Selective sorting of microRNAs into exosomes by phase-separated YBX1 condensates
Figures
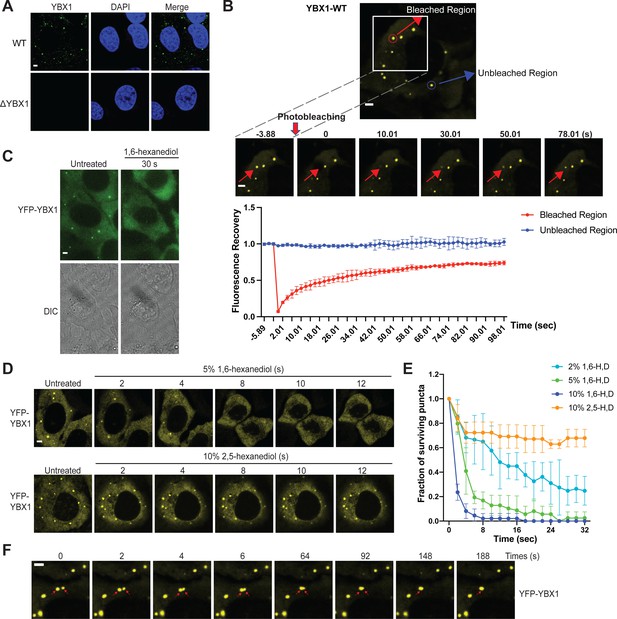
YBX1 forms liquid-like condensates in cells.
(A) Subcellular localization of YBX1 in WT and ΔYBX1 from U2OS cells as visualized by YBX1 antibody. DAPI staining (blue) indicates the location of nuclei. (B) FRAP images show recovery of YFP-YBX1 puncta after photobleaching. U2OS cells with stable expression of YFP-YBX1 was subjected to FRAP analysis. The inset images (middle) are the representative FRAP images. The recovery kinetics of YFP-YBX1 are shown in the bottom. Error bars represent standard errors with n = 3. (C) The effect of 10 % 1,6-hexanediol on YFP-YBX1 puncta in U2OS cells. This image was performed on ECLIPSE TE2000 microscope at room temperature. (D) Fluorescence images of YFP-YBX1 in U2OS cells after treatment with 5 % 1,6-hexanediol or 10 % 2,5- hexanediol. Live cell imaging was performed on an LSM880 microscope with the incubation chamber maintained at 37 °C and 5 % CO2. (See Materials and methods in details). (E) Number of YFP-YBX1 puncta in U2OS cells surviving over time after treatment with 1,6-hexanediol and 2,5-hexanediol. Error bars represent standard errors with n = 3. (F) Representative images of YBX1 puncta coalescence from U2OS cells. This live cell imaging was performed on an LSM880 microscope with the incubation chamber maintained 37 °C and 5 % CO2. Scale bars, 3 µm.
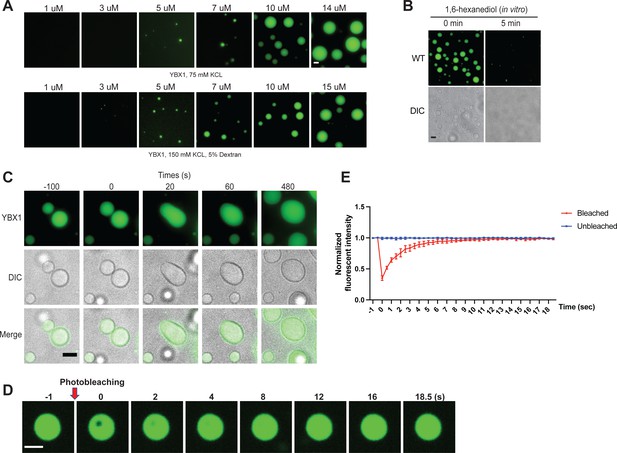
YBX1 forms liquid-like droplets in vitro.
(A) Phase separation of YBX1 at different concentrations with or without addition of a crowding agent. Phase separation was induced by diluting the salt concentration from 500 mM to 75 mM or 150 mM in this assay. (B) The effect of 10 % 1,6-hexanediol on YBX1 droplets in vitro. Phase separation was induced by diluting the salt concentration from 500 mM to 75 mM in this assay. (C) Representative images of YBX1 droplets coalescence in vitro. Phase separation was induced by diluting the salt concentration from 500 mM to 75 mM in this assay. (D, E) Images (D) and quantification (E) of recovery of YBX1 droplets after photobleaching. A representative result of three independent experiments is shown. Phase separation was induced by diluting the salt concentration from 500 mM to 75 mM in this assay. Error bars represent standard errors. Scale bars, 3 µm.
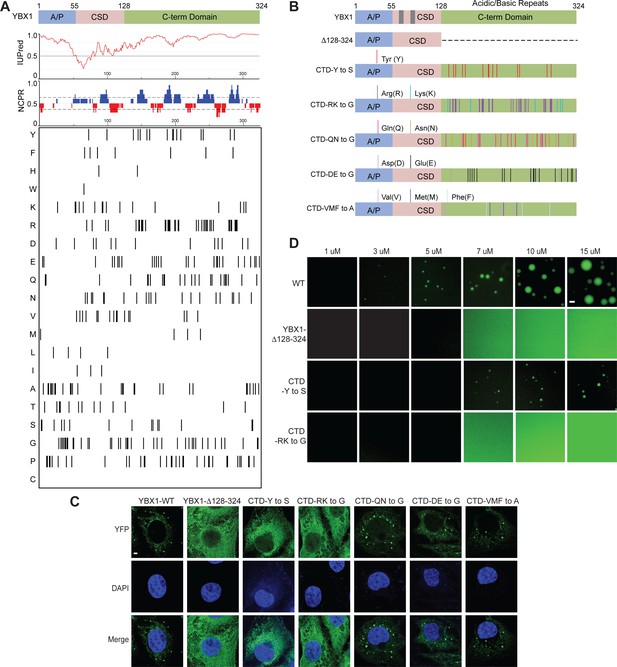
YBX1 phase separation is governed by association of aromatic and basic amino acids in C-terminal IDR.
(A) Structural organization of YBX1. Top, IUPred, prediction of disordered protein regions; Middle, NCPR, net charge per residue with a sliding window of five residues; Net positive, blue, net negative, red; Bottom, visualization outputs for residue plots. (B) Schematic diagrams of different YBX1 mutants with the distribution of mutated amino acids. (C) Truncation mapping and identification of residues in YBX1 C-terminal IDR that are required for YBX1 condensation formation. YFP fused YBX1 wild type and mutants were introduced in ΔYBX1 U2OS cells by transient transfection and visualized by fluorescence microscopy. (D) Phase separation of YBX1 wild type and variants at the indicated concentrations. 6xHis-MBP-mGFP fused YBX1 wild type and variant proteins were purified from insect cells. Phase separation was induced by diluting the salt concentration from 500 mM to 150 mM in this assay. Scale bars, 3 µm.
