DAAM mediates the assembly of long-lived, treadmilling stress fibers in collectively migrating epithelial cells in Drosophila
Figures
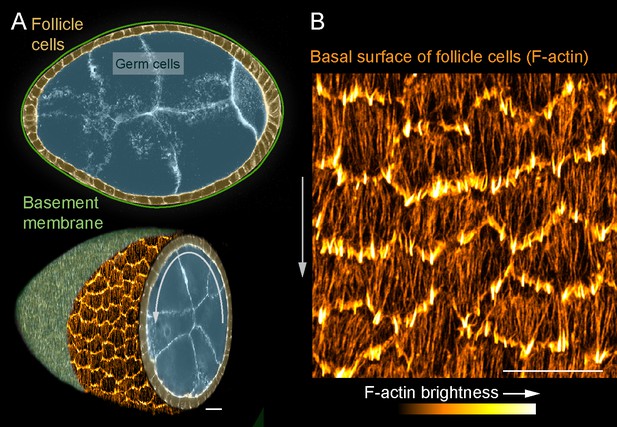
Introduction to the stress fibers (SFs) in collectively migrating follicle cells.
(A) Composite images of an egg chamber (pseudocolored), transverse section above and 3D cutaway view below. Curved arrow shows the rotational migration of the follicle cells as they crawl along the basement membrane extracellular matrix (ECM) (drawn as a line in upper image, from a confocal section of Collagen-IV-GFP in lower image). (B) Image of the basal surface of the follicular epithelium. Each cell has a leading-edge protrusion (yellow) and a parallel array of SFs (orange) oriented in the direction of migration. Experiments performed at stage 7. Gray arrows show migration direction. Scale bars 10 µm.
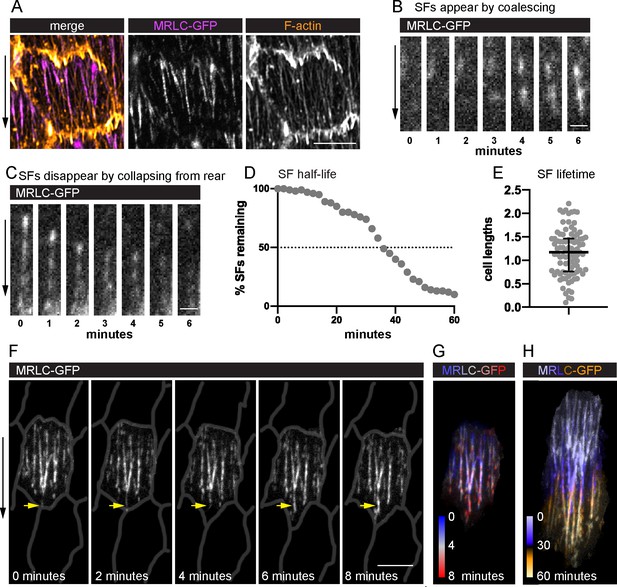
Migrating follicle cells have long-lived, treadmilling stress fibers (SFs).
(A) Image of one cell showing that myosin regulatory light chain (MRLC-GFP) labels SFs, but not leading-edge protrusions. Image shown is representative of five egg chambers. See Figure 2—figure supplement 1 for comparison of live actin labels. Still images from videos showing an SF (B) forming by MRLC-GFP coalescence (n = 64 SF appearances), and (C) disappearing by collapsing from the rear (n = 75 SF disappearances). Quantification of SF lifetimes. (D) Half-life measurement in real time. (E) Lifetimes as a function of how long it took the cell to migrate one cell length. n = 91 SFs from 23 cells in 3 egg chambers. Bars in (E) show median and interquartile ranges. (F) Still images from a video of an optically isolated cell showing an SF tip growing as the cell migrates (arrow). Cell outlines are drawn from CellMask membrane label. See Figure 2—video 1; Figure 2—video 2, and Figure 2—figure supplement 2 for images of the original membrane label and an optically isolated single SF. (G, H) Temporal projections of SFs from the cell in (F). (G) Shows the same period as (F) at 20-s intervals. (H) Shows the period required for the cell to migrate ~1 cell length. Experiments performed at stage 7. Black arrows show migration direction. Scale bars: 5 µm (A, F), 1 µm (B, C).
-
Figure 2—source data 1
Excel file containing the numeric data used to generate the graphs in Figure 2.
Each tab on the spreadsheet corresponds to one graph.
- https://cdn.elifesciences.org/articles/72881/elife-72881-fig2-data1-v1.xlsx
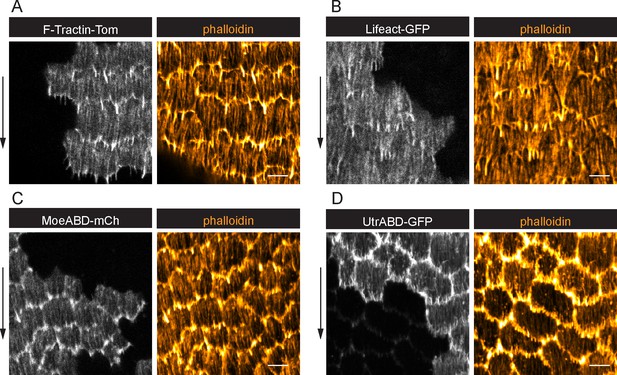
Treadmilling stress fibers (SFs) marked with live F-actin labels.
(A–D) Images of epithelia with clones of cells expressing live F-actin labels. In all cases, labeling of leading-edge protrusions obscures the ends of the SFs. LifeAct and Utr-ABD (B, D) also significantly alter F-actin organization compared to wild-type cells stained with phalloidin. In D, very weak constitutive expression of Utr is seen outside of clones. Images shown are representative of n = 4, 6, 4, 8 egg chambers. Experiments performed at stages 7 and 8. Arrows show migration direction. Scale bars: 5 µm.
