Mg2+-dependent conformational equilibria in CorA and an integrated view on transport regulation
Figures
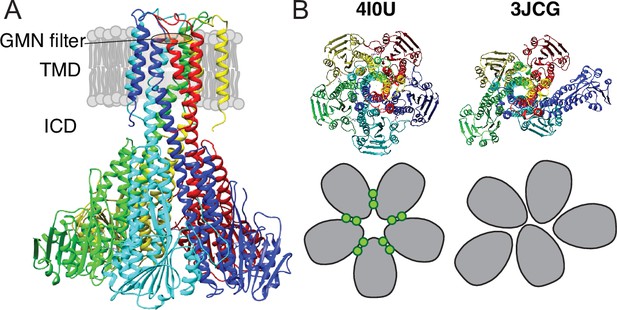
X-ray and cryo-EM structures of CorA.
(A) Side view of symmetric CorA (PDB ID: 4I0U) in presence of Mg2+ (‘closed form’). (B) Top view of the same symmetric state of CorA (PDB ID: 4I0U) side-by-side to one of the asymmetric states observed in the absence of Mg2+ (‘open form’) (PDB ID: 3JCG). A schematic representation of the two forms is shown below their structures, with each monomer shown in gray and Mg2+ ions represented as green circles.
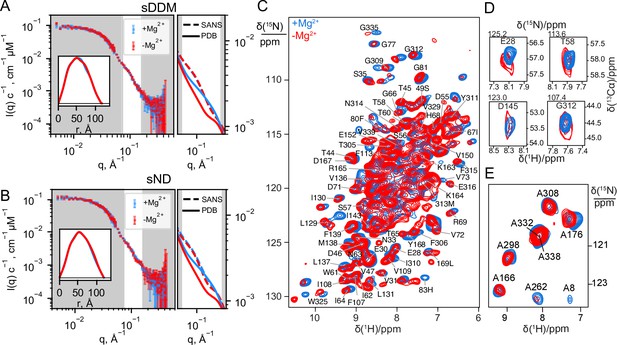
Experimental data on CorA in presence (blue) and absence (red) of Mg2+.
(A + B) Experimental SANS data of CorA embedded in stealth DDM micelles (sDDM) and stealth nanodiscs (sND), respectively, with p(r)-distributions calculated on BayesApp (Hansen, 2014) in the inset. The rightmost plots show zoomed comparisons of the p(r)-fits of experimental data (SANS, dashed lines) with the SANS curves calculated on the X-ray (4I0U) and cryo-EM (3JCG) structures (PDB, full lines). Complete fits based on the PDB structures are shown in Figure 4A. (C) 2D 1H-15N dipolar correlation spectra by MAS NMR of CorA in hydrated DMPC bilayers recorded at 1 GHz 1H Larmor frequency and 107 kHz MAS. (D) 2D sections for a selection of residues obtained from 3D 1H-15N-13Cα spectra recorded at 1 GHz 1H Larmor frequency and 107 kHz MAS. (E) 2D 1H-15N dipolar correlation spectra obtained for 15N-Alanine-labeled CorA recorded at 1 GHz 1H Larmor frequency and 60 kHz MAS. In C and E, site-specific assignments are annotated for resolved resonances.
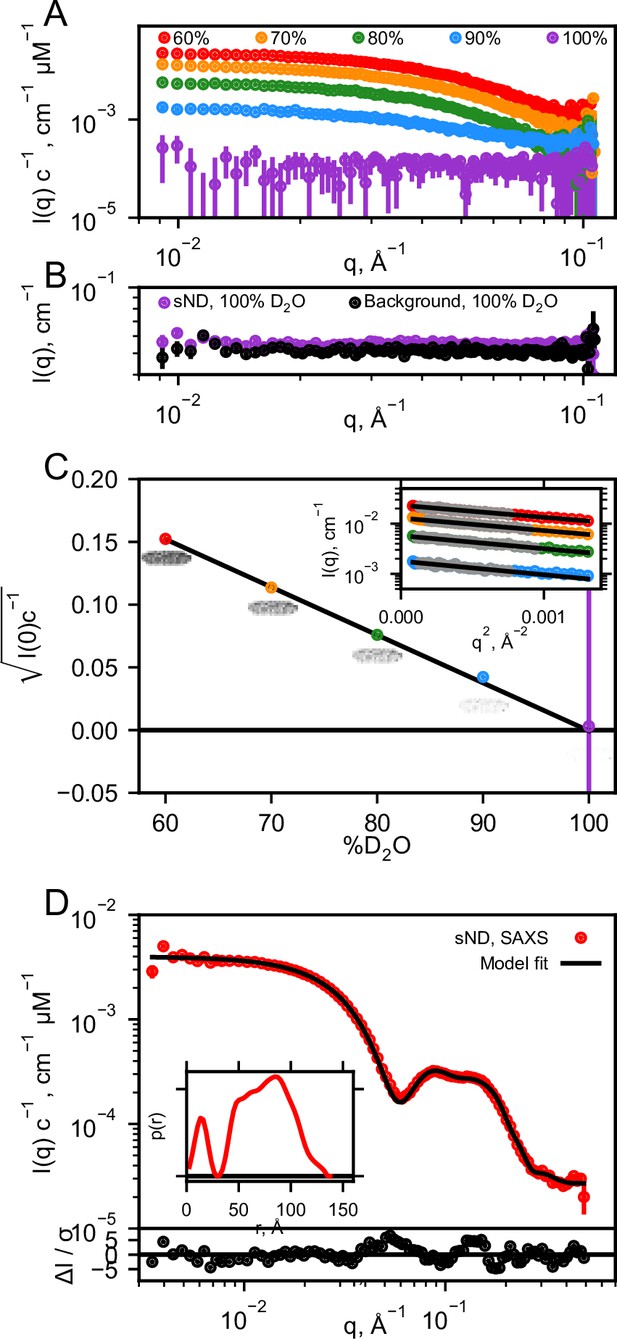
Validation of nanodisc match-out deuteration.
(A) Background subtracted SANS data measured on sNDs in different percentage of D2O. (B) Comparison of unsubtracted SANS data and corresponding background measurement in 100% D2O. (C) Plot of contrast vs D2O to verify match-point of sample (fitted to 99.9% D2O by linear regression). I(0) values were determined using Guinier fits to the low-q data (grey points) as highlighted in the insert. The Guinier fit to the SANS data at 100% D2O has not been shown due to the large errorbars, and the fit was also not included for fitting the contrast vs D2O. (D) SAXS data on sND collected in H2O buffer together with p(r) distribution (insert). A nanodisc model (Skar-Gislinge et al., 2010) was fitted to the data using WillItFit (Pedersen et al., 2013), producing a great fit, as evaluated by the residual plot on the bottom.
