Virophages and retrotransposons colonize the genomes of a heterotrophic flagellate
Figures
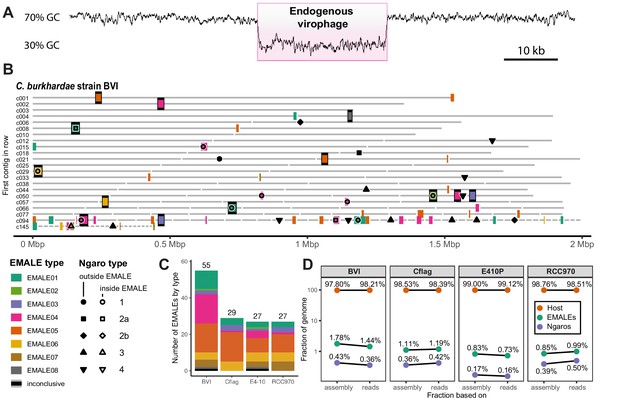
Endogenous virophages in Cafeteria burkhardae.
(A) GC-content graph signature of a virophage element embedded in a high-GC host genome. Shown is a region of contig BVI_c002 featuring an integrated virophage (pink box) flanked by host sequences. (B) Location of partial or complete virophage genomes and Ngaro retrotransposons in the genome assemblies of C. burkhardae strain BVI (see Figure 1—figure supplement 1 for all four strains). Horizontal lines represent contigs of decreasing length ordered from left to right and from top to bottom, with numbers shown for the first contig of each line; colored boxes indicate endogenous mavirus-like elements (EMALEs). Fully assembled elements are framed in black. Ngaro retrotransposon positions are marked by black symbols; open symbols indicate Ngaros integrated inside a virophage element. (C) Graphic summary of the number and types of all EMALEs identified in each of the four C. burkhardae strains. (D) Nucleotide contributions of EMALEs and Ngaros to Cafeteria genomes. Fractions for each strain are computed based on nucleotides in the assembly (left) and nucleotides in the reads (right) mapping to the different parts of the assembly.
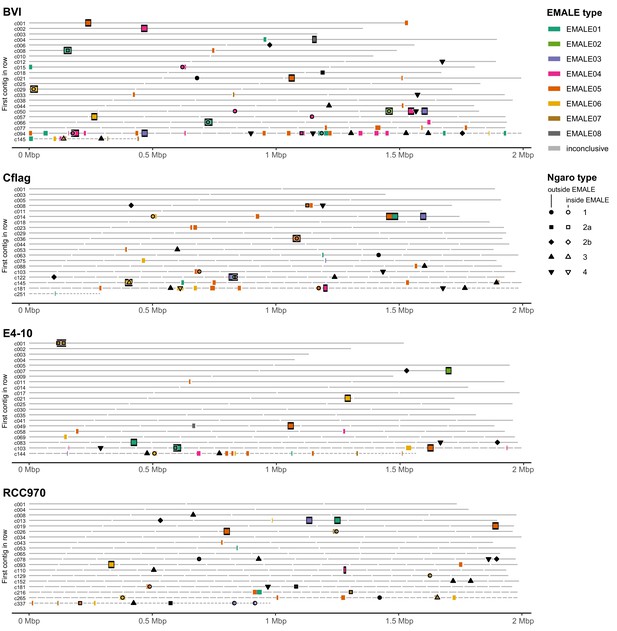
Distribution of EMALE and Ngaro retrotransposon integration sites in the four Cafeteria burkhardae genome assemblies.
Location of partial and complete virophage genomes and Ngaro retrotransposons in the genome assemblies of four C. burkhardae strains. Horizontal lines represent contigs in order of decreasing length; colored boxes indicate the insertion sites of EMALEs. Complete elements are framed in black. Ngaro retrotransposon insertion sites are marked by shapes corresponding to the different Ngaro types, open symbols indicate insertions in EMALEs.
