Regulatory dissection of the severe COVID-19 risk locus introgressed by Neanderthals
Figures
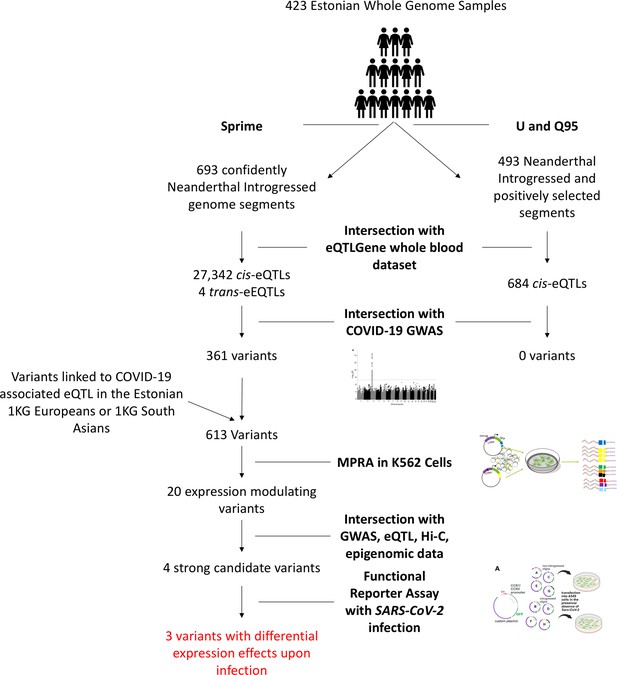
Overview of experimental workflow from whole genome scans for Neanderthal introgression to variant section for the MPRA and SARS-CoV-2 infection reporter assay experiments.
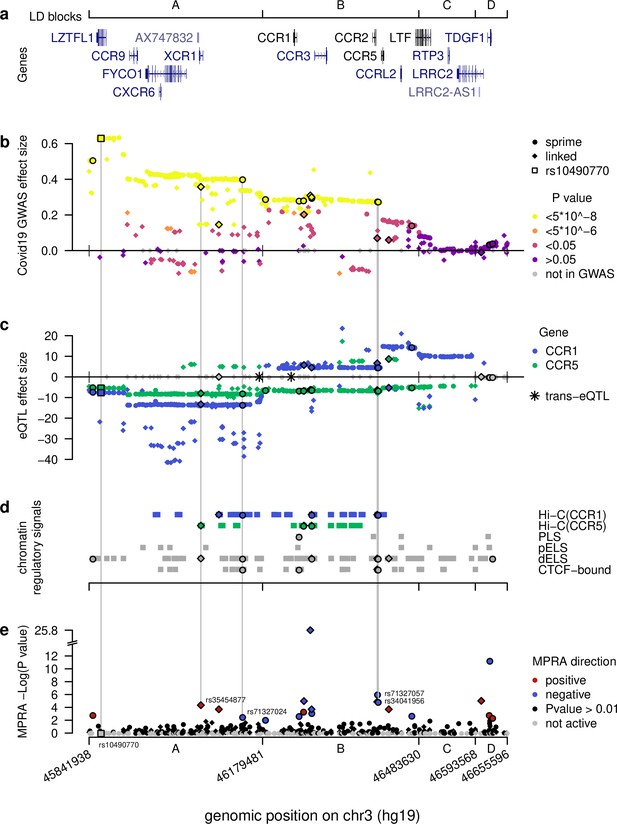
Computational intersections between MPRA emVars and functional genomics datasets across the severe COVID-19 risk locus.
