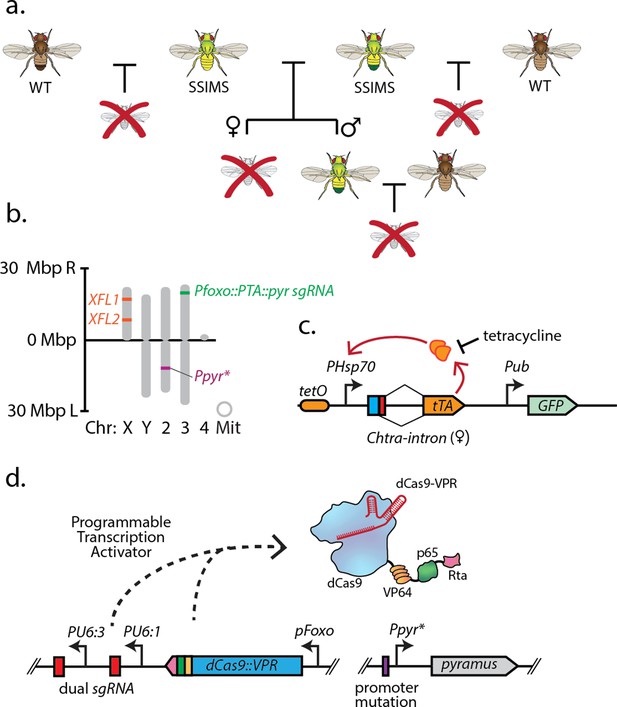
Overall design and implementation of Sex-sorting Incompatible Male System (SSIMS).
(a) Illustration of desired behavior of SSIMS insects (green-yellow) when grown in the absence of Tet. Brown flies represent wild-type, and red crosses represent inviable offspring. (b) Genome-scale location of engineered loci in D. melanogaster SSIMS line. (c,d) Genetic cassette diagrams for the female lethal (FL) locus. The orange oval represents 21 repeats of the tet operator. The blue box represents the start codon. The red box denotes the exon sequence containing multiple stop codons that is retained when an alternative 5’ splice site is utilized in males. Female splicing fuses the start codon to the tTA coding DNA sequence. (d) Genetic cassette diagram for the engineered genetic incompatibility (EGI) loci, respectively. SBOL iconography is used for genetic part representation. FL, X-linked female lethal construct; Mbp, megabasepairs; kb, kilobases; Chr, chromosome; Mit, mitochondrial; PTA, Programmable Transcriptional Activator.
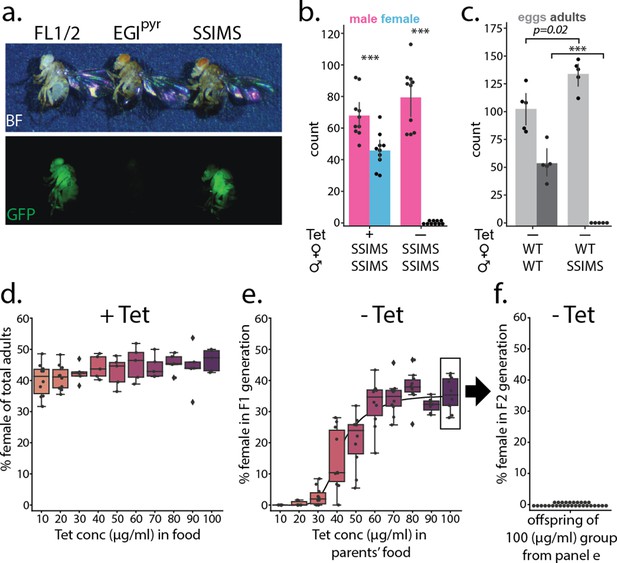
Phenotype of SSIMS flies.