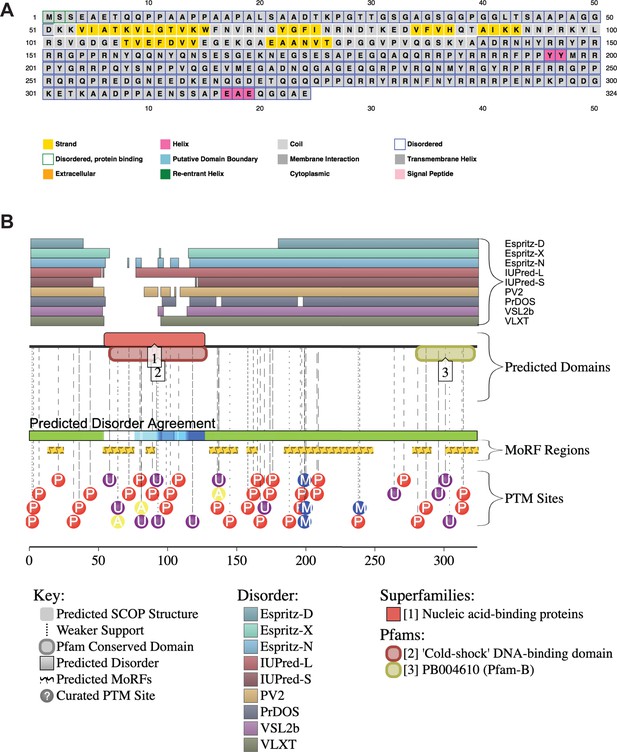
YBX1 amino acid sequences and secondary structure prediction.
(A) Secondary structure of YBX1 was predicted using PSIPRED 4.0 based on amino acid sequences. (B) Secondary structure of YBX1 was predicted using D2P2 (Database of Disordered Protein Prediction, http://d2p2.pro) based on amino acid sequences.
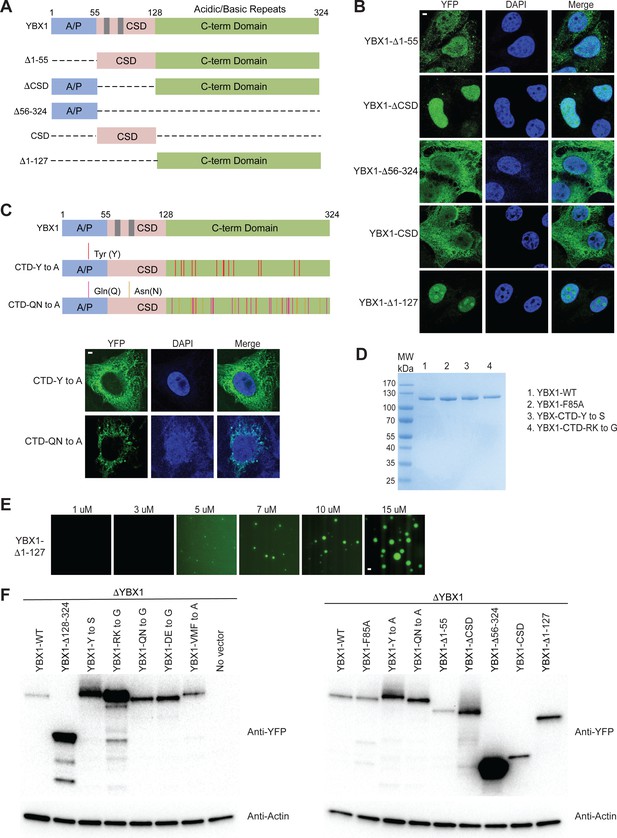
The ability of YBX1 to form LLPS requires C-terminal IDR, likely depending on tyrosine and basic amino acids arginine and lysine.
(A) Schematic diagrams of YBX1 truncation analysis. (B) Analysis of condensation formation for different YBX1 truncations in U2OS cells. YFP- fused YBX1 mutants were introduced in ΔYBX1 U2OS cells by transient transfection and visualized by fluorescence microscopy. (C) Identification of residues in YBX1 C-terminal IDR that are involved in YBX1 condensation formation. (D) SDS-PAGE of YBX1 wild-type and variants tagged with 6xHis-MBP-mGFP. (E) Phase separation of YBX1 N-terminal at the indicated concentrations. (F) Immunoblot analysis of YFP-tagged WT YBX1 and variants transiently expressed in U2OS cells. The blots were probed with antibodies directed against YFP or actin. Scale bars, 3 µm.
-
Figure 3—figure supplement 2—source data 1
Uncropped SDS-PAGE corresponding to Figure 3—figure supplement 2.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig3-figsupp2-data1-v2.zip
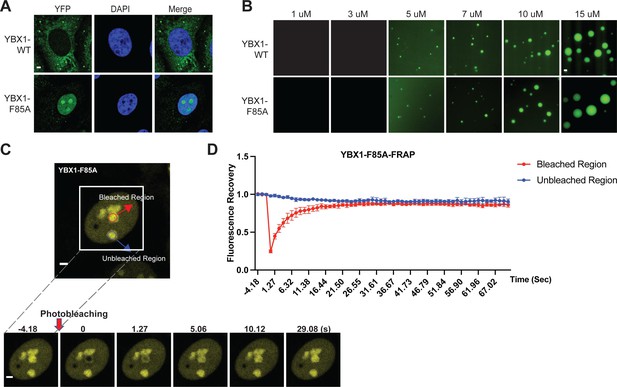
A F85A mutation did not affect YBX1 liquid droplet formation in vitro.
(A) Mutation of F85A caused YBX1 to translocate into nucleus. YFP-fused YBX1 wild type and F85A were introduced in ΔYBX1 U2OS cells by stable transfection and visualized by fluorescence microscopy. (B) YBX1-F85 is not deficient for YBX1 liquid droplet formation. 6xHis-MBP-mGFP fused YBX1 wild type and F85A protein were purified from insect cells. Phase separation was induced by diluting the salt concentration from 500 mM to 150 mM in this assay. (C, D) Images (C) and quantification (D) of recovery of YBX1-F85A signal after photobleaching. A representative result of three independent experiments is shown. Error bars represent standard errors. Scale bars, 3 µm.