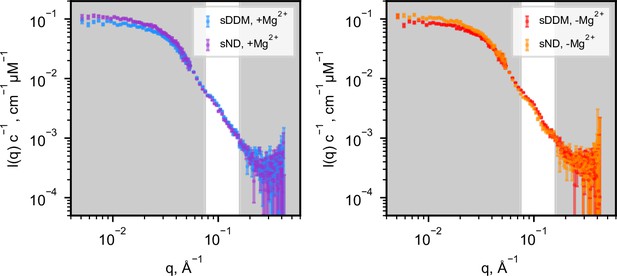
Comparison of SANS data in sDDM and sND.
Same data as shown in Figure 1D–E, but presented here to compare data collected in sDDM vs sND. Despite the slightly higher forward scattering in the sND data, the SANS data from the two different systems overlap in the region of interest, as highlighted. Given that CorA had spent less time and steps in presence of DDM prior to reconstitution in sNDs, the excess signal likely stems from a small number of tightly bound E. coli lipids that do not match out in D2O. By comparing the extrapolated values of I(0) between the sDDM and sND samples, we estimated the number to be on the order of 10 lipids, that is two lipids per protomer. Although the presence of visible lipids would complicate further detailed analysis, the striking identicality of the measured curves with and without Mg2+ in sNDs supports no large structural rearrangements.
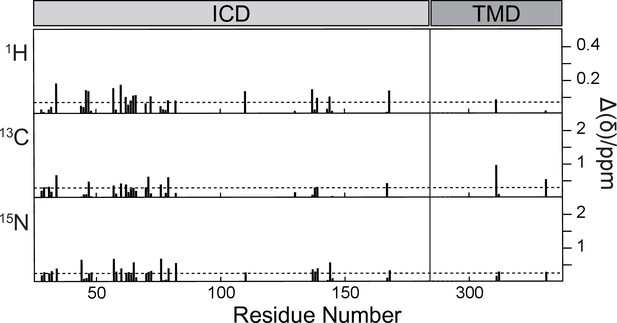
Absolute chemical shift differences in 3D 1H-15N-13Cα spectra of CorA with and without Mg2+, calculated as Δ(δ)= for X=1H,13Cα,15N.
Horizontal dashed lines indicate average values.
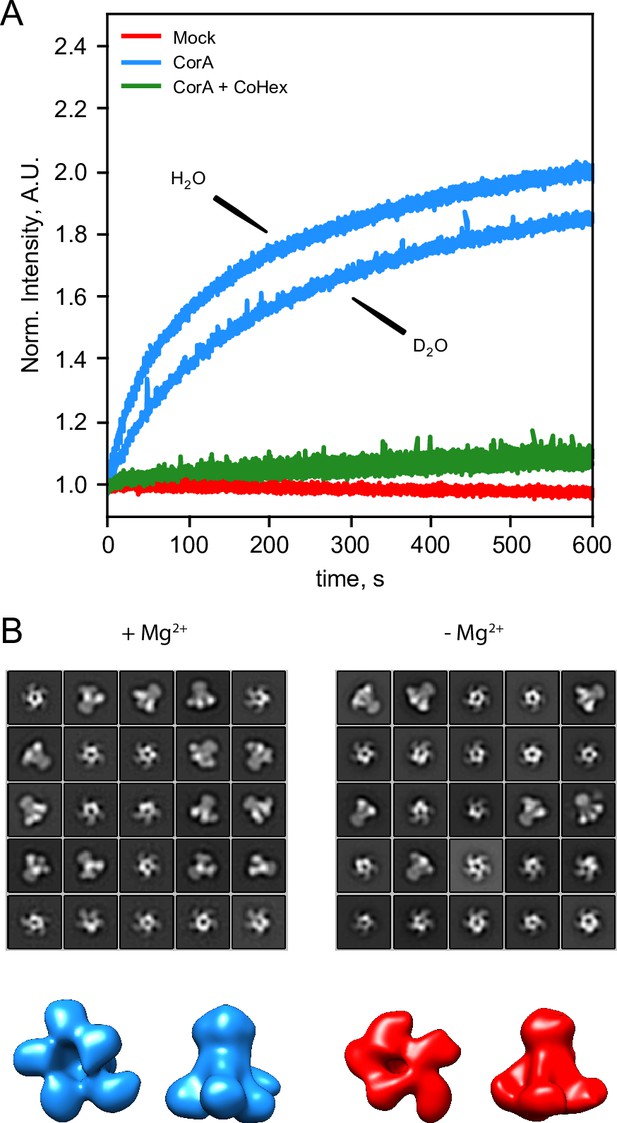
CorA activity in D2O and direct visualization by negative stain EM.
(A) CorA activity in the conditions used for SANS. The traces are the normalized fluorescence signals after adding Mg2+ to either empty POPC liposomes (Mock), CorA-POPC proteoliposomes (CorA), or CorA-POPC proteoliposomes preincubated with the inhibitor Co[NH3]3+ in D2O (CorA+ CoHex). (B) Negative stain EM of CorA in DDM and D2O with and without Mg2+. The 25 most abundant 2D classes are shown for each condition. The box dimensions are 170 × 170 Å2 for scale. (C) Final 3D model for each condition shown from the intracellular side and in side-view, respectively.
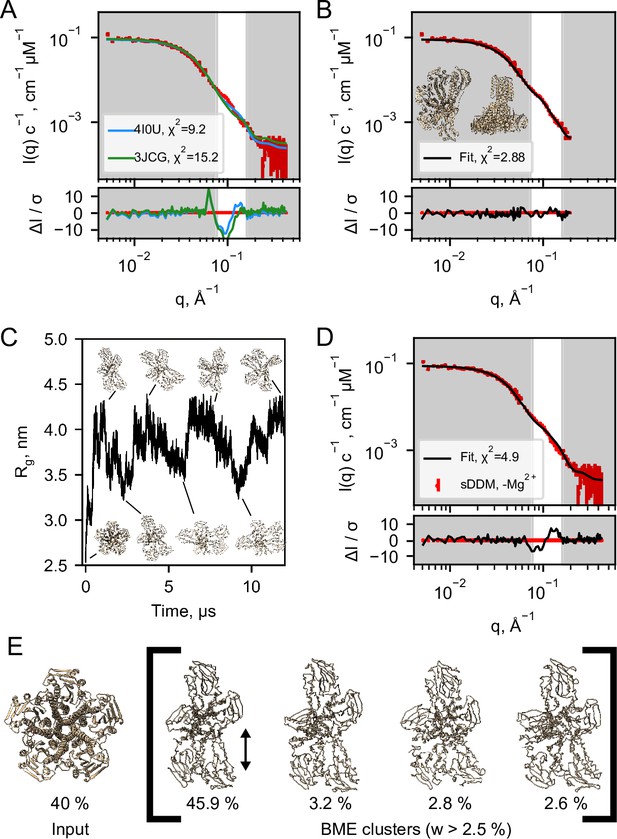
Structural modelling of CorA from SANS data.