(a) Visual markers present in homozygous FL lines (GFP, left) and EGI lines (red eyes, center) are present in SSIMS flies generated via the mating scheme in Appendix 1—figure 1. (b) Counts of male (pink) and female (blue) offspring of SSIMS strain raised in food with 10 μg/ml Tetracyline (Tet) and without Tet. No females offspring were observed when larvae are raised in the absence of Tet. Chi-squared test shows significant difference from expected 50:50 ratio of males:female, n = 10 per group, *** denotes < 0.0001. (c) Counts of egg-lay (light gray) and surviving adults (dark gray) of wild-type females crossed with either wild-type or SSIMS males. SSIMS male mate successfully with wild-type females, producing slightly more eggs, but no adult offspring emerge, n = 5 per group, *** denotes p < 0.0001. (d) Percent female offspring when SSIMS stocks are reared in food with increasing concentrations of Tet, n = 10 per group. (e) Percent female offspring when respective progeny from (d) are reared in food without Tet, n = 7–10 per group. Line behind data shows Hill function that fits data with statistics described in the main text. (f) Percent female offspring when progeny from the 100 μg/ml group in (e) are reared in food without Tet, n = 30. FL, X-linked Female Lethal; EGI , engineered genetic incompatibility targeting pyramus; WT, wild-type; SSIMS, sex-sorting incompatible male system; Tet, Tetracycline.
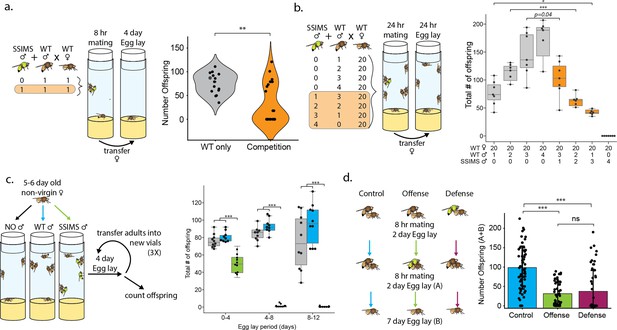
Male mating competition with wild-type D. melanogaster.
(a) Experimental design schema and results of individual male mating competition assay. Violin plots show the number of offspring produced by one WT female crossed to one WT male (gray, n = 15) or one WT female crossed to one WT male and one SSIMS male (orange, n = 26). A Welch two-sample t test was performed between the two groups. (b) Experimental design schema and results of mixed-male mating assay. Box-whisker plots show the median (center line), 25th and 75th percentile boundaries (box), and min/max (whiskers). Student’s t tests were performed between experiments with identical numbers of wild-type males. n = 7 per group. (c) Experimental design schema and results of sperm displacement assay. Box-whisker plots colored to show the addition of no males (gray), wild-type males (blue), or SSIMS males (green) to the non-virgin wild-type females. Student’s t tests were performed for each egg-lay period comparing+ SSIMS male vs both no-male and WT-male groups. n = 10 per group. (d) Experimental design schema and results of offensive and defensive sperm displacement assay. Bar graphs show the number of offspring produced by one WT female mated to a WT male and remated with a second WT male (blue, n = 60), one WT female mated to a WT male and remated with a SSIMS male (green, n = 53), or one WT female mated to a SSIMS male and remated with a WT male (maroon, n = 54). Welch two-sample t tests were performed pairwise between each of the three groups. Statistical significance: ns = not significant, *=p < 0.01, **=p < 0.001, ***=p < 0.0001.
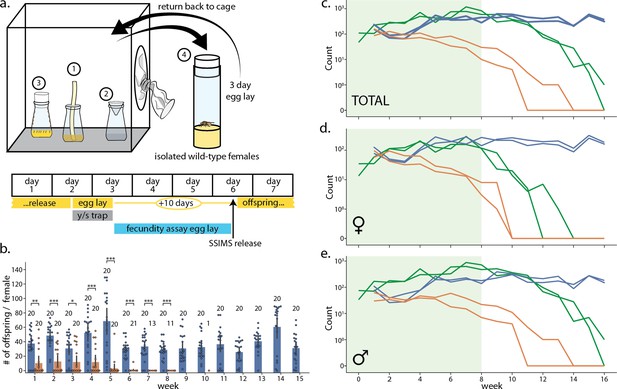
Laboratory cage trial of population suppression of wild-type D. melanogaster.
