Crosstalk with keratinocytes causes GNAQ oncogene specificity in melanoma
Figures
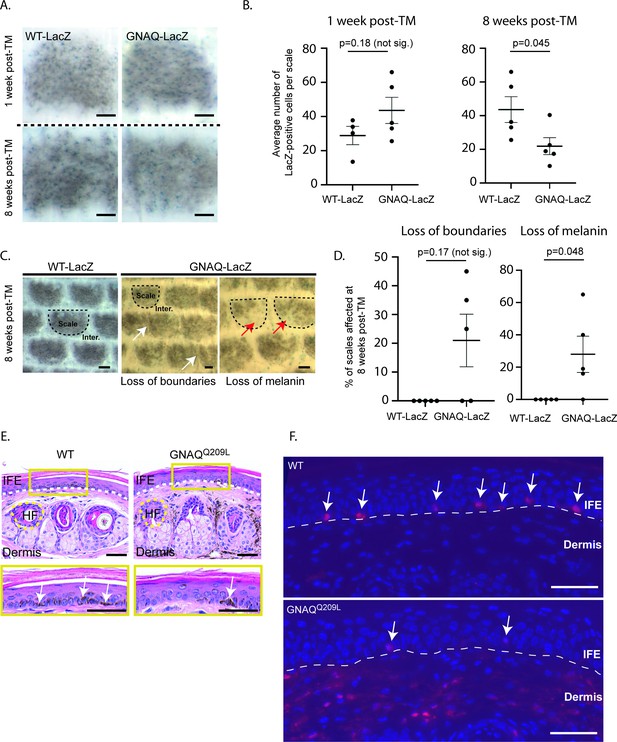
Forced GNAQQ209L signaling reduces the number of melanocytes in the interfollicular epidermis (IFE).
(A) Representative example of LacZ-positive cells within scales of WT-LacZ and GNAQ -LacZ tails at 1 or 8 weeks post-tamoxifen (TM) treatment. (B) Quantification of the average number of LacZ-positive cells per scale at 1 and 8 weeks post-TM treatment. (Each point represents average for one mouse; mean ± SEM; unpaired t-test.) (C) Representative examples of X-gal stained whole mount epidermal tail sheets in WT-LacZ and GNAQ-LacZ mice at 8 weeks post-TM, showing loss of boundaries in scale pigmentation (white arrows) or loss of melanin (red arrows) in GNAQ-LacZ mice. Example scales are outlined in dashed line for reference. (D) Percentage of epidermal scales exhibiting loss of boundaries or loss of melanin in five WT-LacZ and five GNAQ-LacZ mice at 8 weeks post-TM. (Each point represents % for one mouse; Kolmogorov-Smirnov test, mean ± SEM.) (E) H&E stained cross sections of tail skin in wildtype (WT) and GNAQQ209L mice. The yellow box below shows a magnified area of the interfollicular epidermis (IFE). Less melanin was observed in the IFE of GNAQQ209L skin (white arrows point to examples). Dashed lines indicate the boundaries between the IFE, dermis, and an example hair follicle (HF). (F) tdTomato expression (red) in cross sections of tail skin of WT and GNAQQ209L mice at 4 weeks of age showing a reduced number of melanocytes (tdTomato+ cells) in the IFE of GNAQQ209L mice and an abnormal expansion of melanocytes in the dermis. Sections are counterstained with DAPI (blue). Dashed lines indicate the boundaries between the IFE and dermis. White arrows indicate melanocytes located in the IFE. In A and C, scale bars represent 100 µm, while in E and F, scale bars represent 50 μm.