(A) Model fits of the closed crystal structure (4I0U) and open cryo-EM structure (3JCG) to the experimental SANS data obtained in sDDM without Mg2+. The bottom panel shows the error-normalised difference plot of the fits. (B) Model fit of the structure obtained by regularised normal mode analysis (bottom and side views inserted on plot). (C) MetaD molecular dynamics trajectory with representative frames visualized as ribbon structures. (D) Fit of a weighted ensemble obtained from the MD simulation in B together with a 40% contribution from the symmetric CorA. (E) The four highest weighted cluster centroids in the metaD simulation (right bracket) together with the closed symmetric structure (left) which together illustrates the ensemble of CorA structures that produced the best fit to the experimental SANS data.
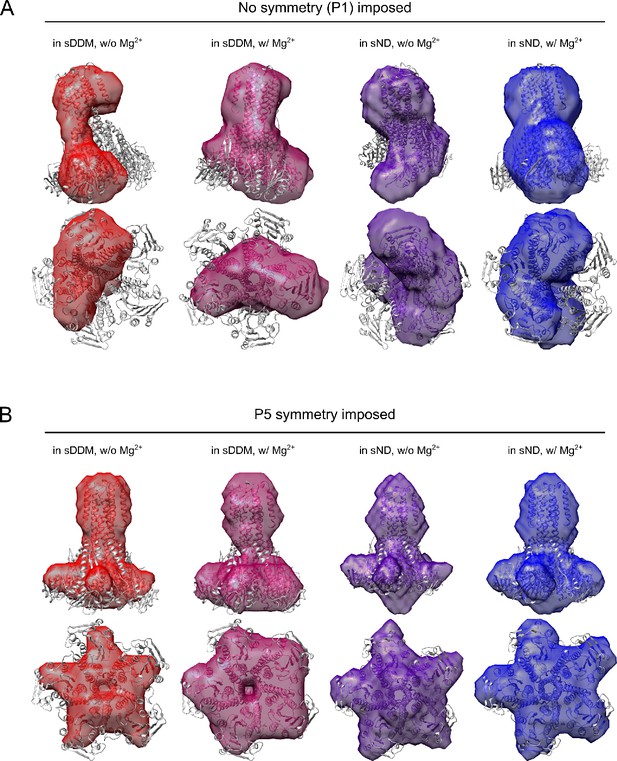
Bead modelling of CorA.
All four SANS data sets were analysed by the DAMMIF pipeline (Franke and Svergun, 2009) available from the ATSAS package (Franke et al., 2017). In total, DAMMIF was run 100 times in slow mode with either P1 (A) or P5 (B) symmetry selected. For P1 symmetry, the normalized spatial discrepancy (NSD) values varied from 1.2 to 1.5, whereas for P5 symmetry, NSD values varied from 1.4 to 1.7. NSD values above one indicate that the calculated models differ systematically from one another.
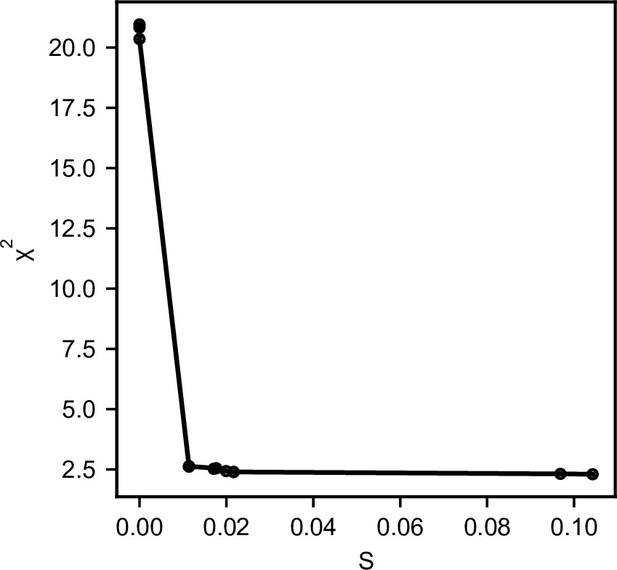
Goodness of fit from NMA generated structures to SANS.
The best compromise between conservation of structure and the best fit to the data is in the ‘elbow’-region, here chosen at S = 0.01 (with α = 600).
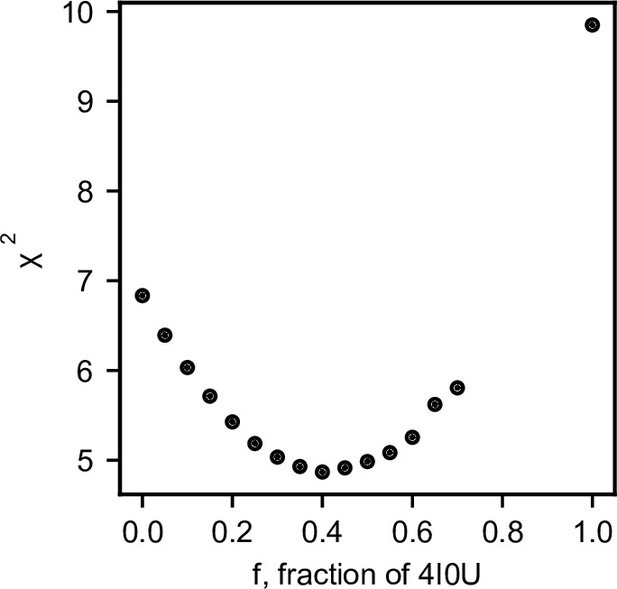
Ensemble fitting to SANS data with symmetric CorA included.
The goodness of fit (χ2) plotted as a function f, the fraction of 4I0U, fixed before BME reweighting of the MetaD generated ensemble to fit the SANS data. The data on the graph were fitted by a quadratic function to yield the minimum with standard deviation at 40% ± 28% 4I0U, where the standard deviation was calculated by an increment in χ2 of 1.