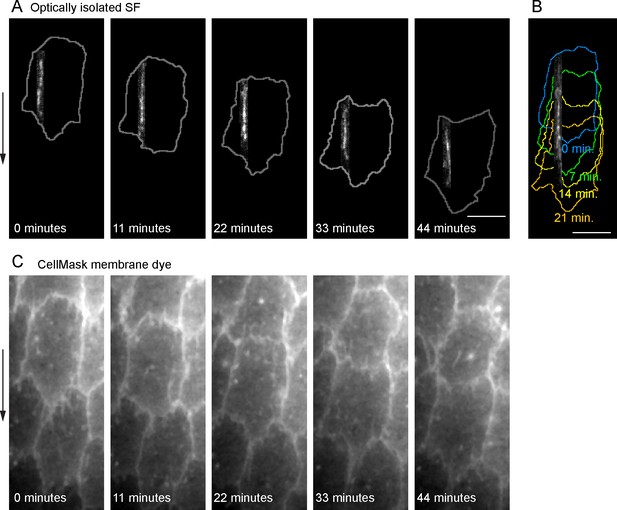
Treadmilling of an optically isolated stress fiber (SF).
(A) Still images from a video showing an optically isolated SF over 44 min; cell outlines are shown in gray. (B) Maximal projection of the same SF as in (Figure 2F), shown over 21 min; colored cell outlines correspond to time intervals shown. (C) Still images from a video showing the original membrane label for Figure 2F. Experiments performed at stages 7 and 8. Arrows show migration direction. Scale bars: 5 µm.
Time-lapse video of a field of follicle cells with plasma membranes labeled with CellMask and stress fibers (SFs) labeled with myosin regulatory light chain (MRLC)-GFP.
Note that individual SFs persist as the cells migrate. Near total internal reflection fluorescence microscopy (near-TIRFM), recorded with Nikon ECLIPSE-Ti with 20-s imaging interval. Playback rate 30 frames/s. Related to Figure 2F.
Time-lapse video of an optically isolated follicle cell with stress fibers (SFs) labeled with myosin regulatory light chain (MRLC)-GFP.
Note MRLC-GFP being added to the tips of existing SFs as the cell migrates. Near total internal reflection fluorescence microscopy (near-TIRFM), recorded with Nikon ECLIPSE-Ti with 20-s imaging interval. Playback rate 30 frames/s. Related to Figure 2F.
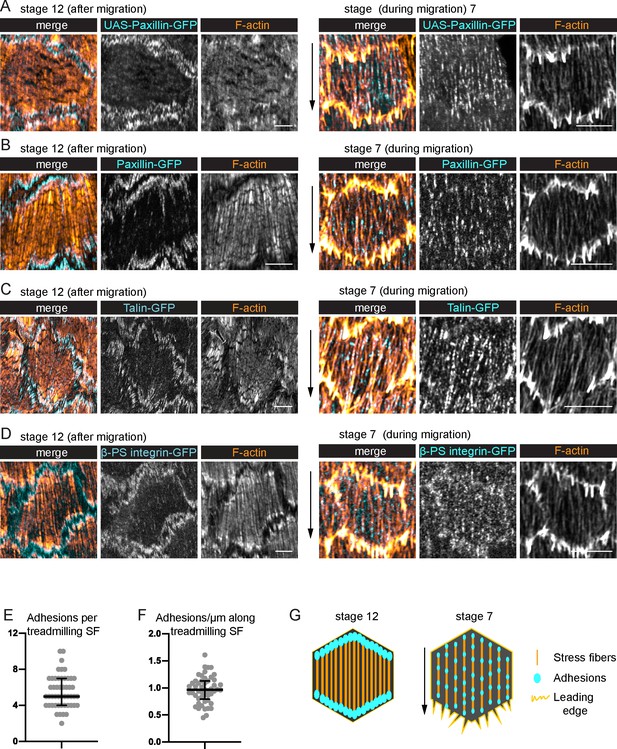
Treadmilling stress fibers (SFs) have many adhesions along their lengths.
(A–D) Images of SFs in single cells with adhesions labeled. After migration stops, there are large adhesions at the ends of the SFs. During migration, there are many smaller adhesions along their lengths. (A) UAS-Paxillin-GFP (images shown representative of 4 stage 12 and 3 stage 7). (B) Paxillin-GFP (n = 4, 5). (C) Talin-GFP (n = 3, 3). (D) βPS-integrin-GFP (n = 4, 3). (B–D) The GFP on the indicated protein is a functional, endogenous tag. (E, F) Quantification of the adhesions associated with individual SFs using Paxillin-GFP. (E) Number. (F) Linear density. n = 277 adhesions from 11 cells in five egg chambers. Bars show medians and interquartile ranges. See Figure 3—figure supplement 1 for method for counting adhesions. (G) Illustration of SF structure in postmigratory and migratory cells. Black arrows show migration direction. Scale bars: 5 µm.
-
Figure 3—source data 1
Excel file containing the numeric data used to generate the graphs in Figure 3.
Each tab on the spreadsheet corresponds to one graph.