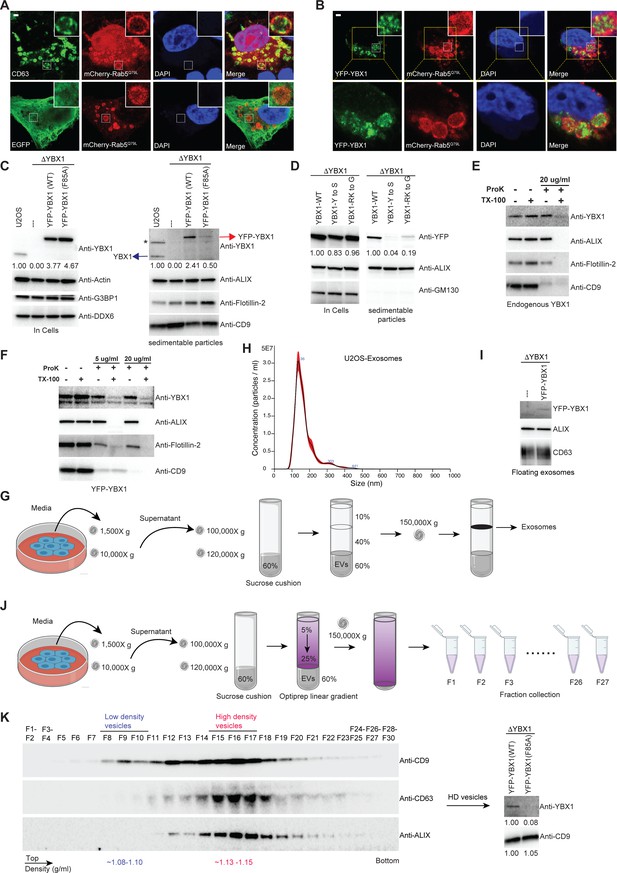
IDR-driven YBX1 phase separation is required for sorting YBX1 into exosomes.
(A) Representative microscope images from U2OS cells expressing mChery-RAB5Q79L. Confocal micrographs of cells expressing mCherry-RAB5Q79L, alone (upper row) or with EGFP (lower row). Cells are stained with anti-CD63 (upper row) or with anti-GFP (lower row). (B) Confocal micrographs of U2OS cells expressing mChery-RAB5Q79L and YFP-YBX1. (C) Over-expression of YBX1 in U2OS cells increased the secretion of YBX1 in EVs. Immunoblots for the indicated protein markers in cells and high-speed pellet fractions. The numbers under the YBX1 blot represent quantification analysis of endogenous YBX1, YFP-YBX1, and YFP-YBX1-F85A in cells and sedimentable particles by Fiji software. ‘*’ is a non-specific band; Blue arrow represents endogenous YBX1; Red arrow represents fusion YBX1 or YBX1-F85A. (D) IDR-driven YBX1 phase separation is required for YBX1 secretion in EVs. Immunoblots for the indicated protein markers in U2OS cells and high-speed pellet fractions. The numbers under the YFP blot represent quantification analysis of endogenous YBX1 and variants in cells and sedimentable particles by Fiji software. (E) Proteinase K protection assay on high-speed pellet fractions from U2OS cells. Triton X-100 (0.5%) was used to disrupt the membranes. Immunoblots for YBX1, ALIX, Flotillin-2, and CD9 are shown. (F) Proteinase K protection assay on high-speed pellet fractions from U2OS cells expressing YFP-YBX1. Triton X-100 (0.5%) was used to disrupt the membranes. Immunoblots for YBX1, ALIX, Flotillin-2, and CD9 are shown. (G) Schematic showing exosome purification with buoyant density flotation in a sucrose step gradient. (H) Nanoparticle tracking analysis (NTA) quantification of exosomes from cultured U2OS cells. (I) YFP-YBX1 detected in sucrose post-flotation fraction from U2OS cells. Immunoblots for YBX1, ALIX, and CD63 from buoyant exosomes are shown. (J) Schematic showing exosome purification with buoyant density flotation in a linear iodixanol gradient. (K) Immunoblots across the iodixanol gradient from U2OS cells for classical exosome markers CD9, CD63 and ALIX (the left panel). Collection of fractions F15-F17 corresponding to high density vesicles and immunoblots for YBX1 and CD9. The numbers under YBX1 blot and CD9 blot represent quantification analysis of YFP-YBX1-WT or YFP-YBX1-F85A and CD9 in HD vesicles, respectively, by Fiji software. Scale bars, 3 µm.
-
Figure 4—source data 1
Uncropped Western blot images corresponding to Figure 4C.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig4-data1-v2.zip
-
Figure 4—source data 2
Uncropped Western blot images corresponding to Figure 4D.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig4-data2-v2.zip
-
Figure 4—source data 3
Uncropped Western blot images corresponding to Figure 4E.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig4-data3-v2.zip
-
Figure 4—source data 4
Uncropped Western blot images corresponding to Figure 4F.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig4-data4-v2.zip
-
Figure 4—source data 5
Uncropped Western blot images corresponding to Figure 4I.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig4-data5-v2.zip
-
Figure 4—source data 6
Uncropped Western blot images corresponding to Figure 4K.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig4-data6-v2.zip
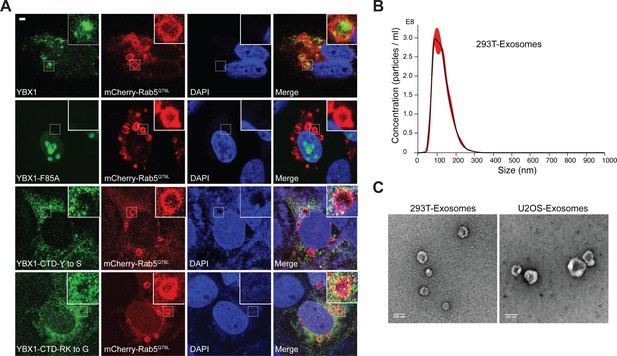
YBX1 entering into ILVs is dependent on IDR-driven phase separation.
(A) Confocal micrographs of U2OS cells expressing mChery-RAB5Q79L and YFP tagged YBX1 variants. Scale bar, 3 µm. (B) Nanoparticle tracking analysis (NTA) quantification of exosomes from cultured HEK293T cells. (C) Representative electron micrographs of negative stained exosomes purified from 10/40 % sucrose interface. Scale bar is 100 nm.