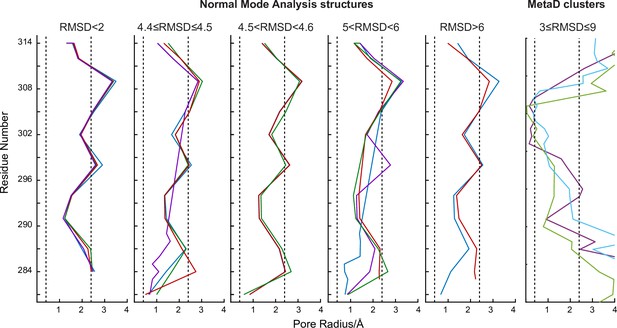
Representation of the pore radius profile in the transmembrane domain obtained by HOLE program out of the structures calculated by Normal Mode Analysis and MetaDynamics.
Left: The NMA structures were clustered according to their global RMSD with respect to the X-ray structure of CorA in the presence of Mg2+ (PDB 4I0U). Right: The analysis of the 6 MetaD clusters revealed only three distinct profiles in the transmembrane region. Dotted lines indicate the radii of dry (0.4 Å) and hexa-hydrated Mg2+ (2.4 Å).
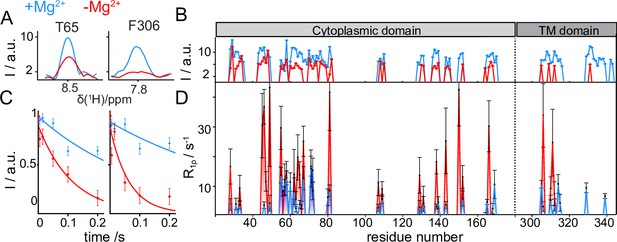
CorA backbone dynamics by MAS NMR in DMPC in presence (blue) and in absence (red) of Mg2+.
(A) Examples of 1D traces of 3D 1H-15N-13Cα peaks for two residues in the ICD (T65) and in the TMD (F306). (B) Comparison of peak intensities in 3D 1H-15N-13Cα spectra over the protein sequence. ICD and TMD are indicated by boxes of different color on top of the plot. C-D: Site-specific 15N R1ρ rates measured with a 15 kHz spin-lock field. (C) Examples of 15N R1ρ relaxation decays together with the corresponding mono-exponential fits for residues T65 and F306. (D) Comparison of site-specific backbone 15N R1ρ rates plotted along the CorA sequence.
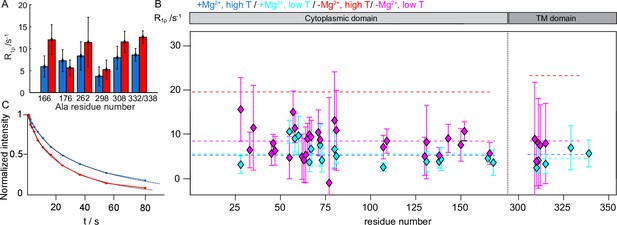
CorA backbone dynamics by MAS NMR: Site-specificity and temperature dependence.
(A) Site-specific 15N R1ρ rates measured with a 15 kHz spin-lock field in the 15N-Ala sample of CorA. (B) Comparison of site-specific 15N R1ρ rates measured with a 15 kHz spin-lock field at ~280 K in the presence (cyan) and in the absence (magenta) of Mg2+. Dashed lines indicate the average relaxation rate values in the ICD and TMD distinctly, at higher (blue and red) and lower (cyan and magenta) temperature. (C) Bulk 15N R1 decay curves measured at 300 K. Experimental curves were fitted with a biexponential decay.
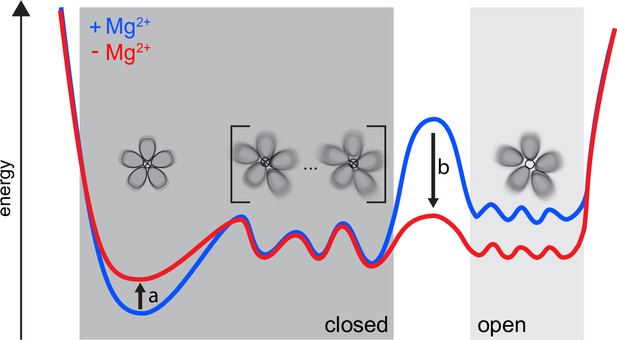
Our proposed dynamic model for CorA.
Both in high and low intracellular Mg2+ levels, a complex ensemble of closed states (left), symmetric as well as asymmetric, are in equilibrium with an ensemble of open states (right). At high Mg2+(blue) the large energy barrier prevents CorA from visiting the ensemble of open states. At low intracellular Mg2+ levels (red) a reduction in the population of the symmetric, closed state (arrow a) is induced. At the same time the energy barrier (arrow b) is lowered toward the ensemble of open states, which then becomes populated.
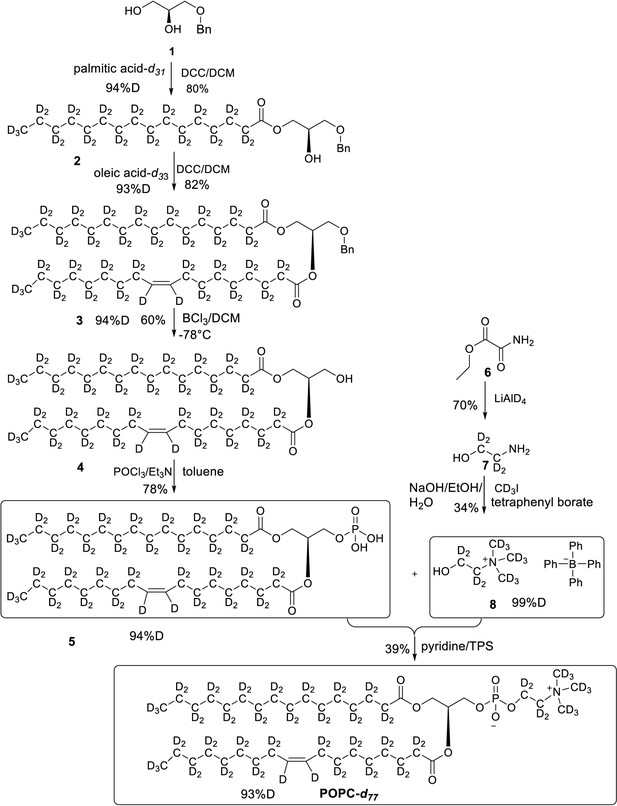
Overall synthesis achieved by following a reported paper, all the intermediates and final POPC-d77 were obtained in similar yields (Yepuri et al., 2016).