- https://cdn.elifesciences.org/articles/72881/elife-72881-fig3-data1-v1.xlsx
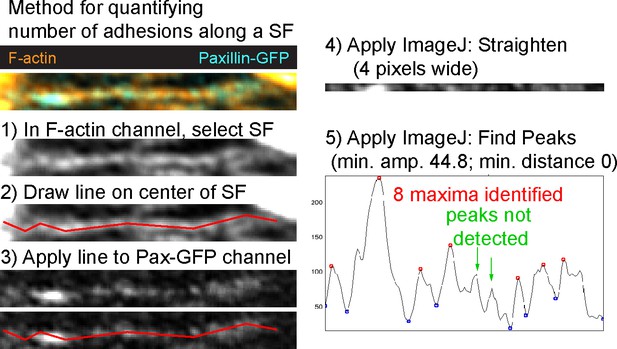
Method for quantifying number of adhesions along a stress fiber (SF).
For the example shown, the method identified seven peaks (maxima), all of which correspond to easily visible adhesions. Two potential maxima did not meet the threshold for inclusion. These peaks correspond to faint possible adhesions (green arrows), which shows that this method provides a conservative estimate of the number of adhesions along a SF.
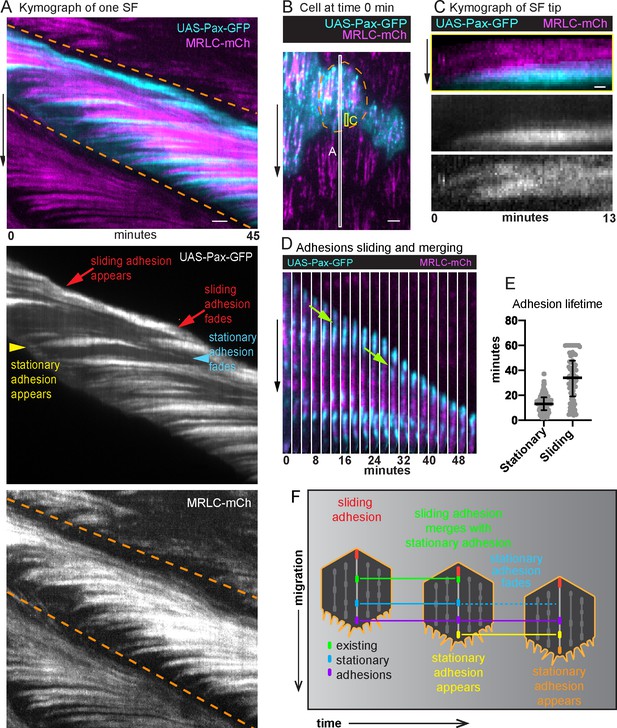
Adhesions are continuously added and removed from treadmilling stress fibers (SFs).
(A) Kymograph of one SF from the white boxed region in (B) showing the appearance and disappearance of adhesions and actomyosin segments over time. The orange dashed lines mark the leading and trailing edges of the cell, respectively, and their diagonal orientation indicates the movement of the cell edges over time. The yellow arrowhead marks the addition of a new adhesion to the front of the SF. The slight rearward curve at the front of the trace suggests that the adhesion matures under tension. The trace then remains largely horizontal showing that the adhesion is stationary relative to the substrate. The trace fades as the cell rear approaches, showing adhesion disassembly. The many horizontal traces show that multiple adhesions are added to the same SF over time. Finally, the red arrows highlight the appearance and disappearance of a distinct adhesion at the rear. The diagonal orientation of this trace indicates that the final adhesion moves with the cell’s trailing edge. The image shown is representative of kymographs for eight SFs. (B) Still image from a video of an epithelium in which all cells express myosin regulatory light chain (MRLC)-mCherry (mCh) and a subset of cells expresses UAS-Paxillin-GFP. Dashed line surrounds one cell. White and yellow boxes correspond to kymographs in (A) and (C), respectively. See Figure 4—video 1. (C) Kymograph of a SF tip from the yellow boxed region in (B), showing that Paxillin-GFP and MRLC-mCh levels increase in synchrony as the SF grows at the front. The image shown is representative of kymographs for seven SFs. (D) Still images from a video showing a sliding adhesion that persists for at least 50 min and merges with three stationary adhesions (green arrows). See Figure 4—video 2. (E) Quantification of adhesion lifetimes. In order on graph, n = 134, 84 adhesions from 23 cells in 3 egg chambers. Bars show medians and interquartile ranges. (F) Illustration summarizing adhesion dynamics in treadmilling SFs. Experiments performed at stage 7. Black arrows show migration direction. Scale bars: 2 µm (A, B), 0.5 µm (C).
-
Figure 4—source data 1
Excel file containing the numeric data used to generate the graph in Figure 4E.
- https://cdn.elifesciences.org/articles/72881/elife-72881-fig4-data1-v1.xlsx
Time-lapse video of a field of follicle cells.
The stress fibers (SFs) are labeled in all cells with myosin regulatory light chain (MRLC)-mCherry (mCh). Adhesions are labeled in a subset of cells by da-Gal4 driving patchy expression of UAS-Pax-GFP. Near total internal reflection fluorescence microscopy (near-TIRFM), recorded with Nikon ECLIPSE-Ti with 10-s imaging interval. Playback rate 30 frames/s. Rotated 14 degrees with bicubic interpolation. Related to Figure 4B.
Time-lapse video of one stress fiber (SF) in which myosin is labeled with myosin regulatory light chain (MRLC)-mCherry (mCh) and adhesions labeled with UAS-Pax-GFP.
Note the sliding behavior of the final adhesions. Near total internal reflection fluorescence microscopy (near-TIRFM), recorded with Nikon ECLIPSE-Ti with 10-s imaging interval. Playback rate 30 frames/s. Rotated 14 degrees with bicubic interpolation. Related to Figure 4D.