(a) Cage trial design showing Bioquip cage with adult feeding apparatus (1), yeast/sugar trap (2), reproduction bottle (3), fecundity assay vials (4), and weekly schematic of the trial, with tasks done each day highlighted below. Two control and two SSIMS treated cages were initiated, see methods for details. (b) Results of the fecundity assay from each week. Each colored dot represents total number of offspring from a single vial with one wild-type female from control cages (blue) and SSIMS treated cages (orange). Sample size of each group per week is illustrated above groups. Statistical tests of significance (Mann-Whitney test) are performed for each week comparing control and +SSIMS treated cages, n = 11–20 per group. (c–e) Total (c), female only (d), and male only (e) counts from the yeast/sugar trap. Light green shading indicates weekly additional SSIMS releases until week 8. Wild-type population in the control cages (blue lines) rise gradually and reach plateau by week 6. Wild-type population in the SSIMS-treated cages (orange line) decreases gradually until wild-type females became undetectable by week 11. SSIMS populations in the treatment cages (green line) rise during weekly release periods, but fall rapidly after week 8 when releases cease. Statistical significance: *=p < 0.01, **=p < 0.001, ***=p < 0.0001.
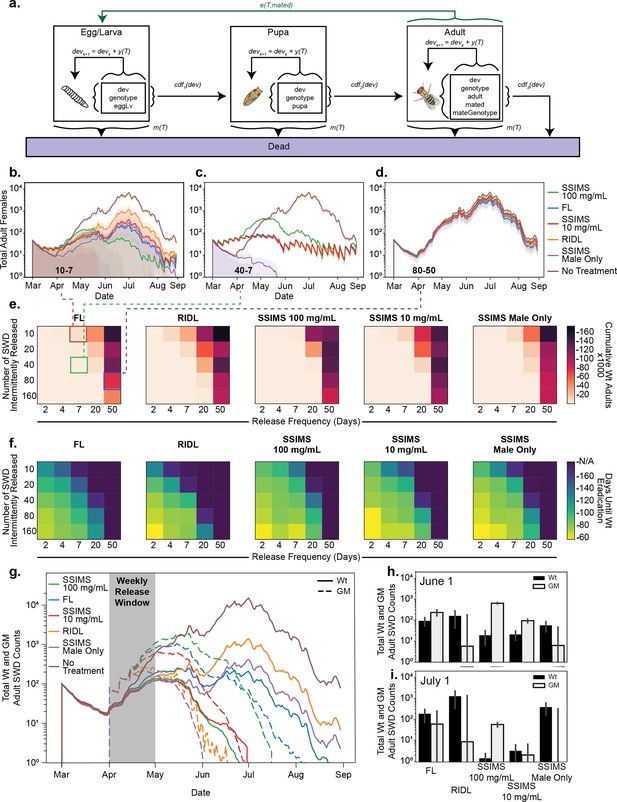
Agent-based modeling.
(a) Schematic of model progression for an individual SWD agent. (b–d) Traces of total adult female populations over a growing season with different biocontrol agents. Shaded area represents one standard deviation among 10 simulations. Numbers inset to bottom left of plots show the insect release numbers and frequency (10–7 for (b), 40–7 for (c), and 80–50 for (d)) and correspond to grid cells identified with colored boxes in (e). (e) Heatmaps representing the cumulative total of WT adults over the course of the season at different release strategies. (f) Heatmaps representing the the average time to Wt eradication. (g) GM agent release was limited to the month of April. Traces of total WT and GM populations were tracked over the season from 10 simulations. (h–i) Total counts of WT and GM populations at June one and July one time points for the April limited release. Error bars show standard deviation of the mean from 10 replicates.