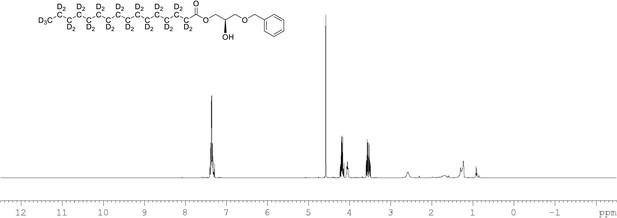
1H NMR of 1-palmitoyl-d31-sn-3-benzyloxy-glycerol (Appendix 1—figure 1, molecule 2) in CDCl3.
(400 MHz, CDCl3), δ residual protons 0.88 (m, 0.22 H), 1.10–17 (m, 1.96 H), 1.57.1.86 (m, 1.55 H), 1.96 (m, 0.54 H), 3.69 (m, 2 H), 4.09 (m, 1 H), 4.22 (m, 2 H) 4.60 (s, 2 H), 7.36 (m, 5 H).
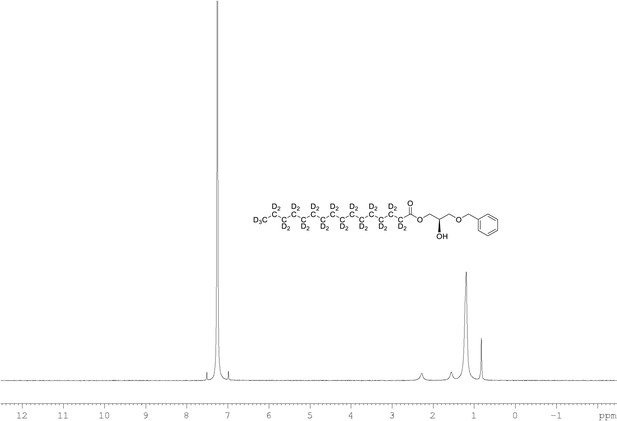
2H NMR of 1-palmitoyl-d31-sn-3-benzyloxy-glycerol (Appendix 1—figure 1, molecule 2) in CDCl3.
(400 MHz, CDCl3), δ 0.82 (m), 1.18 (m), 1.54 (m), 2.27 (m).
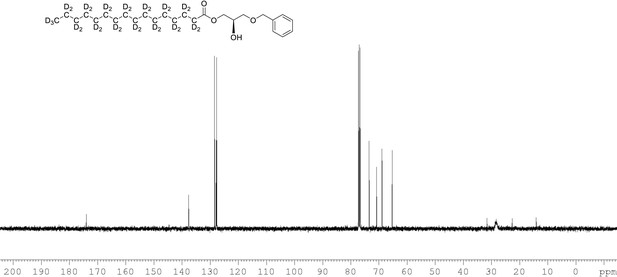
13C NMR of 1-palmitoyl-d31-sn-3-benzyloxy-glycerol (Appendix 1—figure 1, molecule 2) in CDCl3.
(400 MHz, CDCl3), 10.9 (m), 22.09 (m), 28.33 (m), 33.05 (m), 65.33, 68.9, 70.80, 73.5, 127.7, 127.9, 128.5, 137.6, 174.0.
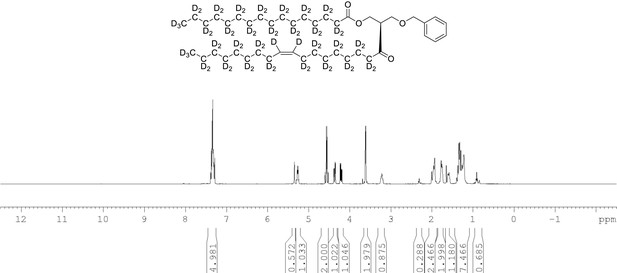
1H NMR of 1-palmitoyl-d31-2-oleoyl-d33-sn-3-benzyloxy-glycerol (Appendix 1—figure 1, molecule 3) in CDCl3.
(400 MHz, CDCl3), δ residual protons 0.90 (m, 3.56 H), 1.29 (m, 5.15 H), 1.98 (m, 0.68 H), 2.29 (m, 1.25 H), 3.61 (d, J = 5.0 Hz, 2 H), 4.21 (m, 1 H), 4.36 (m, 1 H), protonated benzyl protons 4.56 (AB q, J = 12 Hz, 2 H), 5.26 (m, 1 H), 5.35 (s, 0.65 H), 7.34 (m, 5 H).
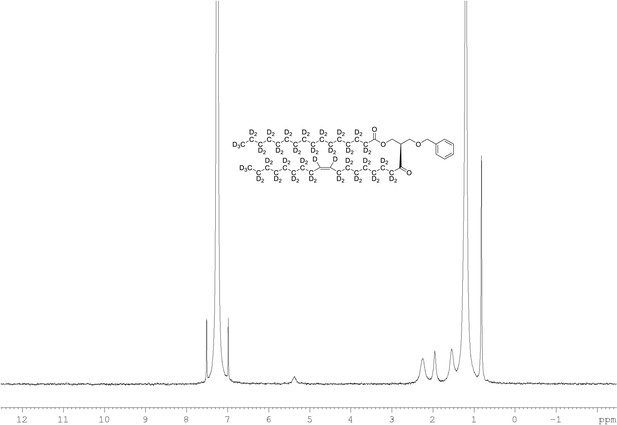
2H NMR of 1-palmitoyl-d31-2-oleoyl-d33-sn-3-benzyloxy-glycerol (Appendix 1—figure 1, molecule 3) in CDCl3.
(400 MHz, CDCl3), δ 0.82 (m, 6D), 1.19 (m, 35.35D), 1.53 (m, 3.74D), 1.94 (m, 3.34D), 2.25 (m, 2.63D), 3.56 (m, 1.71D), 4.27 (m, 1.04D), 5.35 (m, 1.92).
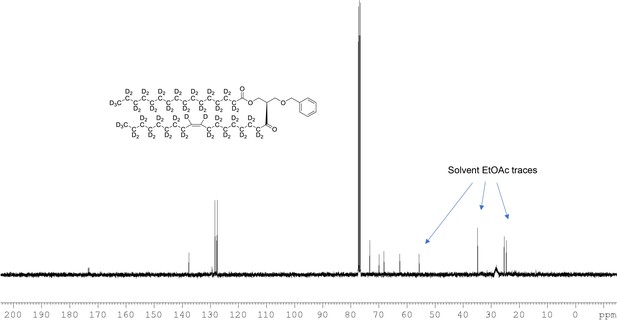
13C NMR of 1-palmitoyl-d31-2-oleoyl-d33-sn-3-benzyloxy-glycerol (Appendix 1—figure 1, molecule 3) in CDCl3.