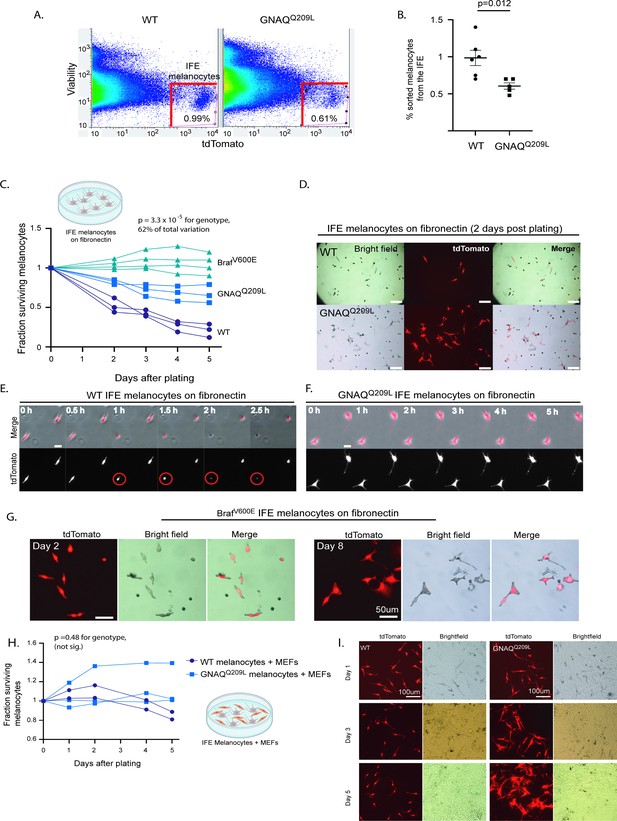
Fluorescent activated cell sorting (FACS) sorted GNAQQ209L and BRAFV600E interfollicular epidermis (IFE) melanocytes cultured on fibronectin have increased survival compared to wildtype (WT).
(A) Representative examples of FACS dot plots of single-cell suspension from the IFE epidermis of WT and GNAQQ209L mice. (B) Average percentage of tdTomato+ cells sorted from the IFE of WT and GNAQQ209L mice, with a significantly smaller percentage in GNAQQ209L. (Each point represents % from one mouse, mean ± SEM; unpaired t test). (C) Fraction of surviving WT, GNAQQ209L, and BrafV600E FACS sorted melanocytes plated on fibronectin (each line represents an independently derived primary culture, two-way ANOVA). (D) Representative images of melanocyte morphology 2 days post-plating on fibronectin-coated wells, showing increased dendrite formation in GNAQQ209L melanocytes. (E) Time lapse microscopy of two WT melanocytes plated on fibronectin. The circled cell adopted a round shape shortly before being lost from view. (F) Time lapse microscopy of two GNAQQ209L melanocytes showing a dendritic cell morphology that remained stable over time. (G) Representative images of BRAFV600E melanocytes at 2 and 8 days post-plating on fibronectin. Scale bars represent 100 μm in D, 20 μm in E and F, and 50 μm in G. (H) Fraction of surviving WT and GNAQQ209L IFE melanocytes co-cultured with mouse embryonic fibroblasts (MEFs), showing initial growth above the baseline for both genotypes before loss began (each line represents an independently derived primary culture, two-way ANOVA). (I) Representative images of FACS sorted WT and GNAQQ209L IFE melanocytes co-cultured with MEFs. While the WT and GNAQQ209L IFE melanocytes had a similar spindle cell morphology at day 1, the GNAQQ209L melanocytes progressively developed large and abnormal shapes.
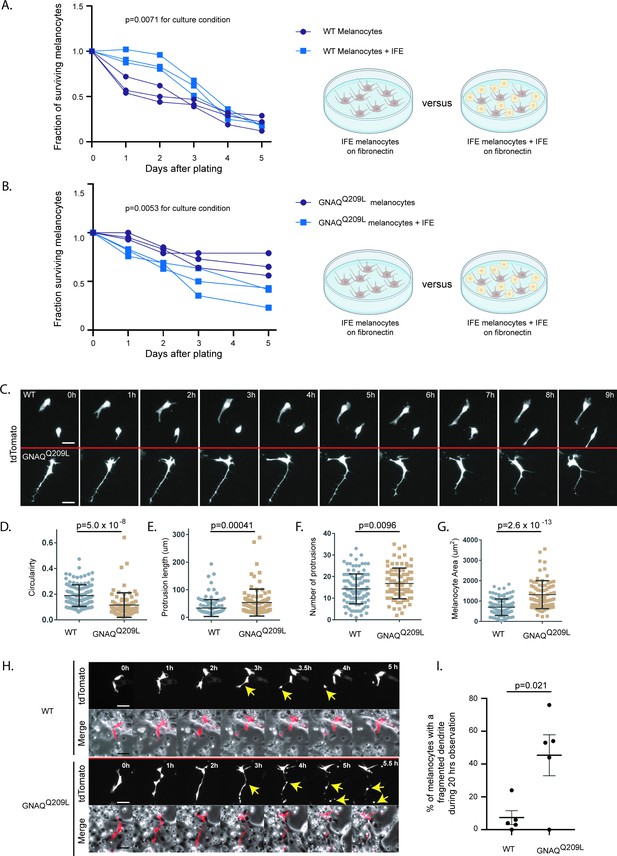
The interfollicular epidermis (IFE) impairs survival and alters pseudopod dynamics in GNAQQ209L melanocytes.