(a) Gene locations across the locus along with boundaries of the four LD blocks (A–D), borders extended to encompass all SNPs in LD (r2 > 0.3) tested with MPRA. (b) Severe COVID-19 GWAS effect sizes from release 5 of the COVID-19 Host Initiative dataset (2021), with strongest genome-wide p-values in yellow spanning the A and B blocks. See key for other color definitions. Dots and diamonds across the panels indicate respectively SNPs identified directly by Sprime (dots) and SNPs in linkage disequilibrium (r2 >0.3) with them (diamonds). (c) eQTL effect sizes across the locus (blue for CCR1, green for CCR5) in whole blood from eQTLGen (Võsa et al., 2018) across the locus. Note the strong down- versus up-regulation of CCR1 for variants in the A versus B blocks, respectively. Grey SNPs are not eQTLs for any of the two genes or were absent from the eQTL study. Asterisks denote trans-eQTLs. (d) Chromatin-based functional annotations across the locus consisting of Hi-C contacts with CCR1 and CCR5 in Spleen, Thymus, or LCL (Jung et al., 2019) and candidate cis-regulatory elements from Moore et al., 2020. (e) -Log p-values for emVars identified using MPRA across the locus. Grey SNPs failed the test for activity in either the archaic or non-archaic form. Vertical lines connect the four putative causal emVars and the most cited tag SNP rs10490770 to functional genomics and genetics data. The four putatively causal variants are unique in having significant hits across all functional genomics and genetics tests.
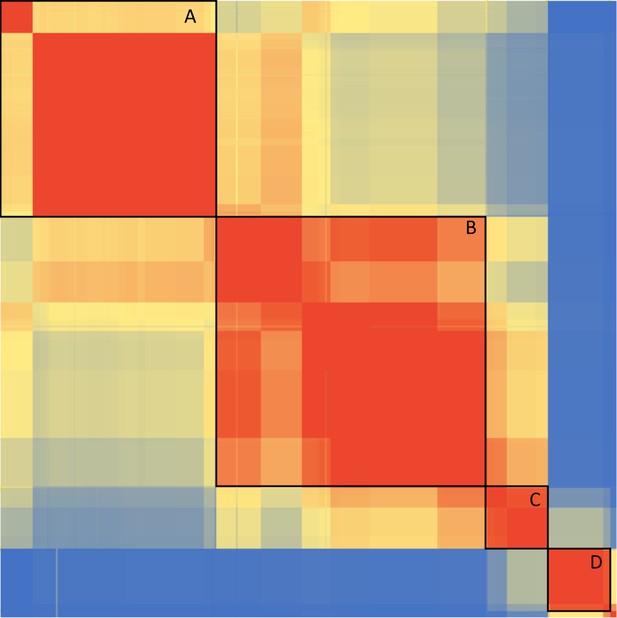
Linkage Disequilibrium (LD) between Sprime identified introgressed variants within the segment (chr3:45,843,242–46,654,616) containing the COVID-19 associated haplotype.
This heatmap shows the pairwise r2 LD between each one of the 361 Sprime identified variants (rows) against every other one of the variants (columns). The color scale is a gradient between blue for r2=0 and red for r2=1. Black boxes indicate the 4 major LD blocks (min r2=0.34) with labels A-D denoting our naming scheme for these blocks. Labeled SNPs represent the boundaries between LD blocks.
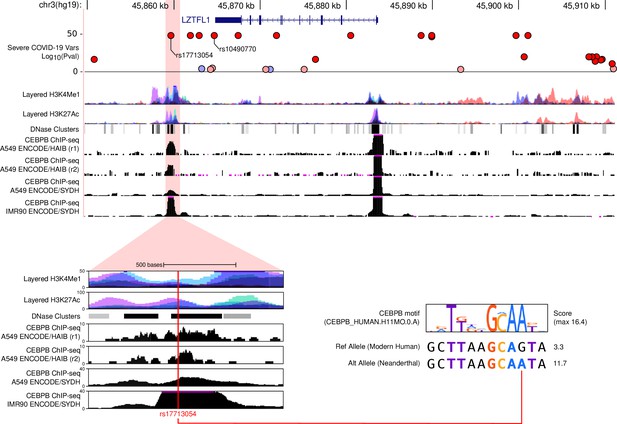
On the top panel we show UCSC Browser tracks for a 61 k region (chr3:45,849,651–45,911,089, hg19) encompassing the leading SNPs on the 3p21.31 COVID-19-risk-associated locus.