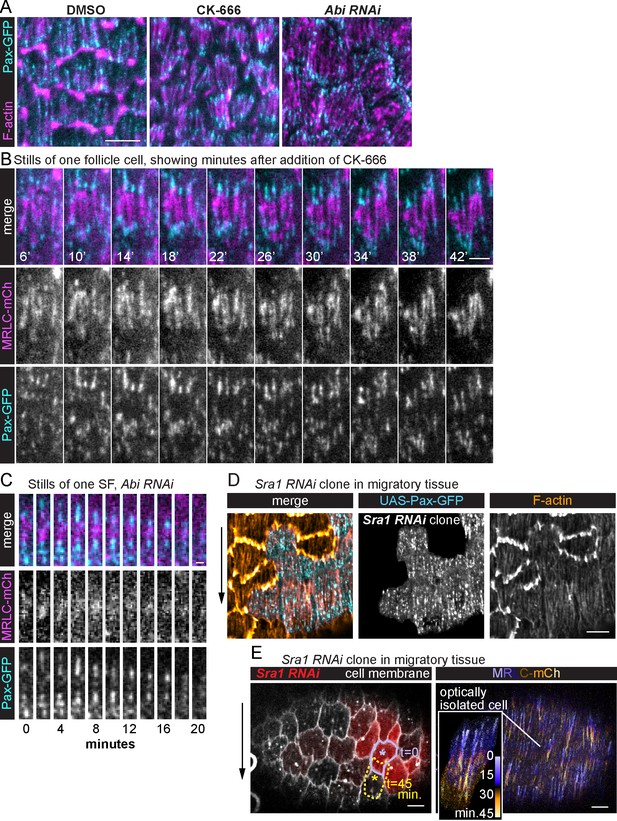
Stress fiber (SF) treadmilling depends on cell migration.
(A) Images of epithelia in which migration has been blocked by eliminating leading-edge protrusions. Adhesions become concentrated at the SF ends. The images shown are representative of n = 28 (Dimethyl Sulfoxide [DMSO]), n = 29 (CK-666), and n = 16 (Abi RNAi) egg chambers that were analyzed. (B) Still images from a video of one cell showing that internal adhesions disappear, and end adhesions grow as addition of CK-666 slowly brings migration to a stop. The image shown is representative of eight egg chambers that were analyzed. See also Figure 5—video 1. (C) Still images from a video showing one SF in an epithelium in which migration has been blocked. The SF shortens and disappears with no new adhesions added to the ends. The images shown are representative of most SFs in eight egg chambers that were analyzed. (D) Image of a migrating epithelium with a clone of cells expressing Sra1 RNAi to eliminate protrusions. The SFs in the clone maintain internal adhesions. The image shown is representative of six egg chambers that were analyzed. (E) Still image from a video of a migrating epithelium with a clone of cells that expresses Sra1 RNAi to eliminate protrusions (left). Outline shows the movement of one cell over 45 min (lavender to yellow asterisks). Temporal projection of the SFs in the same epithelium at 20-s intervals (right). Inset shows SF growth in the Sra1 RNAi cell marked with the asterisk on the left. The image shown is representative of three egg chambers that were analyzed. Experiments performed at stage 7. Black arrows show migration direction. Scale bars: 5 µm (A, D, E), 1 µm (B), 2 µm (C).
Time-lapse video of one follicle cell in which stress fibers (SFs) are labeled with myosin regulatory light chain (MRCL)-mCherry (mCh) and adhesions labeled with endogenously tagged Pax-GFP.
Treatment with the arp2/3 inhibitor CK-666 causes adhesions to become concentrated at SF ends. The time shown reflects minutes after addition of CK-666 (i.e., movie starts at 6 min). Near total internal reflection fluorescence microscopy (near-TIRFM), recorded with Nikon ECLIPSE-Ti with 10-s imaging interval. Playback rate 30 frames/s. Rotated 25 degrees with bicubic interpolation. Related to Figure 5B.
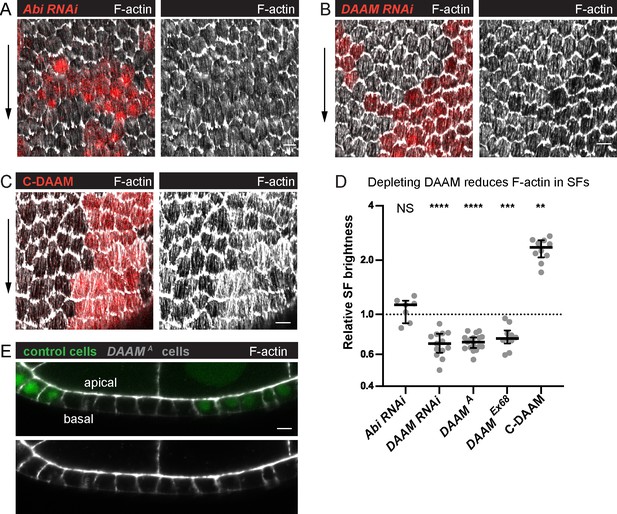
Dishevelled-associated activator of morphogenesis (DAAM) contributes to treadmilling stress fiber (SF) assembly.
(A–C) Images of epithelia with clones of cells expressing various transgenes. (A) Abi RNAi reduces F-actin in protrusions but not SFs. (B) DAAM RNAi reduces F-actin in SFs. (C) An activated form of DAAM (C-DAAM) increases F-actin in SFs. (D) Quantification of the data in (A–C). Each point is the ratio of the mean value for F-actin levels in SFs from 10 experimental cells and 10 nearby control cells in the same egg chamber. In order on graph, n = 7, 13, 17, 10, 10 egg chambers. Bars show medians and interquartile ranges. Two-tailed Wilcoxon matched pairs signed ranks test. NS (not significant) p > 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See Figure 6—figure supplement 1 for tests of additional actin assembly factors. (E) Image of a transverse section through an epithelium with a clone of DAAMA mutant cells, representative of 11 egg chambers. Loss of DAAM does not obviously reduce cortical F-actin on lateral or apical surfaces. Experiments performed at stage 7. Black arrows show migration direction. Scale bars: 5 µm.