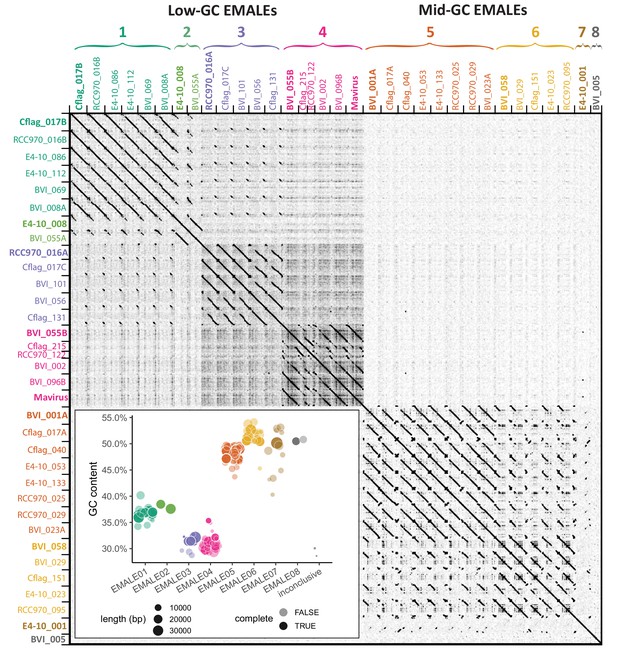
Classification of endogenous virophages based on DNA dot plot analysis.
The self-versus-self DNA dot plot of concatenated sequences of 33 complete EMALE genomes and mavirus reveals two main block patterns, corresponding to EMALEs with low (29–38%) GC-content and medium (47–53%) GC-content. Smaller block patterns define EMALE types 1–8. EMALE identifiers indicate the host strain and contig number where the respective element is found. Multiple EMALEs on a single contig are distinguished by terminal letters. Elements printed in bold represent the type species shown in Figure 3. Inset: GC-content distribution of complete and partial EMALEs labeled ‘complete: TRUE/FALSE’. Some partial EMALEs were too short for type assignment and are thus inconclusive. Retrotransposon insertions, where present, were removed prior to analysis.

Type assignment for incomplete EMALEs.
For partial EMALEs, we assigned types in an automated manner based on blastx hits to type species EMALEs. For each EMALE, blast hits to type species proteins along the genome are shown. Hit positions on the y-axis reflect bitscores, with the highest-scoring hits plotted at the top. Integrated Ngaro retrotransposons are shown as gray boxes.
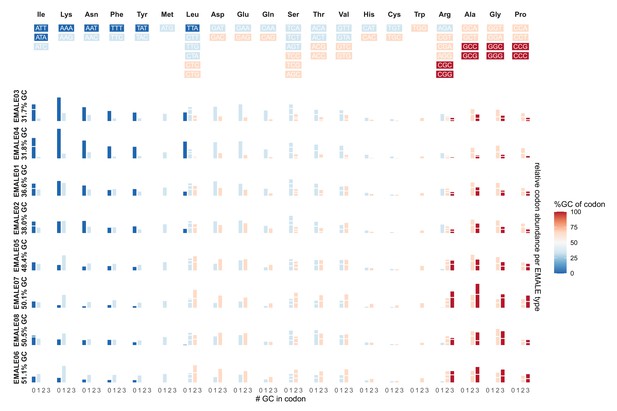
Codon usage with respect to GC-content in different EMALE types.
Each row represents one of the eight EMALE types, ordered by decreasing GC-content. Each column represents a single amino acid. The different codons coding for each amino acid are indicated above the plot, color-coded according to their GC-content. The histograms indicate the relative contributions of each codon with a certain GC-content to the overall amino acid composition of each EMALE. Codons with identical GC-content encoding the same amino acid are represented by stacked bars separated by white lines.
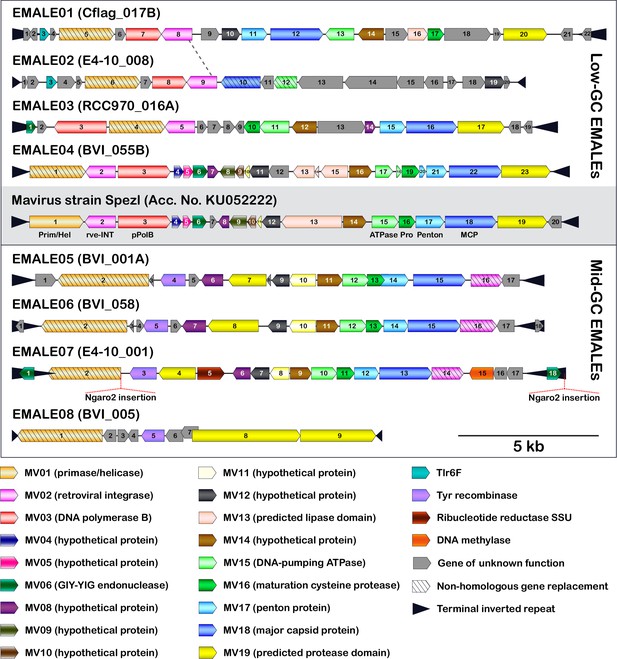
Genome organization of eight EMALE types found in Cafeteria burkhardae.