From top to bottom we show ‘Severe COVID vars’' from ‘COVID GWAS v4’ track, H3K4Me1, H3K27Ac and DNase Clusters ‘ENCODE Regulation’ tracks, and four ‘ENCODE Transcription Factor Binding’ tracks displaying raw ChIP-seq signal for CEBPB. Highlighted in red and enlarged in the bottom-left panel is a regulatory region bound by CEBPB overlapping rs17713054. The red line indicates the position of rs17713054 within the region. On the bottom-right panel, the Neanderthal allele at rs17713054 generates a predicted CEBPB binding motif, possibly affecting the regulation of nearby genes.
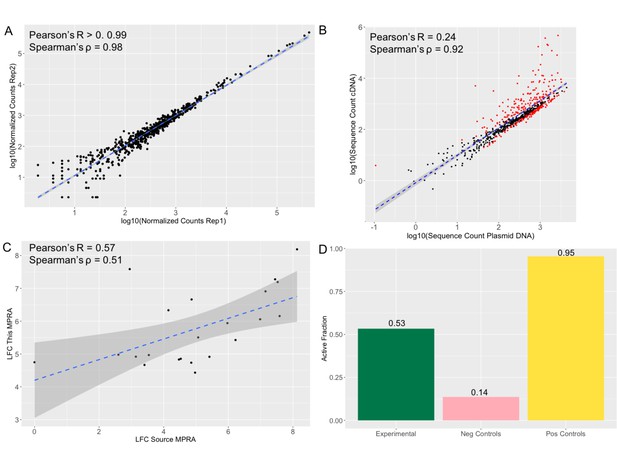
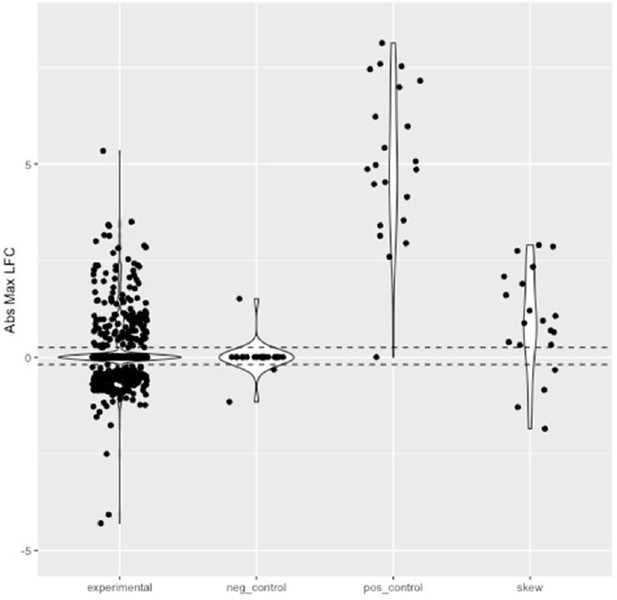
MPRA results show reproducibility and accuracy.
(a) Log normalized counts for each tested sequence in replicate 1 compared with the replicate 2 of the MPRA. Pearson’s R and Spearman’s ρ are extremely high and significant across pairwise replicate comparisons of all four replicates (R>0.99 p-value <2.2 *10–16; ρ=0.98 p-value <2.2 *10–16). (b) Log normalized sequence counts for each tested in the plasmid DNA averaged across the four replicates compared with log normalized average sequence counts in the cDNA averaged across the four replicates. As with other MPRA studies (Tewhey et al., 2016; Uebbing et al., 2021), there is a significant correlation but the plasmid counts do not fully explain the cDNA counts (Pearson’s R=0.24 p-value <2.2 *10–16; Spearman’s ρ=0.92 p-value <2.2 *10–16), suggesting that some of the sequences have an effect on transcription. Sequences determined to be significantly active in the MPRA (methods) are colored in red, non-significant points are black. (c) Activity log fold change (LFC cDNA:pDNA) of positive control sequences in the source MPRA (Jagoda et al., 2021) and in this MPRA. The significant correlation (Pearson’s R=0.57 p-value = 0.006; Spearman’s ρ=0.51 p-value 0.016) suggests that the activity results in this MPRA are accurate. (d) Fraction of sequences tested showing significant activity (LFC cDNA:pDNA corrected p-value >0.01). 95% of positive control sequences tested and 0.14% of negative control sequences tested show activity once again suggesting accuracy in the MPRA results. 53% of experimental sequences show significant activity.