(A) Survival of unsorted wildtype (WT) melanocytes plated with its IFE, compared to sorted IFE WT melanocytes plated onto fibronectin. The presence of IFE increased the survival of WT melanocytes. (Each line represents one independently derived culture, two-way ANOVA.) (B) Survival of unsorted GNAQQ209L melanocytes plated with its IFE, compared to sorted IFE GNAQQ209L melanocytes plated onto fibronectin. The presence of IFE decreased the survival of GNAQQ209L melanocytes. (Each line represents one independently derived culture, two-way ANOVA.) (C) Time lapse images show representative WT and GNAQQ209L melanocytes co-cultured with IFE between 0 and 8 hr. The GNAQQ209L cell exhibits abnormally long dendrites and a less circular (more polygonal) cell body shape. (D–G) Quantification of circularity (D), protrusion length (E), number of protrusions (F), and melanocyte area (G), in WT and GNAQQ209L melanocytes co-cultured with IFE. (Each point represents the measurement of one cell, mean ± SEM; unpaired t test.) (H) Time lapse microscopy showing a representative example of dendrite fragmentation in WT and GNAQQ209L melanocytes. Arrows indicate dendrite breakage points and subsequent fragments that form up into balls. (I) Quantification of the percent of cells experiencing dendrite fragmentation in WT and GNAQQ209L melanocytes cultured with IFE. (Each point represents the measurement from one culture, mean ± SEM; unpaired t test.) Scale bars represent 40 μm in C and H.
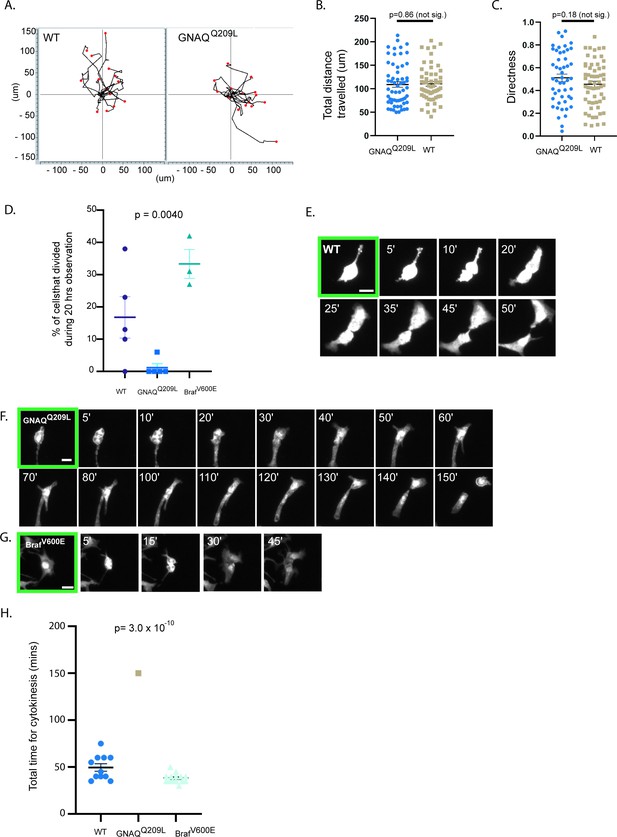
The interfollicular epidermis (IFE) impairs cell division of GNAQQ209L melanocytes.
(A) Migration plots of wildtype (WT) and GNAQQ209L melanocytes co-cultured with IFE over 20 hr. (B) Quantification of total distance traveled in 20 hr. (Each point represents the measurement from one cell, mean ± SEM; unpaired t test.) (C) Quantification of the directness of cell trajectories over 20 hr. Directness = 1 is a straight line cell trajectory. (Each point represents the measurement from one cell, mean ± SEM; unpaired t test.) (D) Percentage of tracked cells undergoing division during 20 hr of time lapse microscopy, when co-cultured with IFE. (Each point represents the measurement from one culture, mean ± SEM; ordinary one-way ANOVA.) (E–G) Representative examples of cell division events from cleavage furrow formation to separation of daughter cells in a WT melanocyte (in E), a GNAQQ209L melanocyte (in F), and a BRAFV600E melanocyte (in G). (H) Quantification of the time taken between cleavage furrow formation to separation into daughter cells in melanocytes co-cultured with IFE. (Each point represents the measurement from one cell, mean ± SEM, ordinary one-way ANOVA.) Scale bar in E–G represents 20 μm.