Shown are schematic genome diagrams of the EMALE type species 1–8; for all 33 complete EMALEs, see Figure 3—figure supplement 1. The reference mavirus genome with genes MV01-MV20 is included for comparison. Homologous genes are colored identically; genes sharing functional predictions but lacking sequence similarity to the mavirus homolog are hatched. Open reading frames are numbered individually for each element. Ngaro retrotransposon insertion sites are indicated where present. The dotted line between EMALE01 and EMALE02 separates a homologous region (left) from unrelated DNA sequences (right) and thus indicates the location of a probable recombination event.
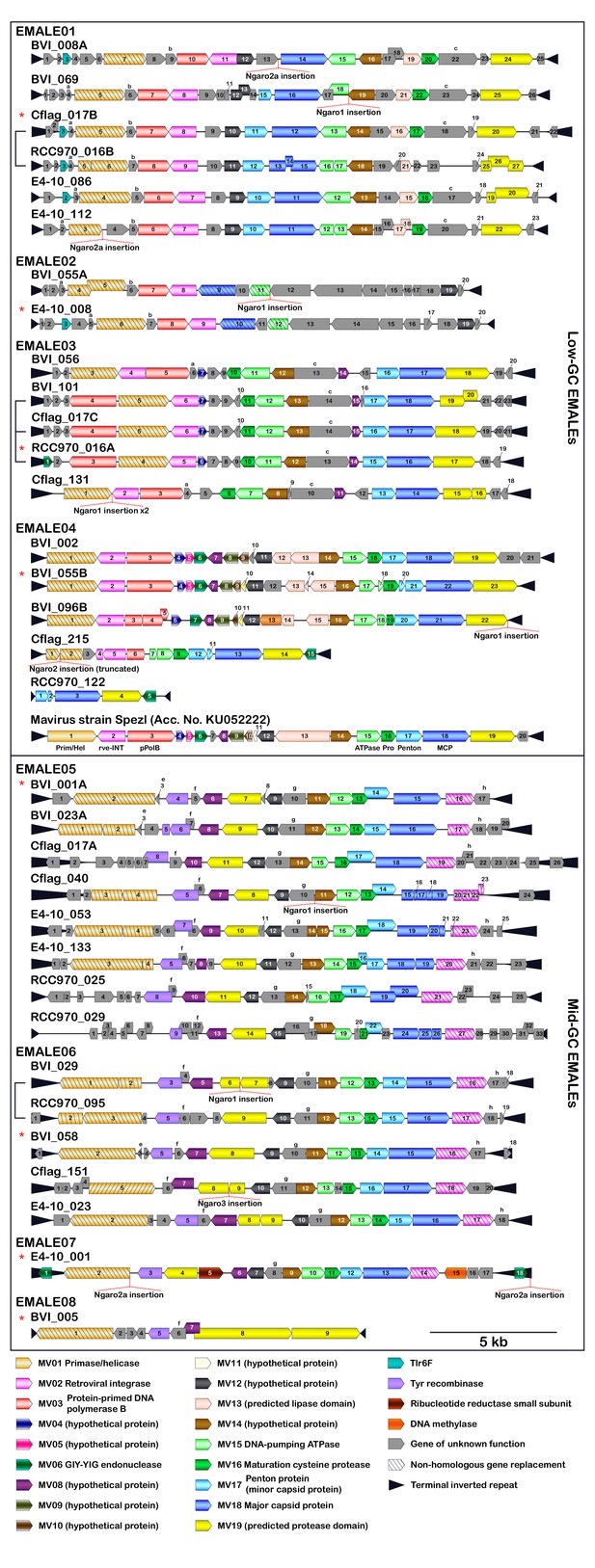
Coding capacity of 33 completely assembled EMALEs in Cafeteria burkhardae.