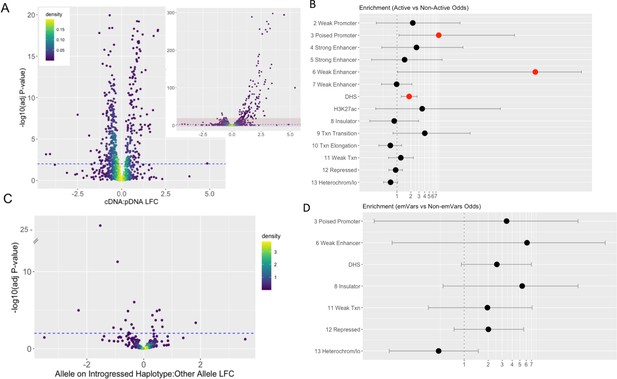
Properties of MPRA-identified active sequences and expression modulating variants.
(a) Log Fold Change of the cDNA count compared to the plasmid DNA for each sequence and the -log10 associated multiple hypothesis corrected p-value. Active sequences are those with a corrected p-value <0.01; this threshold is denoted with a blue dashed line. The larger plot has a y-axis limit of 20; the inset on the right shows the full spread of the data with the light red shaded box denoting the area shown in the larger figure to the left. (b,d) Enrichment of active sequences in K562 genomic features relative to non-active sequences (b) or sequences with emVars relative to sequences without emVars (d). Genomic features indicated with a number represent chromatin states in K562 cells as defined by Ernst and Kellis, 2017. DHS and H3K27ac derive from ENCODE (Butler and Hirano, 2014). Enrichments are reported as Fisher’s odds ratios with lines indicating confidence intervals. Significant enrichments (p<0.05) are colored in red. Missing chromatin states had no overlap with either active sequences (b) or those containing emVars (d). (c) Log Fold Change between active sequences with the allele on the introgressed haplotype compared with the sequence containing the other allele. Expression modulating variants (emVars) are those whose LFC for this measure is significant with a corrected p-value <0.01; this threshold is denoted with a blue dashed line.
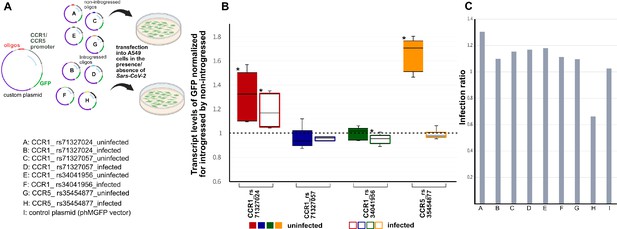
Activity of four emVars in SARS-CoV-2 infected and uninfected A549-ACE2 cells using real-time PCR (qRTPCR).
(a, top) Diagram of construct design and experimental transfection setup. (a, bottom) Constructs tested using in vitro transfection in A549-ACE2 cells whose results are shown in (b) in color-coded fashion. (b) Box-whisker plot graph depicting the transcript level of GFP expression driven by the introgressed allele normalized by the non-introgressed allele for each emVar in mock infection (i.e., absense of SARS-CoV-2) (left, solid color boxes) vs SARS-CoV-2 infected (right, empty color boxes) cells. The ‘*’ depicts significant changes using Wilcoxon statistical test that had a p-value of <0.05 (n=3 per condition). (c) Bar plots showing the infection ratio for each construct.