(400 MHz, CDCl3), δ 13.08 (m), 21.60 (m), 23.96 (m), 26.50 (m), 28.2 (m), 30.60 (m), 33.80 (m), 62.60, 68.39 9, 70.10, 73.1, 127.7, 127.9, 128.5, 137.7, 173.1, 173.4.
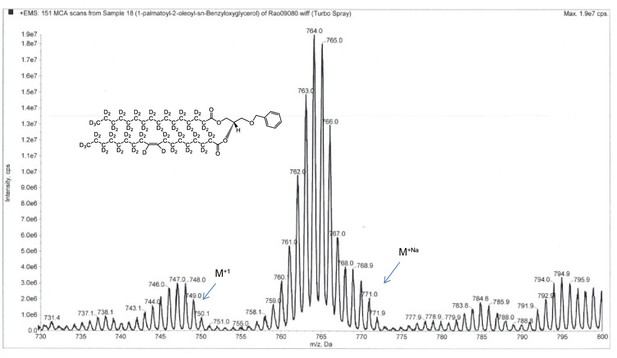
ESI-MS, m/z 749 [M+1]+ of POPC precursor 1-palmtoyl-2-oleoyl-sn-benzyloxyglycerol-d64 (Appendix 1—figure 1, molecule 3).
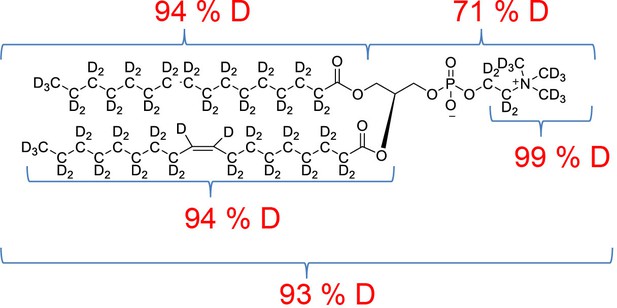
Overall 94%D, isotope distribution d64, 7.8%, d63, 15.1%, d62, 19.5%, d61, 18.0%, d60, 13.1%, d59, 10.2%, d58, 6.5%, d57, 3.9%, d56, 2.7%.
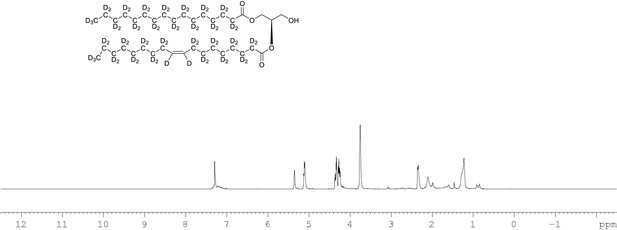
1H NMR of 1-palmitoyl-d31-2-oleoyl-d33-sn-3-glycerol (Appendix 1—figure 1, molecule 4) in CDCl3.
(400 MHz, CDCl3), δ residual protons 0.24 (m, 0.47 H), 1.28 (m, 1.51 H), 1.67 (m, 2.24 H), 2.13 (m, 0.52 H), 2.23 (m, 1.29 H), 3.72 (m, 2 H), 4.30 (m, 2 H), 5.10 (m, 1 H), 5.34 (s, 0.48 H).
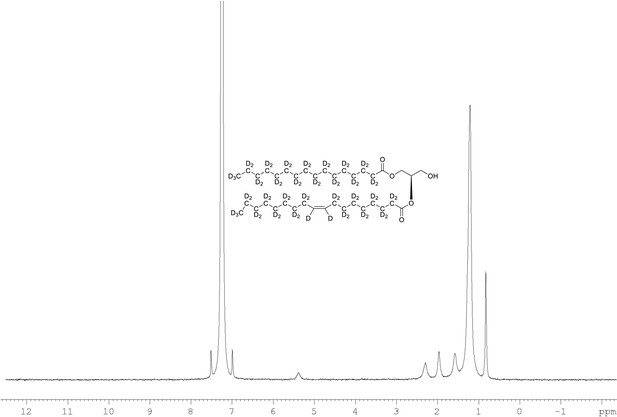
2H NMR of 1-palmitoyl-d31-2-oleoyl-d33-sn-3-glycerol (Appendix 1—figure 1, molecule 4) in CDCl3.
(400 MHz, CDCl3), δ 0.82 (m, 8.6D), 1.19 (m, 49.2D), 1.55 (m, 4.8D), 1.94 (m, 3.46D), 2.28 (m, 3.82D), 5.38 (m, 1.1D).
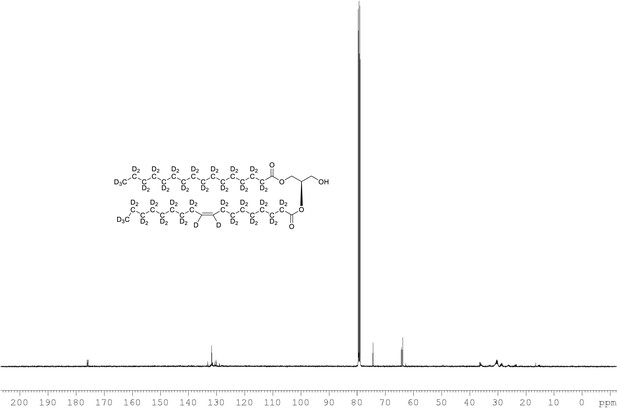
13C NMR of 1-palmitoyl-d31-2-oleoyl-d33-sn-3-glycerol (Appendix 1—figure 1, molecule 4) in CDCl3.
(400 MHz, CDCl3), δ 12.89 (m), 21.39 (m), 22.6 (m), 24.00 (m), 26.25 (m), 28.2 (m), 30.49 (m), 33.86 (m), 65.00, 60.76, 61.60, 71.64, 128.02, 129.14, 129.5, 173.53, 173.93.