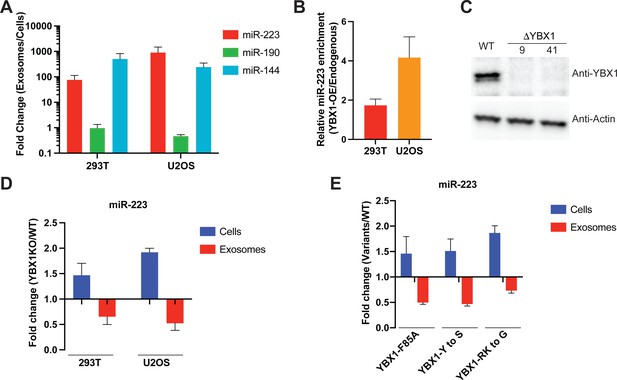
IDR-driven YBX1 phase separation is required for sorting miR-223 into exosomes.
(A) Relative abundance of miRNAs detected in exosomes compared to cellular levels from both HEK293T cells and U2OS cells. Exosomes were purified as in Figure 4G. Fold change of miRNAs in cells and purified exosomes from indicated cells quantified by RT-qPCR. Data are plotted from three independent experiments and error bars represent standard derivations. (B) Overexpression of YBX1 increases sorting of miR-223 into exosomes both in HEK293T cells and U2OS cells. Exosomes were purified as in Figure 4G. Fold change of miR-223 in cells and purified exosomes from indicated cells quantified by RT-qPCR. Relative miR-223 enrichment was calculated by fold change (Exo/cells) of YBX1-OE divided by fold change (Exo/cells) of endogenous YBX1. Data are plotted from three independent experiments and error bars represent standard derivations. (C) Analysis of wild-type and CRISPR/Cas9 genome edited HEK293T clones by immunoblot for YBX1 (top) and actin (bottom). (D) The accumulation of miR-223 in cells and depletion of miR-223 in exosomes derived from ΔYBX1 and WT cells. Exosomes were purified as in Figure 4G. Fold change of miR-223 in cells and purified exosomes from indicated cells quantified by RT-qPCR. Data are plotted from three independent experiments for HEK293T cells and two independent experiments for U2OS cells; error bars represent standard derivations from independent samples. (E) Residues contributing to YBX1 phase separation are required for sorting miR-223 into exosomes. Exosomes were purified from U2OS cells as in Figure 4J. Fold change of miR-223 in cells and purified exosomes from indicated cells quantified by RT-qPCR. All quantifications represent means from three independent experiments and error bars represent standard derivations.
-
Figure 5—source data 1
Uncropped Western blot images corresponding to Figure 5C.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig5-data1-v2.zip
-
Figure 5—source data 2
The numerical data that are represented as graphs in Figure 5.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig5-data2-v2.xlsx
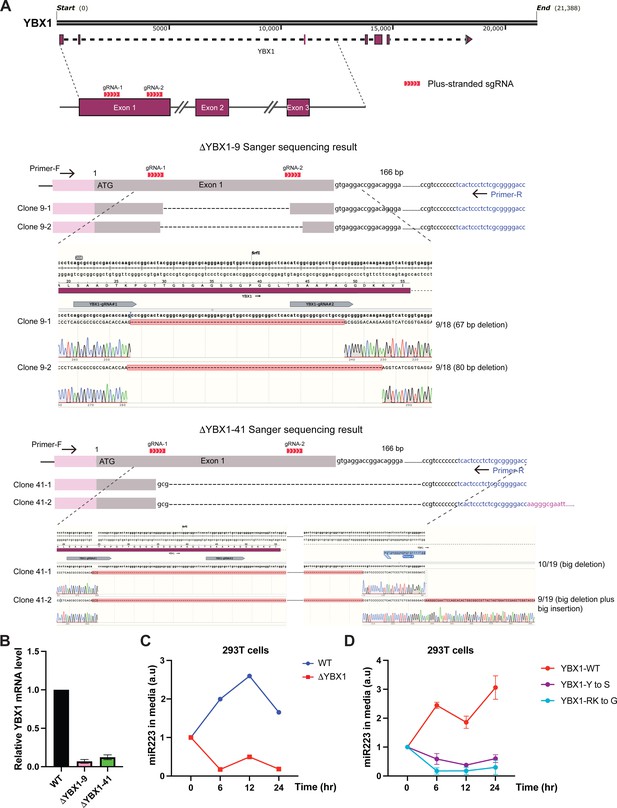
IDR-driven YBX1 phase separation is required for sorting miR-223 into the growth medium.
(A) CRISPR/Cas9-induced mutagenesis results of YBX1 knockout from HEK293T cells. Schematic of the targeted region of YBX1 (up panel) and Sanger sequencing results of YBX1 knockout clones 9 and 41 (bottom panel). Primer-F and primer-R were used to amplify the region around the gRNA recognition site. The PCR product was cloned into vector pCR2.1-TOPO. An M13-reverse primer was used for Sanger sequencing. Representative sequencing alignment of wild-type and YBX1 mutant alleles is shown. YBX1 knockout nine has two unique mutations (67 bp deletion; 80 bp deletion) among 18 sequenced clones. YBX1 knockout 41 has another two subtype mutations (big deletion; big deletion plus big insertion) among 19 sequenced clones. (B) Relative mRNA expression of YBX1 by RT-PCR in YBX1 KO cells generated by CRISPR/Cas9. Beta (β) -actin was used for normalization. Data are plotted as the fold change over WT control, and represent the mean ± SD of three independent experiments. (C) miR-223 secretion into medium in WT and YBX1 KO cells from HEK293T cells. About 200 µl cell culture medium was harvested at each time point and was used to extract RNA after centrifugation at 1500 xg for 15 min to remove debris. The amount of miR-223 was quantified by RT-qPCR. Data are plotted as the fold change over time zero. Different samples are normalized by total cell number. (D) Residues in YBX1-IDR that drive LLPS are required for miR-223 secretion into the growth medium in culture of HEK293T cells. The experiment was performed as (C).