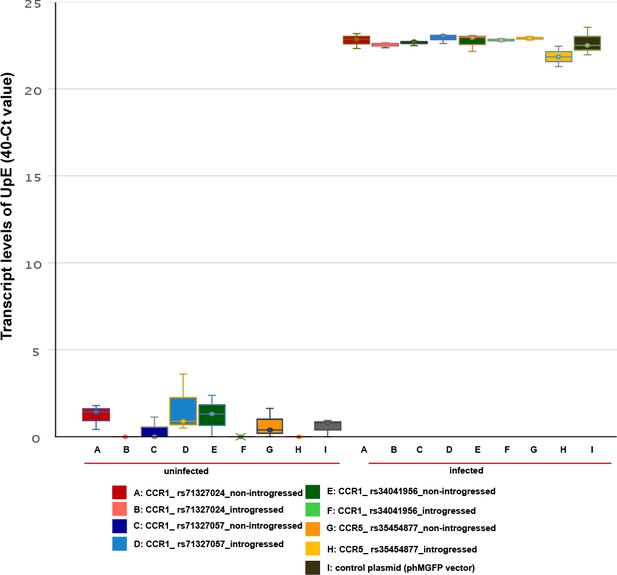
Transcript levels of experimental and control plasmids in the presence and absence of SARS-CoV-2 infection.
Box plot showing the UpE transcript levels (40-Ct values) for the eight custom plasmids (A–H) and the control plasmid (I) for transfections with and without SARS-CoV-2, where increased 40-Ct values indicates higher expression of UpE (n=3 per condition).
Tables
Properties of emVars and prioritization.
This table shows a summary of all the functional data we obtained on the 20 significant emVars identified by the MPRA which we used for prioritization of which these 20 emVars show the most evidence for contributing to the severe COVID-19 phenotype. Specifically, we looked for (1) concordance between the eQTL data (Columns H,I) and the Hi-C Data (K) - specifically for emVars that are eQTLs for a COVID-19 relevant gene with which they also physically interact in an immune tissue; (2) a significant association between the allele and severe COVID-19 in the GWAS data (column J); (3) overlap between the emVar and an ENCODE annotated cCRE (column L) with support in at least one ‘class A’ immune tissue (column M). The 4 variants that met all these criteria are highlighted in bold. For visualization of this data see Figure 1.
| snp | chr:pos_hg19 | Sapiens only Allele/ Allele on Introgressed Hap | Sprime (1) / Linked (0) | LD Block | MPRA LFC (cDNA/pDNA) | MPRA P-value | CCR1 eQTL Direction | CCR5 eQTL Direction | -log10(GWASp) | HI C Data (thymus, spleen, LCL) | cCREs | cCREs Class A Immune Tissue |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs17712877 | 3:45848760 | G/C | 1 | A | 0.30 | 1.46E-03 | - | - | NA | x | dELS | none |
| rs35454877 | 3:46059484 | T/C | 0 | A | 0.31 | 4.60E-05 | - | - | 23.6 | FYCO1,CCR5 | dELS | CD14-positive monocyte female donor ENCDO265AAA |
| rs35657218 | 3:46094462 | A/G | 0 | A | 0.64 | 2.01E-04 | 0 | 0 | 8.2 | CCR1,LZTFL1 | x | x |
| rs71327024 | 3:46140073 | G/T | 1 | A | –0.80 | 2.96E-03 | - | - | 24.7 | CCR1 | dELS,CTCF-bound | CD14-positive monocyte female donor ENCDO265AAA, MM.1S, OCI-LY7, GM12878, PC-9, fibroblast of dermis, B cell adult |
| rs13083881 | 3:46184620 | C/G | 1 | B | –0.53 | 8.20E-03 | 0 | - | 10.3 | x | x | x |
| rs34919616 | 3:46250008 | G/A | 1 | B | –0.36 | 2.11E-03 | 0 | - | 9.5 | x | PLS,CTCF-bound | CD14-positive monocyte female donor ENCDO265AAA, MM.1S, GM12878 |
| rs35617677 | 3:46258740 | C/T | 1 | B | 1.83 | 4.38E-04 | 0 | - | NA | CCRL2,CCR5 | x | x |
| rs34985947 | 3:46260008 | C/T | 0 | B | –2.29 | 1.06E-05 | + | - | 6.6 | CCRL2,CCR5,CCR2 | x | x |