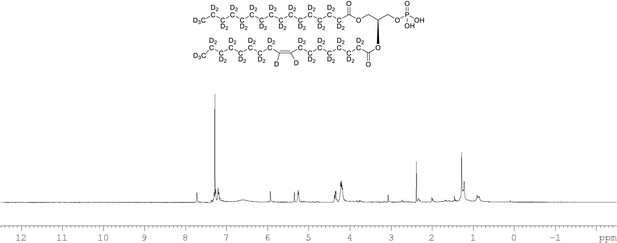
1H NMR of crude product 1-palmitoyl-d31-2-oleoyl-d33-sn-3-glycero-phosphatidic acid (Appendix 1—figure 1, molecule 5) in CDCl3.
This synthesised crude dried product was used in next step without further purification.
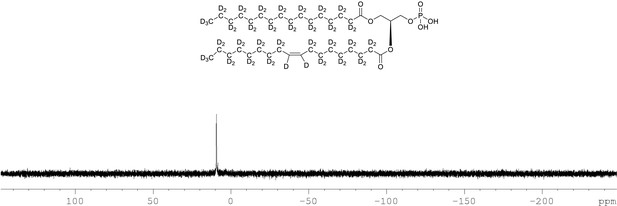
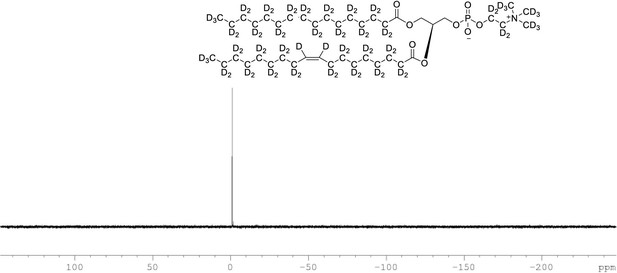
31P NMR of crude product 1-palmitoyl-d31-2-oleoyl-d33-sn-3-glycero-phosphatidic acid (Appendix 1—figure 1, molecule 5) in CDCl3.
This synthesised crude dried product was used in next step without further purification.
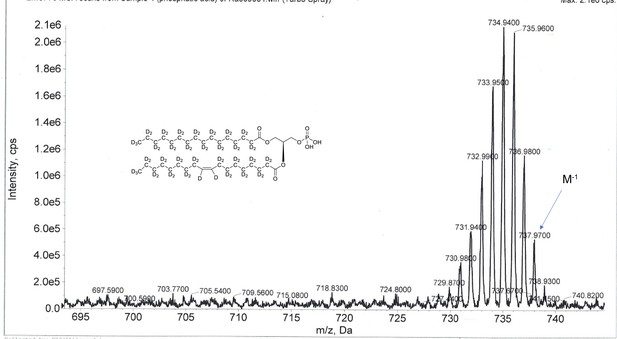
ESI-MS, m/z 737 [M–1]- of crude product of 1-palmitoyl-d31-2-oleoyl-d33-sn-3-glycero-phosphatidic acid (Appendix 1—figure 1, molecule 5).
Overall 94%D, isotope distribution d64, 0.6%, d63, 8.9%, d62, 18.8%, d61, 25.1%, d60, 20.6%, d59, 13.0%, d58, 7.5%, d57, 5.2%, d56, 0.2%, d55, 0.1%.
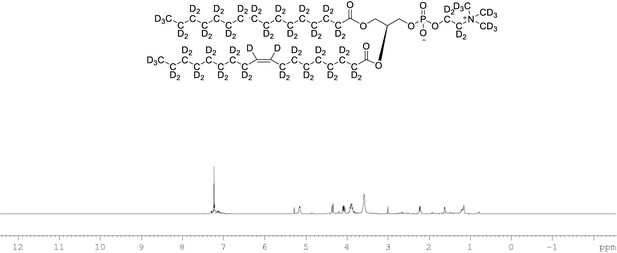
1H NMR of POPC-d77 in CDCl3.
(400 MHz, CDCl3), δ residual protons 0.85 (m, 0.17 H), 1.25 (m, 1.48 H), 1.54 (m, 0.22 H), 1.97 (m, 0.18 H), 2.22 (m, 0.66 H), 3.89 (m, 2 H), 4.08 (m, 1 H), 4.34 (m, 1 H), 5.14 (m, 1 H), 5.27 (s, 0.43 H).
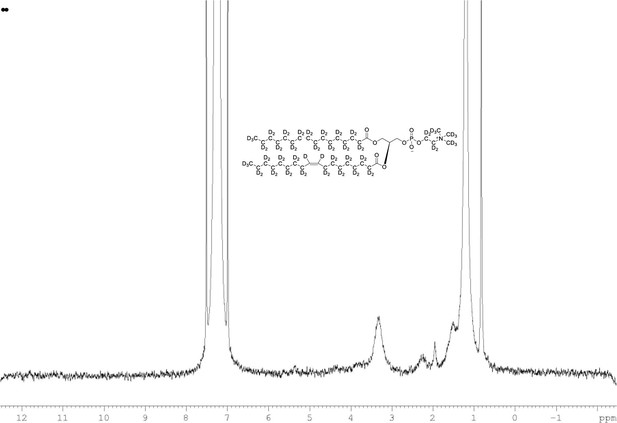
2H NMR of POPC-d77 in CDCl3.
(400 MHz, CDCl3), δ 0.80 (m), 1.19 (m), 2.20 (m), 1.93 (m, 6.0D), 3.35 (m), 3.84 (m), 5.36 (m).
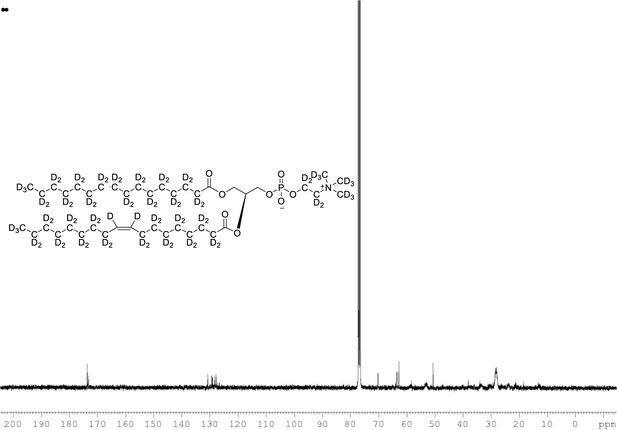
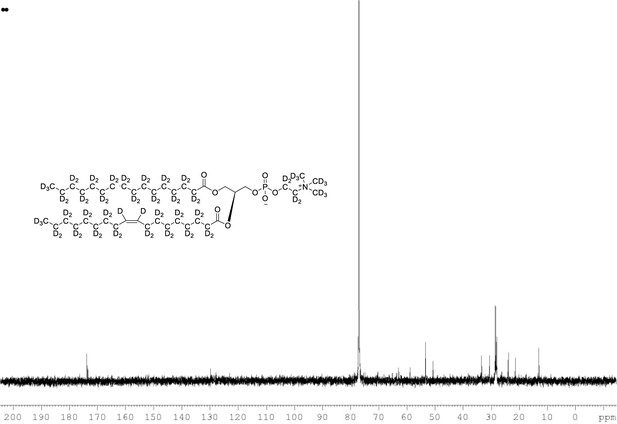
13C NMR of POPC-d77 in CDCl3.