-
Figure 6—source data 1
Excel file containing the numeric data and exact p values used to generate the graphs in Figure 6 and Figure 6—figure supplement 1.
Each tab on the spreadsheet corresponds to one graph.
- https://cdn.elifesciences.org/articles/72881/elife-72881-fig6-data1-v1.xlsx
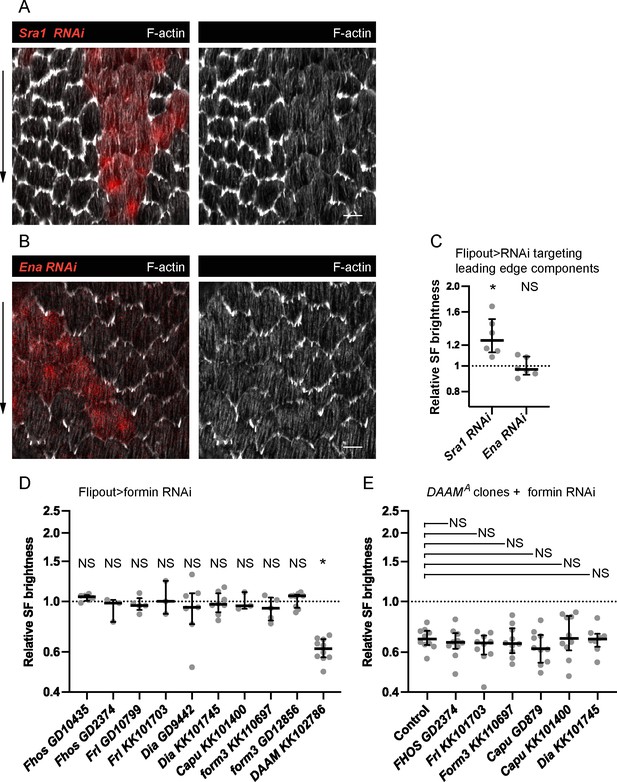
A screen for F-actin assembly factors that mediate treadmilling stress fiber (SF) formation.
Images of epithelia with clones of cells expressing (A) Sra1 RNAi and (B) Ena RNAi. F-actin levels are reduced in leading-edge protrusions but not in SFs. (C) Quantification of the data in (A, B). F-actin levels in SFs are the same or higher than controls for both conditions. In order on graph, n = 6, 6 egg chambers. (D) Quantification of SF F-actin levels in clones of cells expressing RNAi against various formins. In order on graph, n = 6, 3, 5, 3, 7, 8, 3, 5, 9, 10 egg chambers. (E) Quantification of SF F-actin levels in DAAM mutant clones that are within epithelia that also express RNAi against various formins. In order on graph, n = 10, 10, 10, 10, 10, 10, 8 egg chambers. Experiments performed at stage 7. Black arrows show migration direction. Scale bars: 5 µm. (C–E) Each point is the ratio of the mean value for F-actin levels in SFs from 10 experimental cells and 10 control cells in the same egg chamber. Bars show medians and interquartile ranges. NS (not significant) p > 0.05, *p < 0.05. (C, D) Two-tailed Wilcoxon matched pairs signed ranks test. (E) Two-tailed Mann–Whitney compared to control.
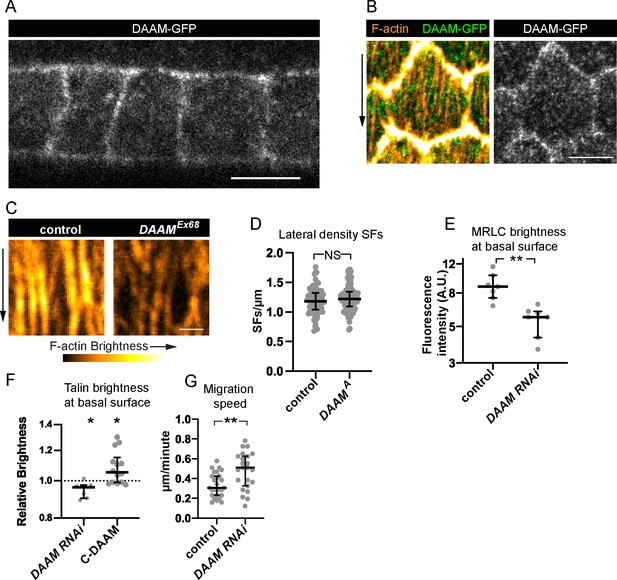
Dishevelled-associated activator of morphogenesis (DAAM) localizes to the cortex and likely strengthens cell–extracellular matrix (ECM) adhesion via stress fibers (SFs).
(A, B) Images of DAAM-GFP (endogenous tag). (A) Transverse section showing that DAAM localizes to the entire cell cortex, image representative of five egg chambers. (B) Basal view of one cell showing DAAM relative to SFs, image representative of five egg chambers. (C) Images of SFs from a control cell and DAAMEx68 cell in the same epithelium stained with phalloidin. SFs in DAAMEx68 cells are similar in number but have reduced F-actin fluorescence. Image representative of three egg chambers. (D) Quantification showing that lateral SF density is normal in DAAMA cells. In order on graph, n = 90, 90 cells from 9 egg chambers with mitotic clones. (E) Quantification showing that myosin regulatory light chain (MRLC) levels are reduced in DAAM RNAi cells. In order on graph, n = 7, 7 egg chambers. (F) Quantification showing that Talin levels are reduced in DAAM RNAi cells. Each point is the ratio of the mean value for Talin levels from 10 experimental cells and 10 control cells in the same egg chamber. In order on graph, n = 7, 14 egg chambers. (G) Quantification of migration rates for control and DAAM RNAi epithelia. In order on graph, n = 27, 23 egg chambers. Experiments performed at stage 7. Black arrows show migration direction. Scale bars: 5 µm (A, B), 1 µm (C). (E–H) Bars show medians and interquartile ranges. NS (not significant) p > 0.05, *p < 0.05, **p < 0.01. (D, E, G) Two-tailed Mann–Whitney test. (F) Two-tailed Wilcoxon matched pairs signed ranks test.