Shown are genome diagrams for 33 EMALEs in four C. burkhardae strains (BVI, Cflag, E4-10, RCC970).The reference mavirus genome with genes MV01-MV20 is included for comparison. EMALE identifiers consist of host strain name, followed by contig number and sometimes letters to distinguish between several EMALEs on the same contig. Directional boxes indicate open reading frames (ORFs) in the respective orientation. Homologous genes that are present in mavirus or have a predicted function are shown in color. Other homologous ORFs are denoted by lowercase letters. Black triangles represent terminal inverted repeats (TIRs). Brackets indicate EMALEs with homologous integration sites in different host strains. Ngaro retrotransposon insertions are shown when present. Asterisks denote type elements as shown in Figure 3.
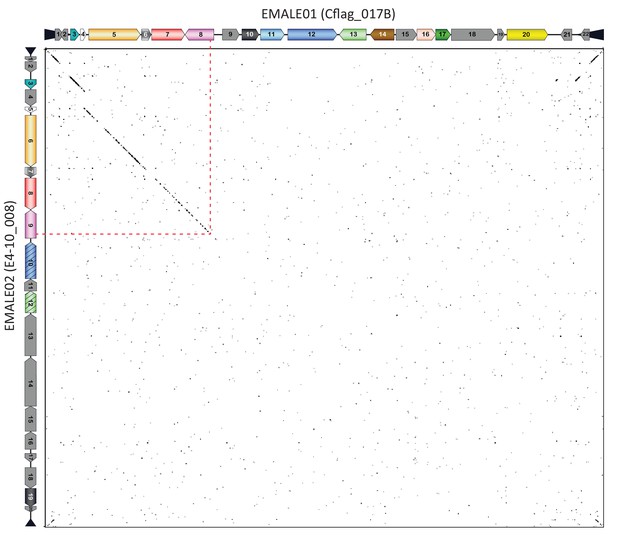
Partial synteny between EMALE01 and EMALE02.
DNA dot plot analysis of EMALE01 Cflag_017B and EMALE02 E4-10_008 showing predicted genes along the axes. The synteny ends within the rve-family integrase (rve-INT) gene, which represents the presumed recombination site (red dotted line). For the open reading frame (ORF) color legend, see Figure 3 and Figure 3—figure supplement 1.
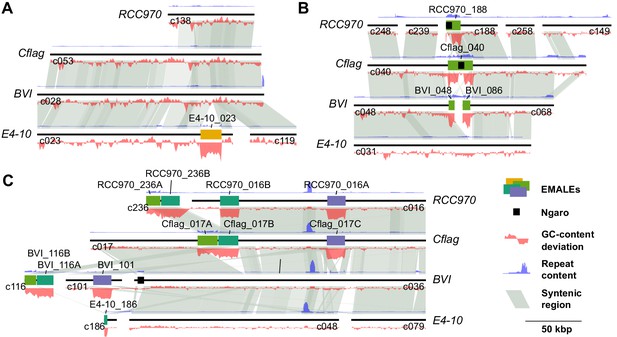
Unique and orthologous EMALE integration loci among four Cafeteria strains.
Synteny plots of three genomic loci illustrate different scenarios of EMALE conservation in Cafeteria burkhardae. Homologous DNA regions are connected by gray shadings. Peaks in the blue curve indicate repetitive regions, red curves represent GC-content. (A) EMALE E4-10_023 represents a unique integration site in host strain E4-10. Syntenic regions in other host strains are well resolved and devoid of EMALEs. (B) Homologs of EMALE Cflag_040 are found in orthologous loci in host strains RCC970 and BVI. The Ngaro retrotransposon in this EMALE apparently caused assembly problems, resulting in premature termination of RCC970 contig 188 and splitting the EMALE onto two contigs in BVI. (C) Comparative analysis of the three EMALEs on Cflag contig 17 reveals a complex situation. The double EMALE Cflag_017 A/B has homologs in BVI and RCC970, albeit as partial elements on short contigs. Most of RCC970 contig 16 is syntenic to Cflag contig 17, except for the shorter flanking region of EMALE RCC970_016B, which is likely caused by mis-assembly of the RCC970 contig at the double-EMALE transition. In the BVI assembly, the flanking regions of EMALE Cflag_017 C are present twice, on the EMALE-containing contig 101 and on the EMALE-free contig 36, suggesting a heterozygous condition in BVI.