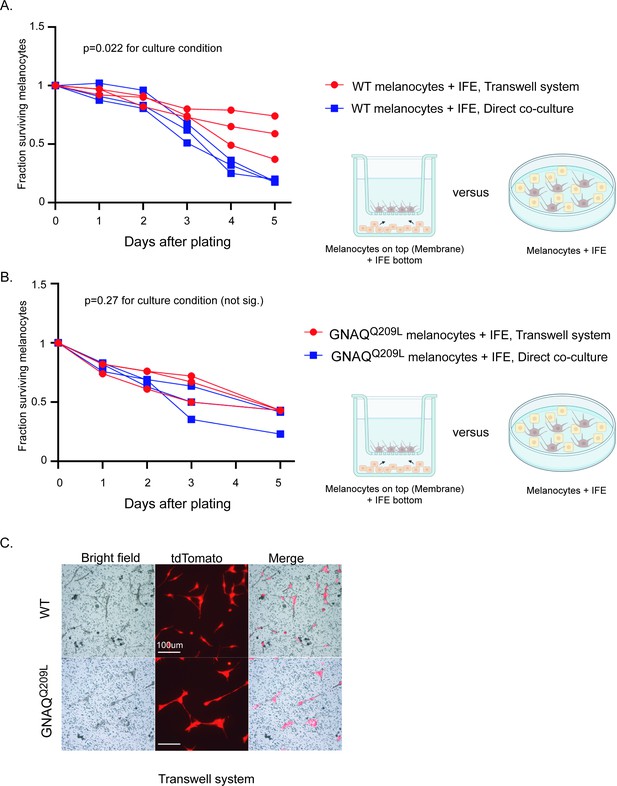
Interfollicular epidermis (IFE) control of melanocytes was maintained in a transwell culture system.
(A) Comparison of survival for wildtype (WT) melanocytes in direct contact with IFE (‘direct co-culture’) versus a transwell system where the two populations are separated by a permeable membrane. In WT melanocytes,there was no significant difference in survival for the first 3 days, after which the transwell melanocytes developed an advantage. (Each line represents one independent culture, mean ± SEM; two-way ANOVA.) (B) Comparison of survival for GNAQQ209L melanocytes as in A. There was no significant difference in survival throughout the culture period. (Each line represents one independent culture, mean ± SEM; two-way ANOVA.) (C) Representative images of WT and GNAQQ209L melanocytes at day 3 on the transwell membrane (no direct cell contact with IFE). GNAQQ209L melanocytes were larger than WT melanocytes, as in direct co-culture. Scale bar represents 100 μm in C.
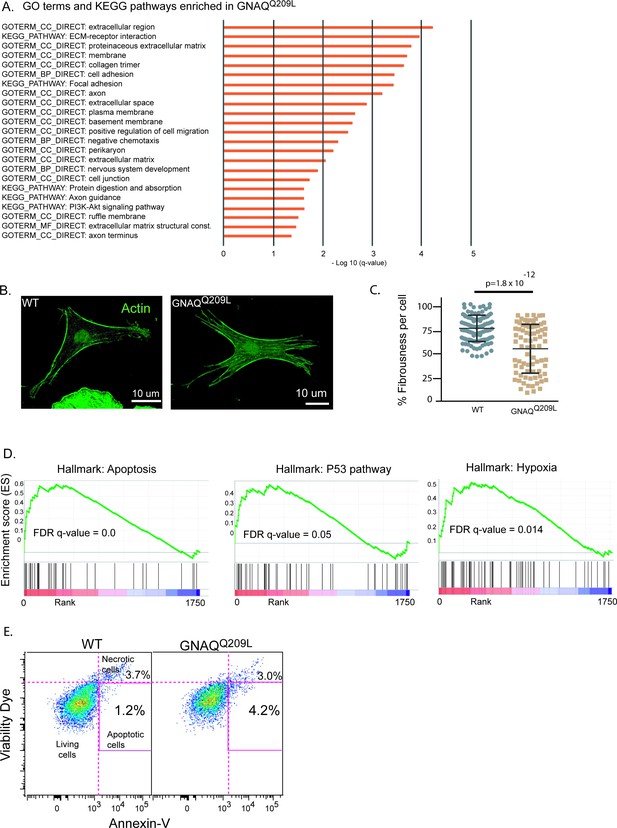
Analysis of GNAQQ209L expressing melanocytes in the interfollicular epidermis (IFE) reveals alterations in the actin cytoskeleton and cell death.