-
Figure 7—source data 1
Excel file containing the numeric data and exact p values used to generate the graphs in Figure 7.
Each tab on the spreadsheet corresponds to one graph.
- https://cdn.elifesciences.org/articles/72881/elife-72881-fig7-data1-v1.xlsx
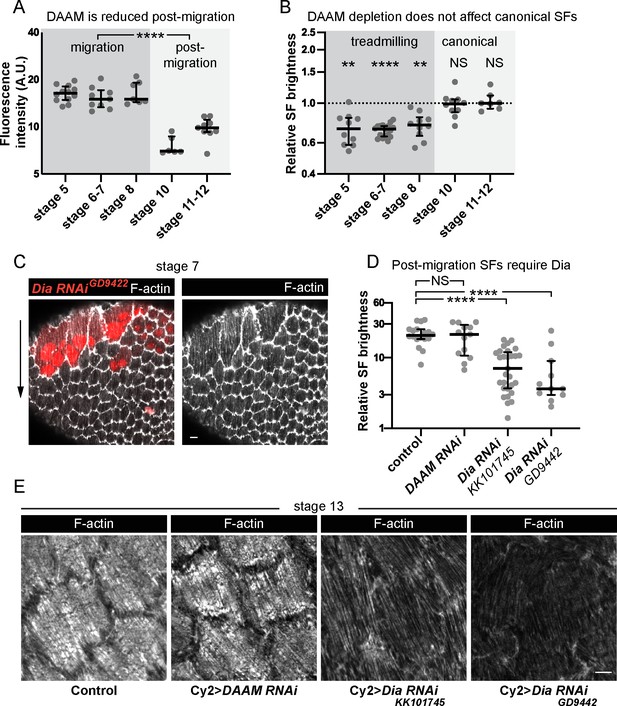
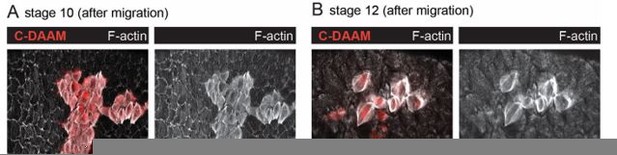
Different formins contribute to treadmilling versus canonical stress fibers (SFs).
(A) Quantification of Dishevelled-associated activator of morphogenesis (DAAM)-GFP levels at basal surface. Levels are higher during migration stages. In order on graph, n = 12, 9, 7, 6, 9 egg chambers. Statistics are on pooled data from migration versus postmigration stages. Two-tailed Mann–Whitney test. (B) Quantification of F-actin levels in SFs. Loss of DAAM reduces F-actin in treadmilling SFs, but not canonical SFs. Each point is the ratio of the mean value for 10 RNAi cells and 10 control cells in the same egg chamber. In order on graph, n = 10, 17, 10, 10, 7 egg chambers. Statistics by stage are two-tailed Wilcoxon matched pairs signed ranks tests. (C) Image of an epithelium with a clone of cells expressing Dia RNAi. Dia RNAi cells have cytokinesis defects shown by multiple red nuclei per cell, but F-actin levels in SFs are normal. (D) Quantification of F-actin levels in SFs at stage 13. When Cy2-Gal4 is used to drive RNAi during postmigration stages, Dia RNAi reduces F-actin in SFs, but DAAM RNAi does not. In order on graph, n = 18, 13, 29, 11 egg chambers. Two-tailed Mann–Whitney tests. (E) Images of cells showing that Dia RNAi reduces F-actin levels in SFs postmigration while DAAM RNAi does not (images selected from intermediate brightness values measured in D). Scale bars: 5 µm. (A, B, D) Bars show medians and interquartile ranges. NS (not significant) p > 0.05, **p < 0.01, ****p < 0.0001.
-
Figure 8—source data 1
Excel file containing the numeric data and exact p values used to generate the graphs in Figure 8.
Each tab on the spreadsheet corresponds to one graph.