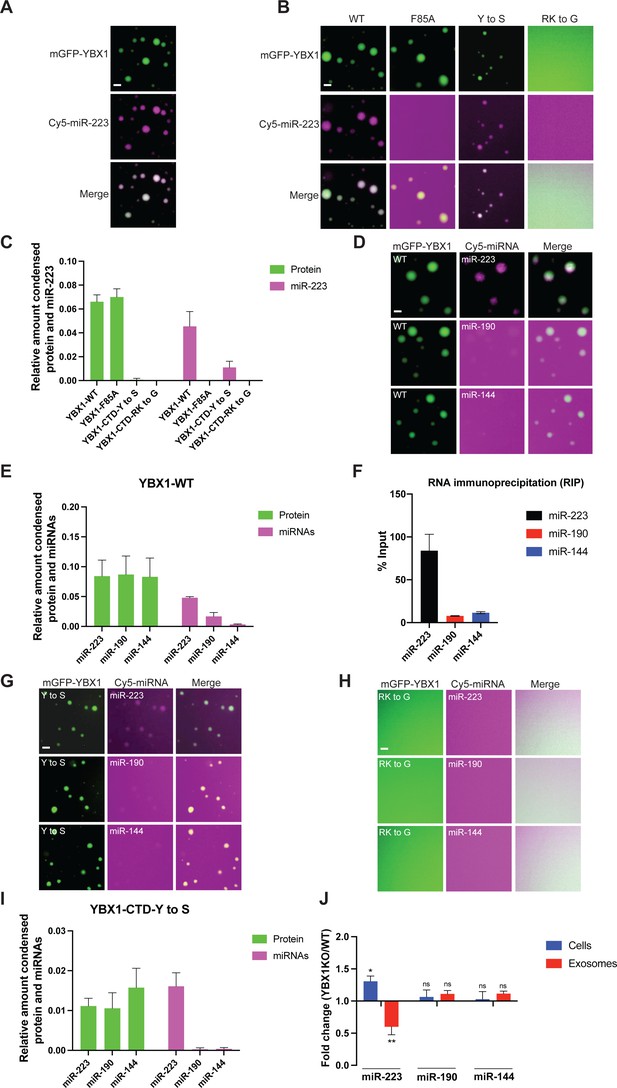
YBX1 phase-separated droplets recruit miRNAs with selectivity correlated with the exosome sorting ability in vivo.
(A) YBX1 phase-separated droplets recruit miR-223. Purified mGFP-YBX1 was incubated with Cy5 labeled miR-223 together with 10 ng/µl total RNA in LLPS buffer and then observed under a microscope. (B, C) The recruitment of miR-223 into YBX1 phase-separated droplets depends on the ability of YBX1 to bind RNA rather than phase separation. Representative images (400 × 400 pixels) (B) and quantification (C) of condensed miR-223 and YBX1 protein. Relative amount condensed protein or miRNAs was calculated as ratio of total intensity of protein inside droplets to total intensity of protein both inside and outside of droplets as quantified using Fiji software. Three images (2048 x 2048 pixels) for per condition were analyzed. The results are plotted as the mean ± the standard deviation (SD). (D, E) YBX1 liquid droplets recruit miRNAs in a selective manner. Purified mGFP-YBX1 was incubated with Cy5 labeled miR-223, miR-190, or miR-144 individually, together with 10 ng/µl total cellular RNA in LLPS buffer and then observed under a microscope. Representative images (400 × 400 pixels) (D) and quantification (E) of condensed miRNAs and YBX1 protein. Relative amounts of condensed protein and RNA were calculated as described in (C). Three images (2048 x 2048 pixels) per condition were analyzed. The results are plotted as the mean ± the standard deviation (SD). (F) RIP assay with GFP-trap beads on YFP-YBX1 expressing HEK293T cell extracts. miRNAs in immunoprecipitated samples were determined by RT-qPCR using Taqman miRNAs assay, and reported as percentage of input sample (% input). Data are plotted as means ± SD of three independent experiments. (G) YBX1-CTD-Y to S mutant recruits miRNAs inefficiently but selectively. Purified mGFP-YBX1-CTD-Y to S was incubated with Cy5 labeled miR-223, miR-190 or miR-144 independently, together with 10 ng/µl total cellular RNA in LLPS buffer and then observed under a microscope. (H) YBX1-CTD-RK to G mutant failed to phase separate and recruit miRNAs. Purified mGFP-YBX1-CTD-RK to G was incubated with Cy5 labeled miR-223, miR-190 or miR-144 independently, together with 10 ng/µl total cellular RNA in LLPS buffer and then observed under a microscope. (I) Quantification of condensed miRNAs and YBX1 protein from (G). Relative amount of condensed protein and RNA were calculated as described in (C). Three images (2048 x 2048 pixels) per condition were analyzed. The results are plotted as the mean ± the standard deviation (SD). (J) YBX1 is required for sorting miR-223 but not miR-190 and miR-144 into exosomes. Exosomes were purified as in Figure 4G. Fold change of miR-223, miR-190, and miR-144 in cells and purified exosomes from indicated cells quantified by RT-qPCR. All quantifications represent means from three independent experiments and error bars represent standard derivations. Statistical significance was performed using unpaired t-test (*p < 0.05, **p < 0.01, and ns = not significant). Scale bars, 3 µm.
-
Figure 6—source data 1
The numerical data that are represented as graphs in Figure 6.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig6-data1-v2.xlsx
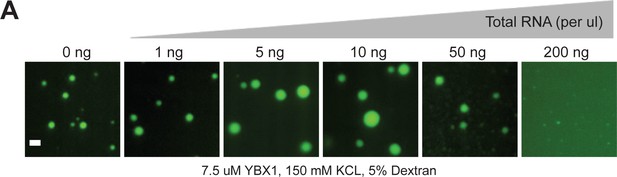
RNA regulates the phase separation behavior of YBX1.
(A) Representative images of purified YBX1 in vitro in the presence of total cellular RNA. Scale bar, 3 µm.
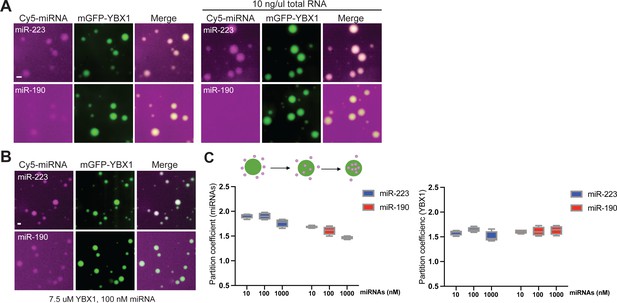
miRNAs differ in affinity to YBX1 phase-separated droplets.
(A) YBX1 phase-separated droplet miRNA recruitment selectivity enhanced upon addition of 10 ng/µl cellular RNA. Purified mGFP-YBX1 was incubated with Cy5-labeled miR-223 or miR-190 individually, together with 10 ng/µl cellular RNA in LLPS buffer and then observed under a microscope. (B) miR-223 selectively partitions into YBX1 phase-separated droplets but miR-190 does not. Purified mGFP-YBX1 was incubated with Cy5-labeled miR-223 or miR-190 at different concentrations individually in LLPS buffer and then observed under a microscope. The representative images for 100 nM miRNAs are shown here. (C) Relative quantification from (B) for partition coefficient. Partition coefficient was calculated as a ratio of mean intensity within droplets to mean intensity outside of droplets using Fiji software. Four images (1024 × 1024 pixels) for per condition were analyzed. Scale bars, 3 µm.