(400 MHz, CDCl3), δ 13.03 (m), 21.48 (m), 23.88 (m), 26.24 (m), 28.36 (m), 30.49 (m), 33.72 (m), 53.17, 58.76 (m), 62.24 (m), 69.84 (m), 127.90 (s), 129.5, 173.31, 173.69.
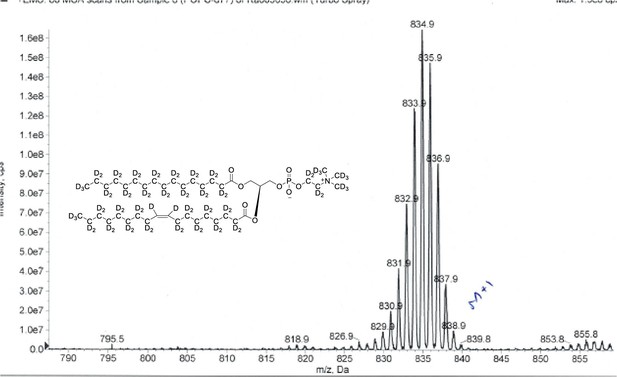
ESI-MS, m/z 838 [M+1]+ of POPC-d77.
Overall 93%D, isotope distribution d77, 0%, d76, 0%, d75, 7.5%, d74, 21.2%, d73, 28.7%, d72, 25.3%, d71, 15.5%, d70, 10.8%.
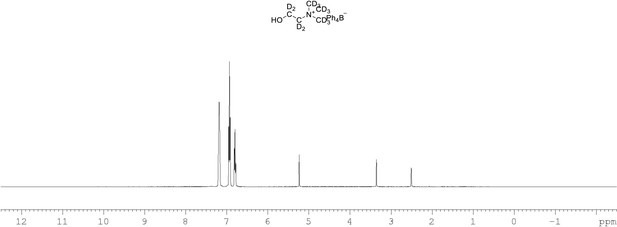
1H NMR of choline-d13 tetraphylborate (Appendix 1—figure 1, molecule 8) in DMSO-d6.
(400 MHz, DMSO-d6) δ 6.85 (m, 4 H), 6.96 (m, 8 H), 7.21 (m, 8 H).
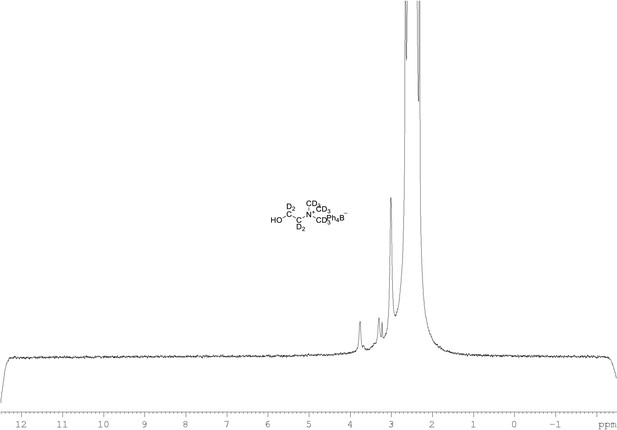
2H NMR of choline-d13 tetraphylborate (Appendix 1—figure 1, molecule 8) in DMSO-d6.
(61.4 MHz, DMSO-d6) δ 3.30 (m, 9D), 3.32 (m, 2D), 3.78 (m, 2D).
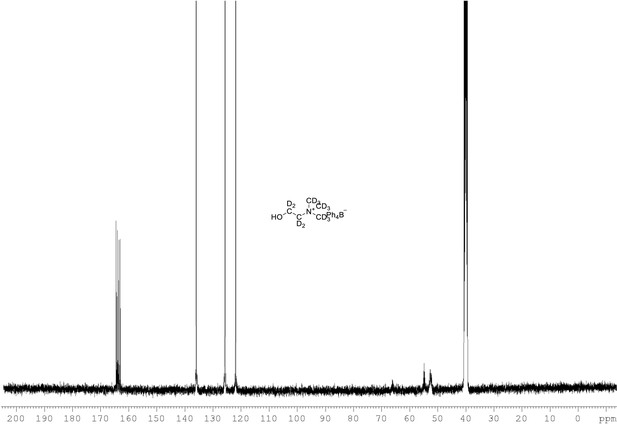
13C NMR of choline-d13 tetraphylborate (Appendix 1—figure 1, molecule 8) in DMSO-d6.
(100 MHz, DMSO-d6) δ 52.5 (m), 54.9 (m), 121.9, 125.9, 136.2, 163.9 (m).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Thermotoga maritima) | corA | Uniprot | Q9WZ31 | |
| Strain, strain background (Escherichia coli) | BL21 star(DE3) | ThermoFisher | C601003 | |
| Software, algorithm | PEPSI-SANS | https://team.inria.fr/nano-d/software/pepsi-sans/ | RRID:SCR_021950 | |
| Software, algorithm | NOLB | Ref. 48 | RRID:SCR_021954 | |
| Software, algorithm | Martini3.0b | Ref. 49 | RRID:SCR_021951 | |
| Software, algorithm | GROMACS-5.1.4 | Ref. 52 | RRID:SCR_014565 | |
| Software, algorithm | PLUMED2.3.0 | Ref. 56 | RRID:SCR_021952 | |
| Software, algorithm | BME | Ref. 58 | RRID:SCR_021953 | |
| Software, algorithm | CcpNmr Analysis | Ref. 64 | RRID:SCR_016984 | |
| Software, algorithm | FLYA | Ref. 65 | RRID:SCR_014229 |
Negative stain EM statistics for 3D model refinement.
| Parameter | 1 mM EDTA | 40 mM MgCl2 |
|---|---|---|
| Pixel size, Å | 3.14 | 3.14 |
| Number of micrographs | 436 | 440 |
| Number of picked particles | 193,606 | 185,577 |
| Final number of particles | 36,176 | 46,544 |
| Resolution, Å | ≈ 15 | ≈ 15 |