- https://cdn.elifesciences.org/articles/72881/elife-72881-fig8-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-GFP directly coupled to Alexa Fluor 488 (rabbit polyclonal) | Invitrogen | Cat # A21311 | (1:400) |
| Chemical compound, drug | CellMask Deep Red Plasma Membrane Stain | Thermo Fisher Scientific | Cat# C10046 | (1:1000) |
| Chemical compound, drug | Alexa Fluor 647 phalloidin | Thermo Fisher Scientific | Cat# A22287 | (1:100) |
| Chemical compound, drug | Alexa Fluor 488 phalloidin | Thermo Fisher Scientific | Cat# A12379 | (1:200) |
| Chemical compound, drug | Schneider’s Drosophila medium | Thermo Fisher Scientific | Cat# 21720-024 | |
| Chemical compound, drug | Fetal bovine serum | Gibco | Cat# 10438-018 | |
| Chemical compound, drug | Recombinant Human Insulin | Millipore Sigma | Cat# 12,643 | |
| Other | Soda Lime Glass Beads, 48–51 µm | Cospheric LLC | Cat# S-SLGMS-2.5 | |
| Chemical compound, drug | Formaldehyde, 16%, methanol free, Ultra-Pure | Polysciences | Cat# 18814-10 | |
| Chemical compound, drug | SlowFade Diamond Antifade mounting medium | Invitrogen | Cat# S36972 | |
| Chemical compound, drug | SlowFade Antifade Kit | Thermo Fisher Scientific | Cat# S2828 | |
| Chemical compound, drug | CK-666 | Millipore Sigma | Cat# SML0006 | |
| Software, algorithm | ImageJ version 2.1.0/1.53c | https://fiji.sc/ | ||
| Software, algorithm | Handbrake 1.3.3 The open source video transcoder | HandBrake Team | https://handbrake.fr/ | |
| Software, algorithm | Zen Blue | Zeiss | ||
| Software, algorithm | Zen Black | Zeiss | ||
| Software, algorithm | MetaMorph | Molecular Devices | ||
| Software, algorithm | Prism Version 8 | Graphpad | ||
| Genetic reagent (D. melanogaster) | w[1118] | Bloomington Drosophila Stock Center | BDSC: 3605; FlyBase ID: FBst0003605; RRID: BDSC_3605 | FlyBase symbol: w[1,118] |
| Genetic reagent (D. melanogaster) | sqh-GFP | Vienna Drosophila Resource Center | VDRC: 318484;FlyBaseID: FBst0492100 | FlyBase symbol:PBac{fTRG00600.sfGFP-TVPTBF}VK00033 |
| Genetic reagent (D. melanogaster) | MRLC-mCh | Laboratory of Eric Wieschaus (Martin et al., 2009) | sqhAx3/FM7;; sqh> sqh-mCh/TM3, Ser, actGFP | |
| Genetic reagent (D. melanogaster) | UAS-Pax-GFP | Laboratory of Denise Montell (He et al., 2010) | ||
| Genetic reagent (D. melanogaster) | daughterless-Gal4 | Bloomington Drosophila Stock Center | BDSC: 55851;FlyBaseID: FBst0055851; RRID:BDSC_55851 | FlyBase symbol: w*; P{GAL4-da.G32}UH1, Sb1/TM6B, Tb1 |
| Genetic reagent (D. melanogaster) | hs-FLP | Bloomington Drosophila Stock Center | BDSC: 8862; FlyBase ID: FBst0008862;RRID:BDSC_8862 | FlyBase symbol: P{ry[+ t7.2] = hsFLP}22, w[*]} |
| Genetic reagent (D. melanogaster) | act5c >> Gal4 | Bloomington Drosophila Stock Center | BDSC: 4780; FlyBase ID: FBst0004780;RRID:BDSC_4780 | FlyBase symbol: y1 w*; P{GAL4-Act5C(FRT.CD2).P}S |
| Genetic reagent (D. melanogaster) | UAS-Ftractin-Tom | Bloomington Drosophila Stock Center | BDSC: 58989;FlyBaseID: FBtp0095457;RRID:BDSC_4780 | P{UASp-F-Tractin.tdTomato}15 A/SM6b; MKRS/TM2 |
| Genetic reagent (D. melanogaster) | UAS-Lifeact-GFP | Bloomington Drosophila Stock Center | BDSC: 35544;FlyBaseID:FBst0035544; RRID:BDSC_35544 | FlyBase symbol: y1 w*; P{UAS-Lifeact-GFP}VIE-260B |
| Genetic reagent (D. melanogaster) | UASMoesinABD-mCh 42c | Laboratory of Brooke McCartney | ||
| Genetic reagent (D. melanogaster) | UAS-Utrophin-ABD-GFP | Laboratory of Thomas Lecuit (Rauzi et al., 2010) | ||
| Genetic reagent (D. melanogaster) | Pax-GFP | Kyoto Stock Center | DGRC: 109971; FlyBaseID:FBst0325098 | FlyBase symbol: w[1,118]; PBac{EGFP-IV}Pax[KM0601] |
| Genetic reagent (D. melanogaster) | Talin-GFP | Laboratory of Hugo Bellen (Venken et al., 2011) | BDSC: 39649; FlyBaseID:FBst0039649;RRID:BDSC_39649 | FlyBase symbol: y1 w*; Mi{PT-GFSTF.0}rheaMI00296-GFSTF.0 lncRNA:CR43910MI00296-GFSTF.0-X |
| Genetic reagent (D. melanogaster) | ß-PS integrin-GFP | Laboratory of Nicholas Brown (Klapholz et al., 2015) | ||
| Genetic reagent (D. melanogaster) | traffic jam-Gal4 | Kyoto Stock Center | DGRC: 104055;FlyBaseID: FBst0302922 | FlyBase symbol: y* w*; P{w + mW.hs=GawB}NP1624/ CyO, P{w-=UASlacZ. UW14}UW14 |
| Genetic reagent (D. melanogaster) | UAS-Abi RNAi | National Institute of Genetics, Japan | NIG: 9749 R-3 | |
| Genetic reagent (D. melanogaster) | UAS-Sra1 RNAi | Bloomington Drosophila Stock Center | BDSC: 38294;FlyBaseID: FBst0038294; RRID:BDSC_38294 | FlyBase symbol: y1 sc* v1 sev21; P{TRiP.HMS01754}attP2 |