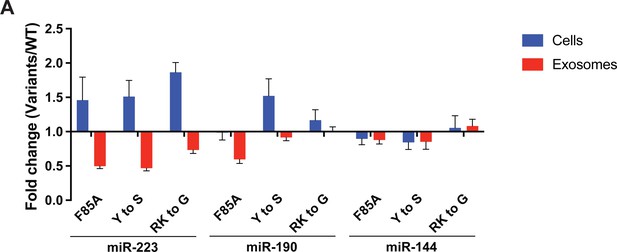
YBX1 condensation is required for sorting miR-223 but not miR-190 and miR-144 into exosomes.
(A) Residues contributing to YBX1 phase separation are required for sorting miR-223 but not miR-190 and miR-144 into exosomes. Exosomes were purified from U2OS cells as in Figure 4J. Fold change of miRNAs in cells and purified exosomes from indicated cells quantified by RT-qPCR. All quantifications represent means from three independent experiments and error bars represent standard derivations. The same data for miR-223 were shown in Figure 5E.
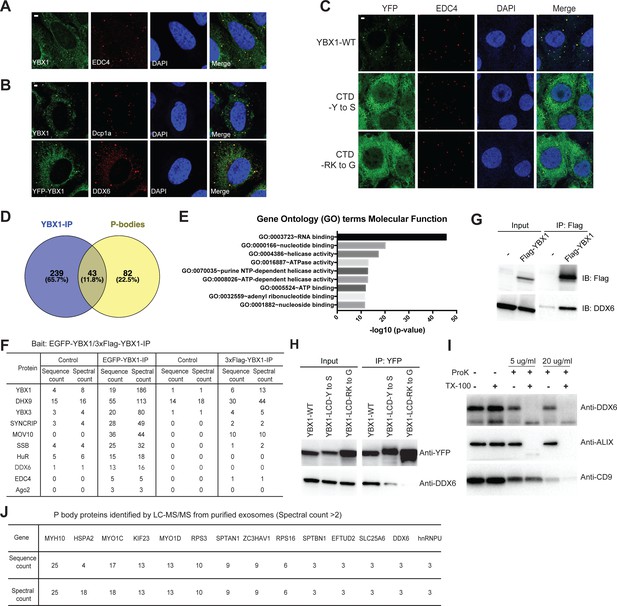
Condensation of YBX1 in PBs is required for sorting miRNAs into exosomes.
(A) YBX1 condensates co-localized with P-body marker EDC4. Indirect immunofluorescence was used to show that YBX1 localized to P-bodies. U2OS cells were stained with anti-YBX1 and anti-EDC4 antibodies. (B) YBX1 condensates co-localized with P-body markers Dcp1a and DDX6. U2OS cells were stained with anti-YBX1 and anti-Dcp1 antibodies (upper row), or with anti-YFP and anti-DDX6 antibodies (lower row). (C) YBX1 condensation into P-bodies dependent on IDR-driven phase separation. YFP-fused YBX1 wild type and variants were introduced in ΔYBX1 U2OS cells by stable transfection and visualized by fluorescence microscopy. Cells were stained with anti-YBX1 and anti-EDC4 antibodies. (D) The Venn diagram shows overlap between YBX1 proteome and previously reported P-body proteome. (E) GO analysis (molecular function) of genes associated with YBX1. (F) Proteins identified by either 3xFlag-YBX1-IP or mGFP-YBX1-IP, coupled with mass spectrometry. (G) Coimmunoprecipitation of DDX6 with YBX1 from HEK293T cells. (H) Residues in YBX1-IDR that drive LLPS are required for its interaction with DDX6 in HEK293T cells. (I) DDX6 resides in exosomes. Proteinase K protection assay for DDX6 using exosomes that were isolated by buoyant density flotation from HEK293T cells. Triton X-100 (0.5%) was used to disrupt the membranes. Immunoblots for DDX6, ALIX, and CD9 are shown. (J) Identification of P-body components in purified exosomes from HEK293T cells by LC-MS/MS. Exosomes were purified as in Figure 4G. Scale bars, 3 µm.
-
Figure 7—source data 1
The proteins identified by YBX1 immunoprecipitation coupled with mass spec analysis and P bodies proteome.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig7-data1-v2.xlsx
-
Figure 7—source data 2
The proteins identified in exosomes from HEK293T cells and P bodies proteome.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig7-data2-v2.xlsx
-
Figure 7—source data 3
Uncropped Western blot images corresponding to Figure 7G.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig7-data3-v2.zip
-
Figure 7—source data 4
Uncropped Western blot images corresponding to Figure 7H.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig7-data4-v2.zip
-
Figure 7—source data 5
Uncropped Western blot images corresponding to Figure 7I.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig7-data5-v2.zip
-
Figure 7—source data 6
The proteins identified in exosomes from HEK293T cells and stress granules proteome.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig7-data6-v2.xlsx
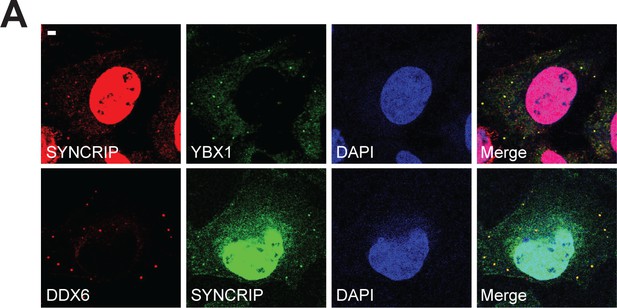
SYNCRIP forms condensates and co-localizes with YBX1 and P-body marker DDX6.
(A) SYNCRIP condensates co-localized with YBX1 and P-body marker DDX6. Indirect immunofluorescence was performed. U2OS cells were stained with anti- SYNCRIP, anti-YBX1, and anti-DDX6 antibodies. Scale bar, 3 µm.
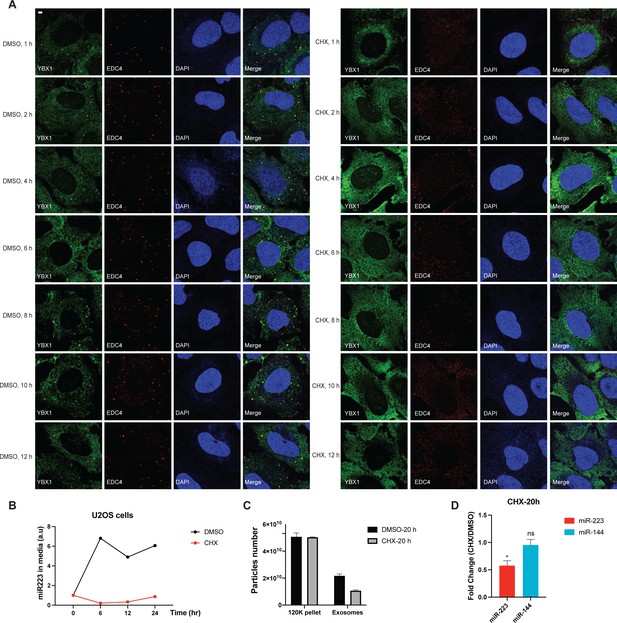
Disruption of P-body formation by CHX decreases miR-223 sorting into exosomes.