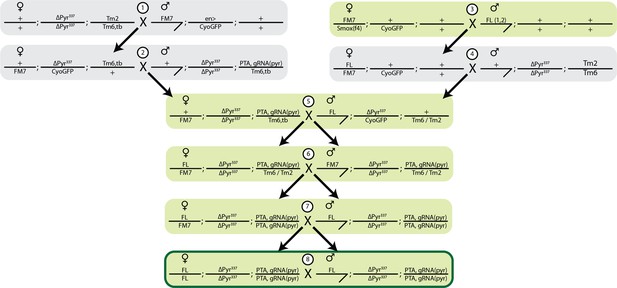
Cross strategy to generate SSIMS line.
Female lethal (FL) and EGI (pyr337; PTA, gRNA-pyr) were crossed to balancer chromosomes, and subsequent males and females were isolated indicated by the black arrow. FL chromosomes contains 2 insertions on the X chromosomes, homozygous females contain 4 copies of the transgene. PTA+gRNA-pyr is a single insertion site containing foxo driven dead-Cas9::VPR and 2 guide RNAs driven by U6:3 and U6:1 promoters. Green shading indicates crosses done in the presence of 100 g/ml Tet. FL was tracked by GFP, PTA+gRNA was tracked by red eyes (mini white), and the pyr337 promoter mutation was tracked using balancers.
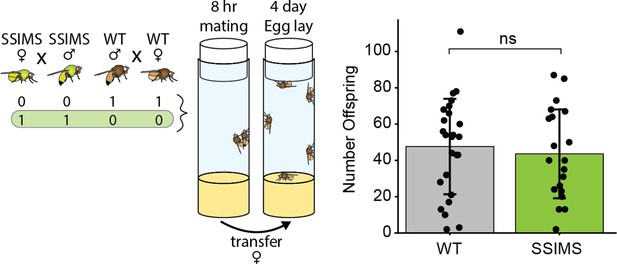
Fecundity of SSIMS flies.
Experimental design schema and results of fecundity assay. Bar graphs show the number of offspring produced by one WT male crossed to one WT female (gray, n = 25) or with one SSIMS male crossed to one SSIMS female (green, n = 20). Welch two-sample t test was performed between the two groups. ns = not significant.
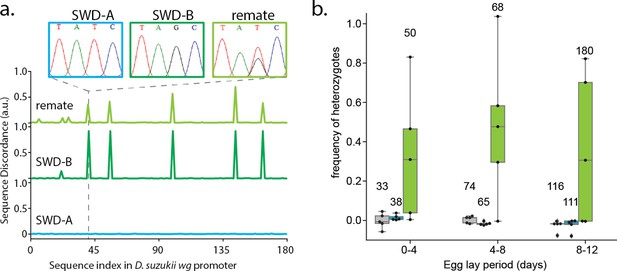
Polyandry in Spotted Wind Drosophila.
(a) Representative data highlighting the calculation of sequence discordance when compared to the wg promoter sequence of inbred strain SWD-A (blue). Inset Sanger sequence chromatographs highlight the T41G SNP in inbred strain SWD-B (dark green). The remating trace (light green) is representative of many samples from (b). (b) Heterozygote frequency when non-virgin SWD-A flies were co-housed with no males (grey), SWD-A males (blue), or SWD-B males (light green). Numbers above box-whisker plots show total number of offspring collected from each experimental group. A one-tailed Welch’s T-test of significance was performed for each egg-lay period comparing+ SWD B male vs+ SWD A male groups (n = 5 per group), and each produced a p-value between.01 and.05.
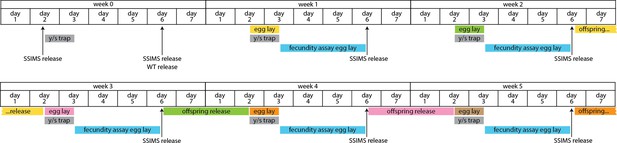
Extended timeline of cage trial design.
In this multi-week view of the cage trial data, the timing between egg lay and offspring release is shown by coloring corresponding events. During the 10 days between egg lay and offspring release, the bottles were kept in cages, but were capped with a foam plug to prevent caged flies from entering and becoming stuck in the cornmeal food.