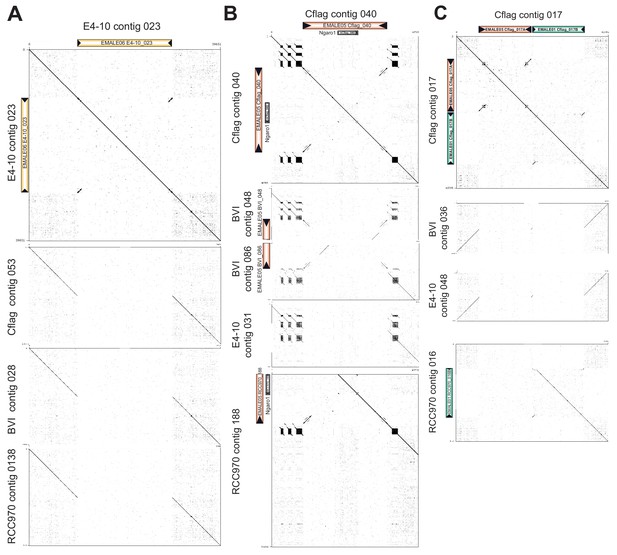
DNA dot plots of selected EMALE loci as shown in Figure 3—figure supplement 3.
Shown are DNA dot plots of EMALE(s) including 10 kb of flanking host DNA versus itself and versus syntenic regions in other host strains. Black triangles represent EMALE terminal inverted repeats (TIRs). (A) EMALE E4-10_023 is integrated in non-repetitive host DNA and represents a unique insertion in strain E4-10, with EMALE-free loci in the other three strains. (B) EMALE Cflag_040 is integrated in a cluster of complex host repeats and has homologous integration sites in strains BVI and RCC970. (C) The double EMALE Cflag_017 A/B likely caused mis-assembly in strain RCC970.
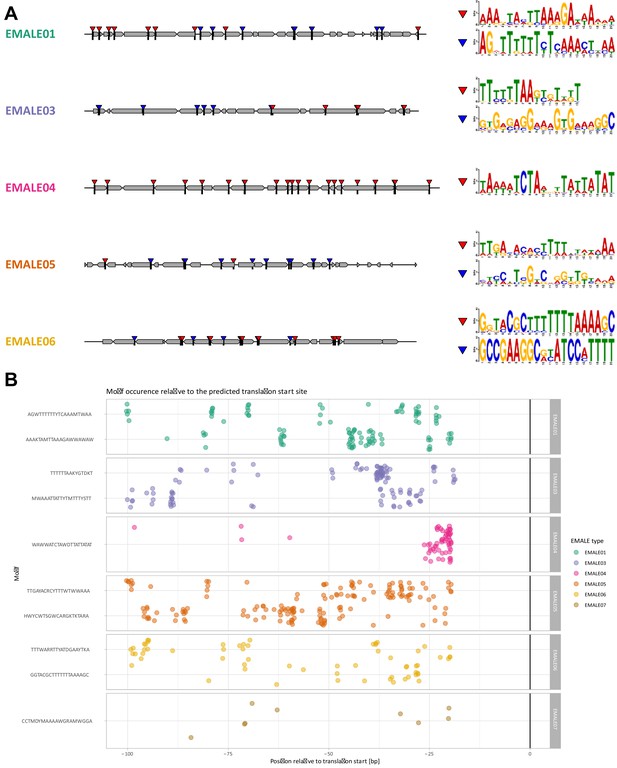
Putative promoter motifs in EMALE genomes.
(A) Sequence logos of high-scoring motifs predicted with MEME in immediate upstream regions of coding sequences and their positions in EMALE genomes. Character height at each position of a sequence logo corresponds to the frequency of the respective nucleotide at that position. (B) EMALE promoter motif occurrences relative to predicted translation start sites. Each dot corresponds to the start of a predicted promoter motif plotted relative to the ATG start codon of the downstream gene. Motifs are grouped by the EMALE type in which they were initially predicted.
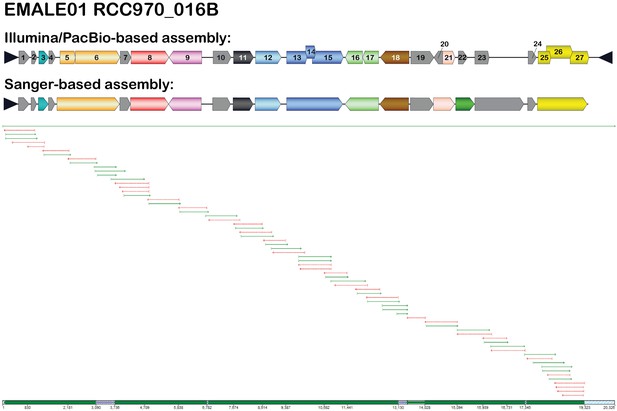
Correction of Illumina/PacBio-based assemblies by PCR and Sanger sequencing.