(A) P-bodies rapidly disassembled in cells treated with 10 µg/ml CHX. U2OS cells were stained with anti-EDC4 and anti-YBX1 antibodies. (B) miR-223 secretion into medium from U2OS cells with or without 10 µg/ml CHX treatment. About 200 µl cell culture medium was harvested at each time point and used to extract RNA after centrifugation at 1500 xg for 15 min to remove debris. The amount of miR-223 was quantified by RT-qPCR. Data are plotted as the fold change over time zero. Samples were normalized by total cell number. (C) Nanoparticle tracking analysis (NTA) quantification of exosomes and particles in high-speed pellet fractions from cultured U2OS cells. Exosomes were purified as in Figure 4G. (D) Quantification of miR-223 and miR-144 by RT-qPCR in purified exosomes from U2OS cells with or without 10 µg/ml CHX treatment. Exosomes were purified as in Figure 4G. Samples were normalized by total cell number. Scale bar, 3 µm.
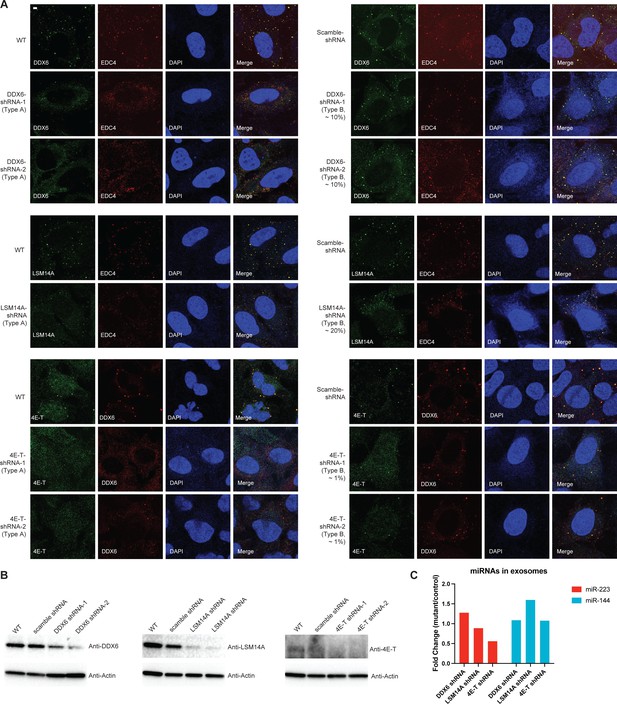
Effects of shRNA knockdown of selected P body proteins on miR-223 sorting into exosomes.
(A) P-bodies were analyzed by indirect immunofluorescence with anti-EDC4 or DDX6 antibodies, along with antibodies against the knockdown proteins in U2OS cells. Two types of cells were observed in individual knockdown cells. Type A cells: fewer and small granules; Type B cells: more abundant and big granules. (B) Proteins were analyzed by immunoblot with indicated antibodies in parental, scramble shRNA and shRNA expressing U2OS cells (Two shRNAs for DDX6 and 4E-T; one shRNA for LSM14A). (C) Quantification of miR-223 and miR-144 by RT-qPCR in purified exosomes from U2OS shRNA cells. Exosomes were purified as in Figure 4G. Mutants are cells with DDX6, LSM14A, or 4E-T shRNA; Control are cells with scrambled shRNA. Samples were normalized by total cell number. Scale bar, 3 µm.
-
Figure 7—figure supplement 3—source data 1
Uncropped Western blot images corresponding to Figure 7—figure supplement 3B.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig7-figsupp3-data1-v2.zip
-
Figure 7—figure supplement 3—source data 2
Oligo sequences used for shRNA cloning for DDX6, 4E-T, and LSM14A.
- https://cdn.elifesciences.org/articles/71982/elife-71982-fig7-figsupp3-data2-v2.docx
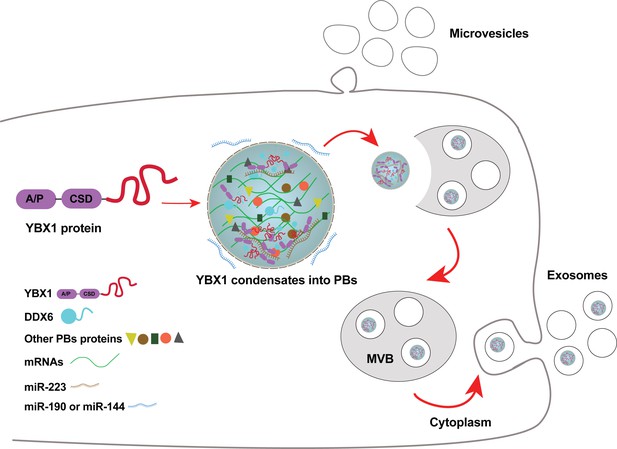
Diagram representing a working model of miRNA selectively sorted into exosomes by phase-separated YBX1 condensates.