| rs1542755 | 3:46272440 | G/T | 0 | B | –1.53 | 1.68E-26 | 0 | - | 10.8 | CCR2 | x | x |
| rs13096905 | 3:46274766 | G/A | 1 | B | –0.43 | 7.23E-04 | 0 | - | 9.3 | CCR5,CCR1,CCRL2,CCR2,CCR2 | dELS | none |
| rs10510750 | 3:46274886 | T/C | 0 | B | –0.64 | 2.01E-04 | + | - | 9.5 | CCR5,CCR1,CCRL2,CCR2 | dELS | none |
| rs762789 | 3:46402627 | G/A | 0 | B | 0.46 | 1.34E-05 | + | - | 1.6 | CXCR6,CCR1 | dELS,CTCF-bound | CD14-positive monocyte female donor ENCDO265AAA |
| rs71327057 | 3:46402645 | A/C | 1 | B | –0.33 | 9.19E-07 | + | - | 7.9 | CXCR6,CCR1 | dELS,CTCF-bound | CD14-positive monocyte female donor ENCDO265AAA |
| rs34041956 | 3:46402734 | G/A | 1 | B | –0.55 | 1.34E-05 | + | - | 7.8 | CXCR6,CCR1 | dELS,CTCF-bound | CD14-positive monocyte female donor ENCDO265AAA |
| rs6777875 | 3:46425092 | G/T | 0 | B | 0.42 | 2.01E-04 | - | + | NA | x | dELS | B cell adult, MM.1S, OCI-LY7 |
| rs34245951 | 3:46469384 | T/C | 1 | B | –1.06 | 1.88E-03 | + | - | 2.7 | LZTFL1 | x | x |
| rs73072002 | 3:46605033 | C/T | 0 | D | 0.55 | 9.64E-06 | 0 | 0 | 0.1 | ALS2CL,LTF | x | x |
| rs55649212 | 3:46620020 | A/G | 1 | D | 0.66 | 1.46E-03 | 0 | 0 | 0.3 | x | x | x |
| rs2293021 | 3:46621117 | A/G | 1 | D | –0.92 | 5.35E-12 | 0 | 0 | 0.3 | x | x | x |
| rs73073752 | 3:46626599 | C/T | 1 | D | 0.35 | 4.07E-03 | 0 | 0 | 0.3 | x | dELS | none |
Additional files
-
Supplementary file 1
This is an excel file containing supplementary data in the form of 13 tables/sheets called (a-m).
(a) presents the African samples used as outgroup for our Sprime introgression scan. (b) presents the full results from our Sprime scan. (c) presents Neanderthal matching Sprime segments. (d) presents results from our U and Q95 scan. (e) presents cis-eQTLs from our U and Q95 Scan. (f) presents cis-eQTLs from our Sprime scan. (g) presents trans-eQTLs from our Sprime scan. (h) presents expression of genes responsive to cis-eQTLs within the introgressed region in RNA-seq studies of infection by SARS-CoV-2 and related viruses. (i) presents expression of genes responsive to trans-eQTLs within the introgressed region in RNA-seq studies of infection by SARS-CoV-2 and related viruses. (j) presents data on all MPRA tested variants. (l) presents data from our ChIP-seq analyses. (l) presents our analyses on ChIP-seq data and predicted transcription factor binding sites (TFBS). (m) presents our construct sequences used in transfection studies.
- https://cdn.elifesciences.org/articles/71235/elife-71235-supp1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/71235/elife-71235-transrepform1-v1.docx