EMALE01 RCC970_016B was re-evaluated by PCR analysis and subsequent Sanger sequencing of PCR products. The resulting assembly is compared to the Illumina/PacBio-based assembly (top). The bottom part of the figure shows a Sequencher screenshot of the assembled Sanger reads. The long green arrow represents the Illumina/PacBio sequence, the shorter green and red arrows represent individual Sanger reads.
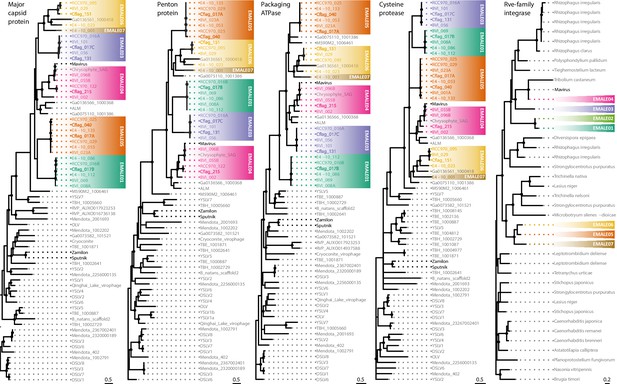
Phylogenetic reconstruction of conserved EMALE proteins.
Unrooted maximum likelihood trees were constructed from multiple sequence alignments of the four virophage core proteins major capsid protein (MCP), penton protein (PEN), ATPase, and protease (PRO), as well as of the retroviral integrase. Nodes with bootstrap values of 80% or higher are marked with dots. EMALEs are color-coded by type; cultured virophages are printed in bold. ALM, Ace Lake Mavirus; DSLV, Dishui Lake virophage; OLV, Organic Lake virophage; RVP, rumen virophage; TBE/TBH, Trout Bog Lake epi-/hypolimnion; YSLV, Yellowstone Lake virophage. Metagenomic sequences starting with Ga and M590 are derived from Paez-Espino et al., 2019.
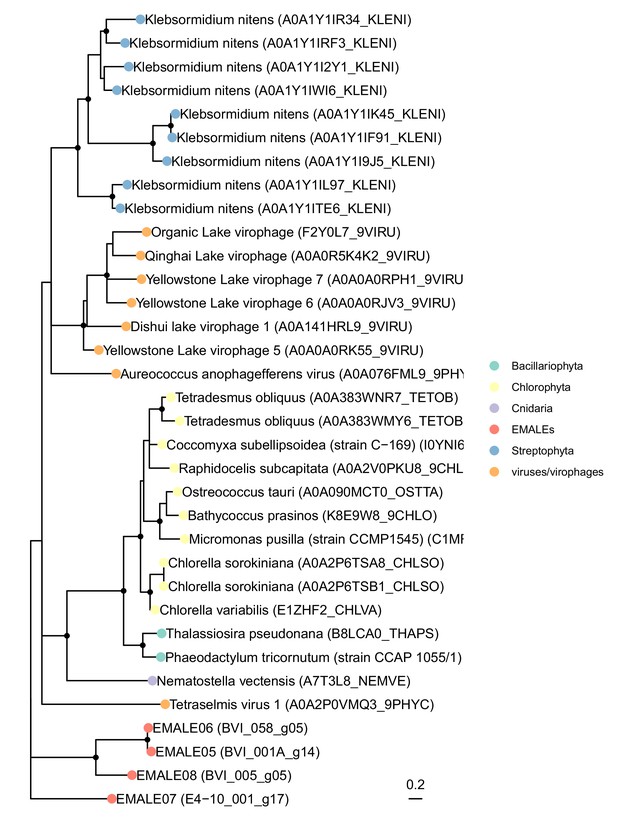
Maximum likelihood reconstruction of EMALE tyrosine recombinase phylogenies.
Shown is an unrooted ML phylogenetic tree of tyrosine recombinases comparing EMALE proteins (red dots) to their most similar sequences in UniProt (color-coded at the phylum level). Nodes with <50% bootstrap support were collapsed; nodes with >80% bootstrap support are marked by solid black dots.