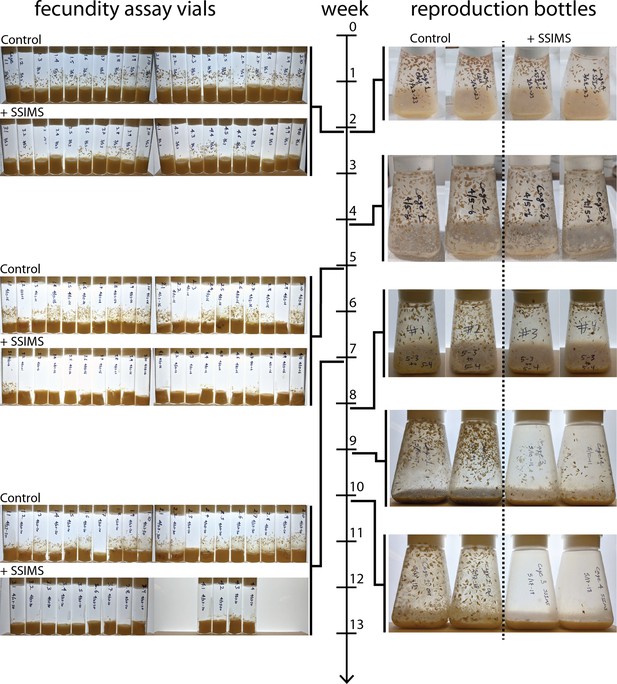
Sample pictures from cage trial.
Timeline of cage trial shown in middle. Jagged lines indicate the week when images were taken. Fecundity assay vials from control cages and SSIMS treated cages to the left of the timeline. Images of the reproduction bottles from the control and SSIMS treated cages to the right of the timeline.
Tables
Key parameters in SWD model.
| Parameter | Value | Reference |
|---|---|---|
| Maximum Egg/Female | 150 | Tochen |
| Egg/Larval Degree Days - mean | 140.785 | Asplen |
| Egg/Larval Degree Days - SD | 20.49 | Asplen |
| Pupal Degree Days - mean | 93.22 | Asplen |
| Pupal Degree Days - SD | 6.18 | Asplen |
| Adult Degree Days - mean | 1,050 | Asplen |
| Adult Degree Days - SD | 40.41 | Asplen |
Key to genotype nomenclature in the model.
| Allele | Definition |
|---|---|
| b | wildtype promoter for EGI target locus |
| B | resistant promoter for EGI target locus |
| d | wildtype allele at location of PTA integration |
| D | PTA targetting wildtype promoter b |
| p | wildtype promoter for EGI target locus |
| P | resistant promoter for EGI target locus |
| t | wildtype promoter for EGI target locus |
| T | PTA targetting wildtype promoter p |
| l | wildtype locus at the site of Female Lethal transgene integration |
| L | Female Lethality transgenic cassette |
| W | wildtype allele at locus of Gene Drive integration |
| G | Homing gene-drive allele |
| R | Gene Drive resistance allele with functional target locus |
| I | Gene Drive resistance allele that inactivates target locus |
| X | female sex chromosome |
| Y | male sex chromosome |
| q | promoter conversion of p (which behaves like P but is tracked independently) |
| r | natural resistance mutation in p that provides protection to T |
| z | promoter conversion of b (which behaves like B but is tracked independently) |
| c | natural resistance mutation in b that provides protection to D |
Genotypes of wildtype or biocontrol agents released in simulations described in this study.
| Genotype in model | Definition |
|---|---|
| bbddppttllWWXX | wildtype female |
| bbddppttllWWXY | wildtype male |
| bbddPPTTllWWXX | EGI female |
| bbddPPTTllWWXY | EGI male |
| bbddppttLLWWXX | female-lethal female |
| bbddppttLLWWXY | female-lethal male |
| bbddppttLLWWXX* | RIDL female |
| bbddppttLLWWXY* | RIDL male |
| bbddPPTTLLWWXX | SSIMS female |
| bbddPPTTLLWWXY | SSIMS male |
-
*
Removed "and self.sex==’female’" from line 236