Cytosolic RBP YBX1 forms liquid-like condensates in cells and liquid droplets in vitro. Phase separation of YBX1 is governed by a C-terminal IDR, most likely through the association of aromatic amino acid tyrosine and basic amino acids arginine or lysine. Phase-separated YBX1 recruits miRNAs in a selective manner through N-terminal CSD- mediated specific protein-RNA interaction. YBX1 condensation increases its local concentration and the affiliation with P body components (such as DDX6), which further facilitates YBX1 and its cognate miRNAs sorting into exosomes. Segregation of RNA and RBPs for capture by invagination into an endosome occurs at the level of granule formation or by sorting of selected RNAs and RBPs from larger, more heterogeneous granules.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo-sapiens) | YBX1 | Addgene | RRID:Addgene_19878 | |
| Cell line (Homo- sapiens) | U-2 OS cells | PMID:27174937 | Gift of Dr. Pavel Ivanov lab | |
| Cell line (Homo- sapiens) | U-2 OS ΔYBX1 cells | PMID:27174937 | Gift of Dr. Pavel Ivanov lab | |
| Cell line (Homo- sapiens) | HEK293T cells | Other | Cell culture facility at UC Berkeley | |
| Cell line (Homo- sapiens) | HEK293T ΔYBX1 cells | This study | Obtained by CRISPR-Cas9, cell line maintained in Schekman lab | |
| Cell line (Homo- sapiens) | U-2 OS cells | Other | Cell culture facility at UC Berkeley | |
| Cell line (Spodoptera frugiperda) | Sf9 cells | Other | Cell culture facility at UC Berkeley | |
| Recombinant DNA reagent | mCherry-Rab5CA(Q79L)(plasmid) | Addgene | RRID:Addgene_35138 | |
| Recombinant DNA reagent | EGFP-YBX1(plasmid) | This study | EGFP tagged YBX1, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | 3xFlag-YBX1(plasmid) | This study | 3xFlag tagged YBX1, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1(plasmid) | This study | YFP tagged YBX1, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-F85A(plasmid) | This study | YFP tagged YBX1-F85A, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-(128-324)-Y to S/A(plasmid) | This study | YFP tagged YBX1-(128-324)-Y to S/A, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-(128-324)-RK to G(plasmid) | This study | YFP tagged YBX1-(128-324)-RK to G, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-(128-324)-QN to G/A(plasmid) | This study | YFP tagged YBX1-(128-324)-QN to G/A, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-(128-324)-DE to G(plasmid) | This study | YFP tagged YBX1-(128-324)-DE to G, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-(128-324)-VMF to A(plasmid) | This study | YFP tagged YBX1-(128-324)-VMF to A, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-Δ (1-55)(plasmid) | This study | YFP tagged YBX1-Δ (1-55), plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-Δ (56-127)(plasmid) | This study | YFP tagged YBX1-Δ (56-127), plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-Δ (1-127)(plasmid) | This study | YFP tagged YBX1-Δ (1-127), plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-Δ (56-324)(plasmid) | This study | YFP tagged YBX1-Δ (56-324), plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-(56-127)(plasmid) | This study | YFP tagged YBX1-(56-127), plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | YFP-YBX1-Δ (128-324)(plasmid) | This study | YFP tagged YBX1-Δ (128-324), plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | His6-MBP-3C-mGFP-TEV-NotI-ccdB-AscI-stop-HindIII cassette(plasmid) | Addgene | RRID:Addgene_118890 | Gift of Dr. Anthony A. Hyman lab |
| Recombinant DNA reagent | His6-MBP-mGFP-YBX1(plasmid) | This study | To express YBX1 in insect cells, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | His6-MBP-mGFP-YBX1-F85A(plasmid) | This study | To express YBX1-F85A in insect cells, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | His6-MBP-mGFP-YBX1-(128-324)-Y to S(plasmid) | This study | To express YBX1-(128-324)-Y to S in insect cells, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | His6-MBP-mGFP-YBX1-(128-324)-RK to G(plasmid) | This study | To express YBX1-(128-324)-RK to G in insect cells, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | His6-MBP-mGFP-YBX1-Δ (128-324)(plasmid) | This study | To express YBX1-Δ (128-324) in insect cells, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | His6-MBP-mGFP-YBX1-Δ (1-127)(plasmid) | This study | To express YBX1-Δ (1-127) in insect cells, plasmid maintained in Schekman lab | |
| Recombinant DNA reagent | pX330-Venus(plasmid) | Other | Gift of Dr. Robert Tjian lab | |
| Recombinant DNA reagent | PLKO.1-Puro (plasmid) | Addgene | RRID:Addgene_10878 | Pol III based shRNA backbone |
| Antibody | anti-YBX1 (Rabbit polyclonal) | Cell Signaling Technology | RRID:AB_1950384 | WB (1:1000) |
| Antibody | anti-YBX1 (Rabbit polyclonal) | Abcam | RRID:AB_2219278 | WB (1:1000)IF (1:100) |
| Antibody | anti-CD9 (Rabbit monoclonal) | Cell Signaling Technology | RRID:AB_2798139 | WB (1:3000) |
| Antibody | anti-ALIX (Mouse monoclonal) | Santa Cruz Biotechnology | RRID:AB_673819 | WB (1:1000) |
| Antibody | anti-flotillin-2(Mouse monoclonal) | BD Biosciences | RRID:AB_397766 | WB (1:1000) |
| Antibody | Anti-CD63 (Mouse monoclonal) | Thermo Fisher Scientific | RRID:AB_2572564 | WB (1:1000)IF (1:100) |
| Antibody | Anti-Actin (Mouse monoclonal) | Abcam | RRID:AB_449644 | WB (1:3000) |
| Antibody | Anti-DDX6 (Rabbit polyclonal) | Bethyl | RRID:AB_2277216 | WB (1:1000)IF (1:100) |
| Antibody | anti-EDC4 (Mouse monoclonal) | Santa Cruz Biotechnology | RRID:AB_10988077 | WB (1:1000)IF (1:50) |
| Antibody | anti-G3BP1 (Mouse monoclonal) | Santa Cruz Biotechnology | RRID:AB_1123055 | WB (1:1000) |
| Antibody | anti-GFP (Mouse monoclonal) | Santa Cruz Biotechnology | RRID:AB_627,695 | WB (1:1000)IF (1:50) |
| Antibody | anti-GFP (Rabbit polyclonal) | Torrey Pines Biolabs | RRID:AB_10013661 | WB (1:1000)IF (1:100) |
| Antibody | anti-GM130 (Mouse monoclonal) | BD Biosciences | RRID:AB_398142 | WB (1:1000) |
| Antibody | anti-Flag (Mouse monoclonal) | Sigma-Aldrich | RRID:AB_439698 | WB (1:3000) |
| Antibody | anti-hDcp1a (Mouse monoclonal) | Santa Cruz Biotechnology | RRID:AB_2090408 | IF (1:50) |
| Antibody | anti-LSM14A (Rabbit polyclonal) | Proteintech | RRID:AB_10644339 | WB (1:1000)IF (1:100) |
| Antibody | anti-4E-T (Mouse monoclonal) | Santa Cruz Biotechnology | sc-393788 | WB (1:1000)IF (1:50) |
| Software, algorithm | Fiji | NIH | RRID:SCR_002285 | https://fiji.sc/ |
| Software, algorithm | GraphPad Prism | Other | RRID:SCR_002798 | https://www.graphpad.com |
| Software, algorithm | IUPred | PMID:15955779 | RRID:SCR_014632 | http://iupred.enzim.hu/ |
| Software, algorithm | Sigmaplot 12.5 | Other | RRID:SCR_003210 | http://www.sigmaplot.com/products/sigmaplot/ |
| Software, algorithm | NCPR | Other | http://www.bioinformatics.nl/cgi-bin/emboss/charge |