| Genetic reagent (D. melanogaster) | act5c >> Gal4, UAS-RFP | Bloomington Drosophila Stock Center | BDSC: 30558; FlyBase ID: FBst0030558; RRID:BDSC_30558 | FlyBase symbol: w[1,118]; P{w[+mC] = GAL4-Act5C(FRT.CD2).P}S, P{w[+mC] = UAS RFP.W}3/TM3, Sb (Ballestrem et al., 2001) |
| Genetic reagent (D. melanogaster) | UAS-pTWFlag-C-DAAM | Laboratory of József. Mihály (Matusek et al., 2006) | ||
| Genetic reagent (D. melanogaster) | hsFLP RFP FRT 19A | Bloomington Drosophila Stock Center | BDSC: 31418;FlyBaseID: FBst0031418; RRID: BDSC_31418 | FlyBase symbol: Ubi-mRFP.nls, w*, hsFLP neoFRT19A |
| Genetic reagent (D. melanogaster) | DAAMA FRT 19A | Bloomington Drosophila Stock Center | BDSC: 52348;FlyBaseID: FBst0052348; RRID: BDSC_52348 | FlyBase symbol: y1 DAAMA w* P{neoFRT}19 A/FM7c, P{GAL4-Kr.C}DC1, P{UAS-GFP.S65T}DC5, sn+ |
| Genetic reagent (D. melanogaster) | DAAMEx68 | Laboratory of József. Mihály (Dollar et al., 2016) | ||
| Genetic reagent (D. melanogaster) | 19A FRT | Bloomington Drosophila Stock Center | BDSC: 1709;FlyBaseID: FBst0001709; RRID: BDSC_1709 | FlyBase symbol: P{ry[+ t7.2] = neoFRT}19A; ry[506] |
| Genetic reagent (D. melanogaster) | DAAMEx68 FRT 19A | Recombination only, this study | DAAMEx68 from J. Mihály; and 19A FRT from BDSC: 1,709 | |
| Genetic reagent (D. melanogaster) | RFP FRT 19A | Bloomington Drosophila Stock Center | BDSC: 31416; FlyBase ID:FBst0031416; RRID: BDSC_31416 | FlyBase symbol: P{w[+ mC] = Ubi mRFP.nls}1, w[1,118], P{ry[+ t7.2] = neoFRT}19A |
| Genetic reagent (D. melanogaster) | UAS-DAAM RNAi | Vienna Drosophila Resource Center | VDRC: 103921;FlyBase ID: FBst0475779 | FlyBase symbol:P{KK102786}VIE-260B |
| Genetic reagent (D. melanogaster) | UAS-Dia RNAi | Vienna Drosophila Resource Center | VDRC: 103914;FlyBase ID: FBst0475772 | FlyBase symbol:P{KK101745}VIE-260B |
| Genetic reagent (D. melanogaster) | UAS-Dia RNAi | Vienna Drosophila Resource Center | VDRC: 20518;FlyBase ID: FBst0453738 | FlyBase symbol: w1118; P{GD9442}v20518 |
| Genetic reagent (D. melanogaster) | UAS-FHOS RNAi | Vienna Drosophila Resource Center | VDRC: 45,838 (line has been discontinued) | w[1,118]; +; P{GD10435}v145838 |
| Genetic reagent (D. melanogaster) | UAS-FHOS RNAi | Vienna Drosophila Resource Center | VDRC: 34034; FlyBase ID: FBst0460422 | FlyBase symbol: w1118; P{GD10435}v34034 |
| Genetic reagent (D. melanogaster) | UAS-Frl RNAi | Vienna Drosophila Resource Center | VDRC: 34413;FlyBase ID: FBst0460614 | FlyBase symbol: w1118; P{GD10799}v34413 |
| Genetic reagent (D. melanogaster) | UAS-Frl RNAi | Vienna Drosophila Resource Center | VDRC: 110438; FlyBase ID: FBst0482010 | FlyBase symbol:P{KK101703}VIE-260B |
| Genetic reagent (D. melanogaster) | UAS-Capu RNAi | Vienna Drosophila Resource Center | VDRC: 110404; FlyBase ID: FBst0481976 | FlyBase symbol: P{KK101400}VIE-260B |
| Genetic reagent (D. melanogaster) | UAS-Capu RNAi | Vienna Drosophila Resource Center | VDRC: 34278; FlyBase ID: FBst0460552 | FlyBase symbol: w1118; P{GD879}v34278 |
| Genetic reagent (D. melanogaster) | UAS-Form3 RNAi | Vienna Drosophila Resource Center | VDRC: 45594; FlyBase ID: FBst0466234 | FlyBase symbol: w1118; P{GD12856}v45594 |
| Genetic reagent (D. melanogaster) | UAS-Form3 RNAi | Vienna Drosophila Resource Center | VDRC: 107473; FlyBase ID: FBst0479293 | FlyBase symbol: P{KK110697}VIE-260B |
| Genetic reagent (D. melanogaster) | UAS-Ena RNAi | Vienna Drosophila Resource Center | VDRC: 43058; FlyBase ID: FBst0464896 | FlyBase symbol: w1118; P{GD8910}v43058/CyO |
| Genetic reagent (D. melanogaster) | w[1118] DAAM-GFP | Laboratory of József. Mihály (Molnár et al., 2014) | ||
| Genetic reagent (D. melanogaster) | UAS-Dcr | Bloomington Drosophila Stock Center | BDSC: 24651; FlyBase ID: FBst0024651; RRID:BDSC_24651 | FlyBase symbol: w1118; P{UAS-Dcr-2.D}10 |
| Genetic reagent (D. melanogaster) | Cy2 Gal4 | Laboratory of Nir Yakoby (Queenan et al., 1997) | FlyBase ID: FBti0007266 | FlyBase symbol: Dmel\P{GawB}CY2 |
Additional files
-
Supplementary file 1
Experimental genotypes.
Detailed list of the genotype corresponding to each figure panel, also showing the temperature at which females were matured on yeasted food prior to dissection.
- https://cdn.elifesciences.org/articles/72881/elife-72881-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/72881/elife-72881-transrepform1-v1.docx