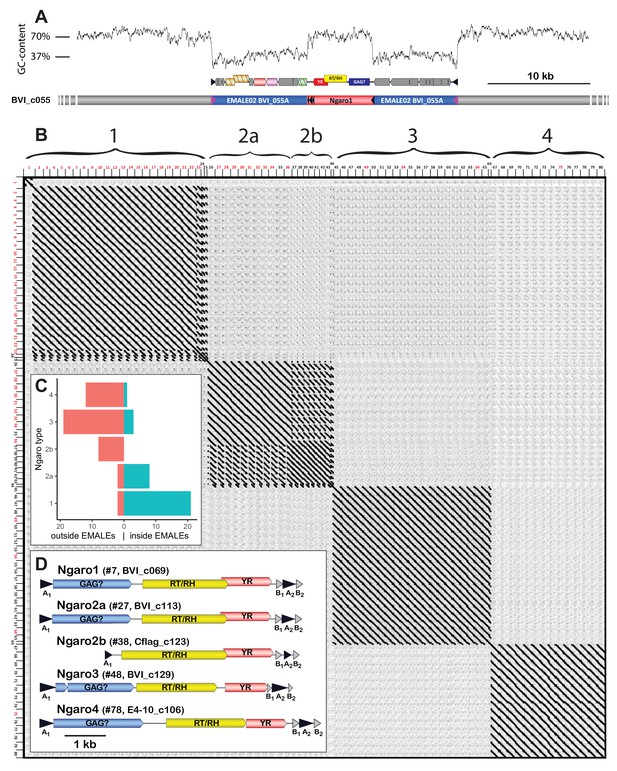
Ngaro retrotransposons in Cafeteria burkhardae.
(A) Genomic profile of an EMALE-integrated Ngaro element showing a GC-content graph (top), open reading frame (ORF) organization of EMALE and Ngaro (middle), and a schematic overview of the three genomic entities (bottom; host: gray, EMALE: blue, Ngaro: red). (B) Self-versus-self DNA dot plot of 80 concatenated Ngaro sequences. Block patterns define Ngaro types 1–4. Ngaros are numbered according to Supplementary file 1, with red numbers indicating retrotransposons inserted in EMALEs. (C) Distribution of Ngaro integration loci in EMALE and host DNA. Ngaro types 1 and 2a show a clear preference for EMALE loci, in contrast to Ngaro types 2b, 3, and 4 that are mostly found in host loci. (D) Coding potential of C. burkhardae Ngaro retrotransposons, shown for one example per type with their host strain and contig numbers listed. Triangles indicate direct repeats. GAG, group specific antigen; RT, reverse transcriptase; RH, ribonuclease H; YR, tyrosine recombinase.
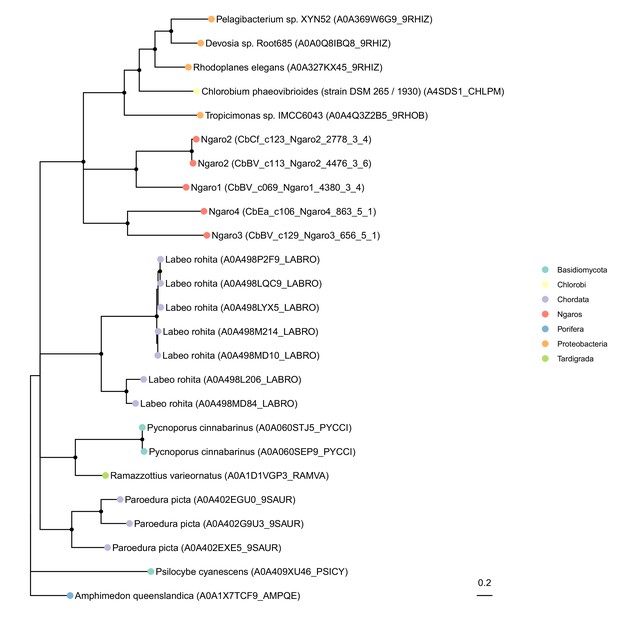
Phylogenetic placement of Ngaro tyrosine recombinases.
Shown is an unrooted maximum likelihood phylogenetic tree of tyrosine recombinases comparing Cafeteria Ngaro-encoded proteins (red dots) with their most similar sequences in UniProt (color-coded at the phylum level). Nodes with <50% bootstrap support were collapsed; nodes with >80% bootstrap support are marked by black dots. The distribution across a very broad range of phyla (fish, fungi, sponges, tardigrades, and bacteria) indicates that this integrase is likely associated with highly mobile transposons that have broad host ranges, confirming previous studies on these elements (Poulter and Goodwin, 2005).