(A) Significant terms identified by Gene Ontology analysis for differentially expressed (DE) genes (log2 fold change [LFC] >2 or <−2) in GNAQQ209L melanocytes. All were enriched in the GNAQQ209L melanocytes (red). (B) Representative examples of phalloidin staining for f-actin in wildtype (WT) and GNAQQ209L melanocytes co-cultured with IFE. The actin is less organized in the GNAQQ209L cell. (C) Reduced fibrousness in GNAQQ209L cells indicates disorganization of the actin cytoskeleton. (Each point represents a measurement from one cell, mean ± SEM; unpaired t test). (D) Gene set enrichment plots for apoptosis, P53 pathway, and hypoxia hallmarks enriched in GNAQQ209L melanocytes. (E) tdTomato+ cells identified by fluorescent activated cell sorting (FACS) from 4-week-old tail IFE and sorted for Annexin-V and viability. N = 2 WT and 2 GNAQQ209L mice, melanocytes pooled. Early apoptotic cells are positive for Annexin-V and negative for the viability dye. Note, the necrotic cells are cells that had a disrupted membrane that allowed entry of the viability dye. They should not be used to assess apoptosis because Annexin-V is able to enter damaged cells and bind phosphatidylserine on the inner leaflet, creating a false positive.
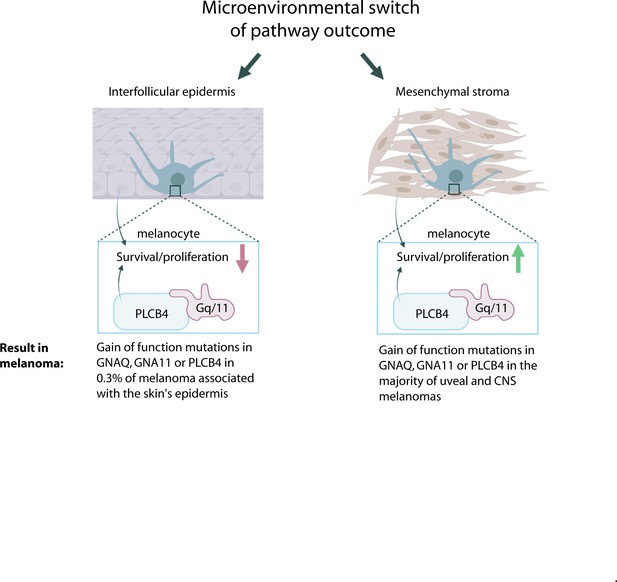
Model for microenvironmental control of the Gαq-PLCB4 signaling outcome and how this leads to GNAQ oncogene specificity in melanoma.
G alpha q (GNAQ) and G alpha 11 (GNA11) are classic signaling components that transmit G protein-coupled receptor (GPCR) activation to phospholipase C-beta (PLC-B) inside the cell. Either GNAQ, GNA11, or PLCB4 is activated by a gain-of-function (GOF) hotspot mutation in the majority of uveal (ocular) and CNS melanomas, but these mutations are almost never found in melanomas arising in the epidermis of the skin. We propose that the reason for this is that paracrine signaling from the epidermis reversibly switches Gαq/11 signaling from promoting growth to inhibiting melanocyte survival and proliferation. GNAQQ209L, in combination with signaling from the interfollicular epidermis (IFE), stimulates dendrite extension, leads to actin cytoskeleton disorganization, inhibits proliferation, and promotes apoptosis in melanocytes.
Videos
Wildtype (WT) melanocytes co-cultured with interfollicular epidermis (IFE), 625 min time lapse.
Cell protrusions, cell movements, and cell division events can be seen in the melanocytes in this video. Cell protrusions are shorter in WT cells than in GNAQQ209L. One of the cell division events in this video occurs at around 9 s in a cell near the center of the field.
GNAQQ209L melanocytes co-cultured with interfollicular epidermis (IFE), 625 min time lapse.