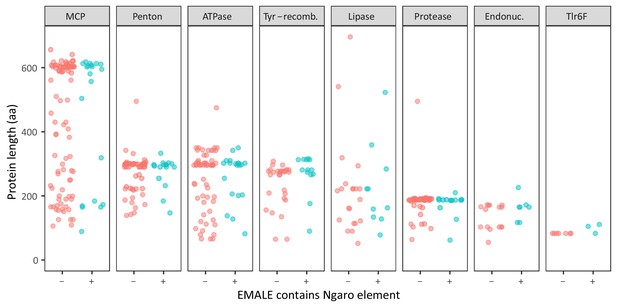
Protein length distributions in EMALEs with and without retrotransposons.
Integration of Ngaro retrotransposons might lead to the inactivation of EMALEs, thereby promoting their degeneration and the pseudogenization of their genes. To test this hypothesis, we compared the predicted protein lengths for conserved genes in EMALEs without (red) and with transposons (blue). Gene length here serves as a proxy for pseudogenization because this process leads to the emergence of premature stop codons and frameshifts, which in turn leads to shorter annotated genes.
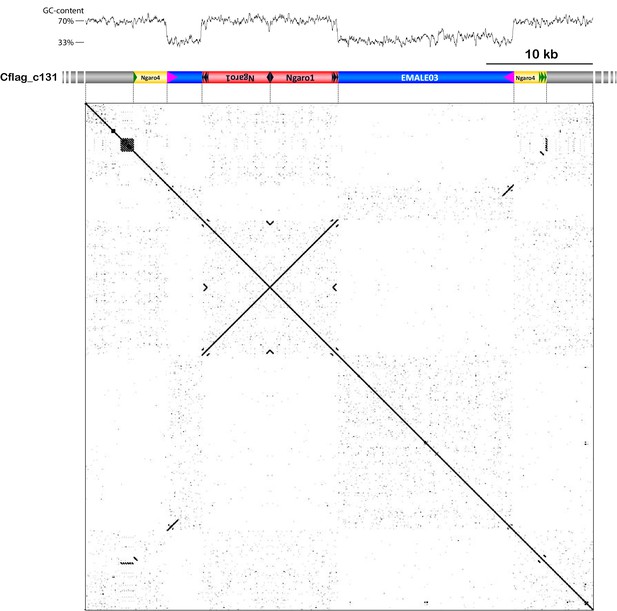
Nested integration scenario involving one EMALE and three Ngaro retrotransposons.
EMALE03 Cflag_131 (blue) is inserted into a type 4 Ngaro retrotransposon (yellow) and contains two insertions of type 1 Ngaros (red). The nested integration scenario is depicted by a GC-content graph (top), a graphic representation of the respective genomic region on contig 131 of Cafeteria burkhardae strain Cflag (middle), and a self-versus-self DNA dot plot (bottom). Host sequence is shown in gray; terminal repeats are represented by colored triangles.
Additional files
-
Supplementary file 1
Sheet 1: Endogenous mavirus-like element (EMALE) statistics. This dataset contains information on each of the 138 EMALEs in four Cafeteria burkhardae strains, including their exact location in the host assembly, length, presence of terminal inverted repeats, type score, and Ngaro insertions.
Sheet 2: EMALE integration sites. This dataset lists information for each of the 33 fully assembled EMALEs regarding orthologous integration loci in all four host strains, target site duplications, and host genomic context of the integration loci.
Sheet 3: Ngaro statistics. This dataset contains information for 80 Ngaro retrotransposons identified in the four C. burkhardae assemblies, including their exact location in the host assembly, length, type, and insertion locus (EMALE or eukaryotic chromatin).
Sheet 4: Primer sequences. List of oligonucleotides used as PCR and sequencing primers for the validation of EMALE01 RCC970_016B. Numbers in the last column refer to the sequenced PCR products of the assembly diagram shown in Figure 3—figure supplement 6, starting with number 1 in the top left corner and ending with number 66 in the bottom right corner.
- https://cdn.elifesciences.org/articles/72674/elife-72674-supp1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/72674/elife-72674-transrepform1-v1.pdf