Cell protrusions and cell movements can be seen in the melanocytes in this video. Protrusions are longer in GNAQQ209L melanocytes than in wildtype (WT) and some cells take on usual shapes. The bottom right cell shows an example of dendrite breakage, with the fragmented piece balling up and drifting away.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain background (Mus musculus domesticus, males and females) | C3HeB/FeJ | Jackson Laboratories | Jackson Labs: strain #000658 | |
| Gene(Homo sapiens) | GNAQ | GenBank | GenBank Gene ID: 2776 | |
| Genetic reagent(Mus musculus domesticus males and females) | Tg(Mitf-cre) 7114Gsb | Alizadeh et al., 2008 | PMID:18353144RRID:MGI:5702900 | |
| Genetic reagent(Mus musculus domesticus males and females) | Tg(Tyr-cre/ ERT2)13Bos/J | Jackson Laboratories | Jackson Labs: strain #012328 | |
| Genetic reagent(Mus musculus domesticus males and females) | Gt(ROSA) 26Sortm1(GNAQ*)Cvrk | Huang et al., 2015 | PMID:26113083RRID:MGI:5702877 | |
| Genetic reagent(Mus musculus domesticus males and females) | Gt(ROSA) 26Sortm14(CAG-tdTomato)Hze | Jackson Laboratories | Jackson Labs: strain #007914 | |
| Genetic reagent(Mus musculus domesticus males and females) | Gt(Rosa) 26Sortm1Sor/J | Jackson Laboratories | Jackson Labs: strain #003474 | |
| Genetic reagent(Mus musculus domesticus males and females) | Braftm1Mmcm | Jackson Laboratories | Jackson Labs: strain #017837 | |
| Commercial assay or kit | Annexin-V- FLUOS Staining Kit | Millipore Sigma | MilliporeSigma SKU:11858777001 | |
| Commercial assay or kit | DNeasy Blood and Tissue kit | Qiagen | Qiagen ID: 69504 | |
| Commercial assay or kit | HotStar Taq DNA polymerase | Qiagen | Qiagen ID: 203207 | |
| Chemical compound, drug | Tamoxifen | Millipore Sigma | MilliporeSigma catalog:T5648 | |
| Chemical compound, drug | 4-Hydroxyt amoxifen | Millipore Sigma | Millipore Sigma catalog:H6278 | |
| Software, algorithm | Custom Matlab scripts | Haage et al., 2018 | PMID:30485809 | https://github.com/Tanentzapf-Lab/ActinOrganization_CellMorphology_Haage |
| Other | eBioscience Fixable Viability Dye eFluor 450 | ThermoFisher | ThermoFisher catalog : 501128817 | |
| Other | RiboLock RNase inhibitor | ThermoFisher | ThermoFisher catalog :EO0381 | |
| Other | Fibronectin | Millipore Sigma | MilliporeSigma catalog:F0895 | Liquid, 0.1% solution |
Additional files
-
Supplementary file 1
Excel file providing more details of the differentially expressed genes in GNAQQ209L melanocytes sorted from the IFE.
(a) Genes with FPKM >0.1 in wildtype (WT) and/or GNAQQ209L melanocytes. (b) Differentially expressed genes down-regulated in GNAQQ209L interfollicular epidermis (IFE) melanocytes, sorted by Z_score. (c) Differentially expressed genes up-regulated in GNAQQ209L IFE melanocytes, sorted by Z_score. (d) Genes supporting pathway analysis terms related to cell adhesion, focal adhesion, and the extracellular matrix. (e) Genes supporting pathway analysis terms related to axon guidance, nervous system development, and axon cellular component.
- https://cdn.elifesciences.org/articles/71825/elife-71825-supp1-v2.xlsx
-
Supplementary file 2
Word file providing more details on gene expression in mouse tail IFE melanocytes, frequency of oncogenic mutations in GNAQ and GNA11 in human cutaneous melanomas, and statistical tests used in the studies.
(a) Top 20 most highly expressed genes in mouse wildtype (WT) interfollicular epidermis (IFE) melanocytes. (b) Identification of GNAQ hotspot mutations among human malignant melanomas potentially arising in the epidermis. (c) Identification of GNA11 hotspot mutations among human malignant melanomas potentially arising in the epidermis. (d) Information on statistical tests in Figures 1—6.
- https://cdn.elifesciences.org/articles/71825/elife-71825-supp2-v2.doc
-
Supplementary file 3
Prism file with source data used to create graphs and calculate statistics in Figures 1—6.
- https://cdn.elifesciences.org/articles/71825/elife-71825-supp3-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/71825/elife-71825-transrepform1-v2.pdf





