α-/γ-Taxilin are required for centriolar subdistal appendage assembly and microtubule organization
Figures
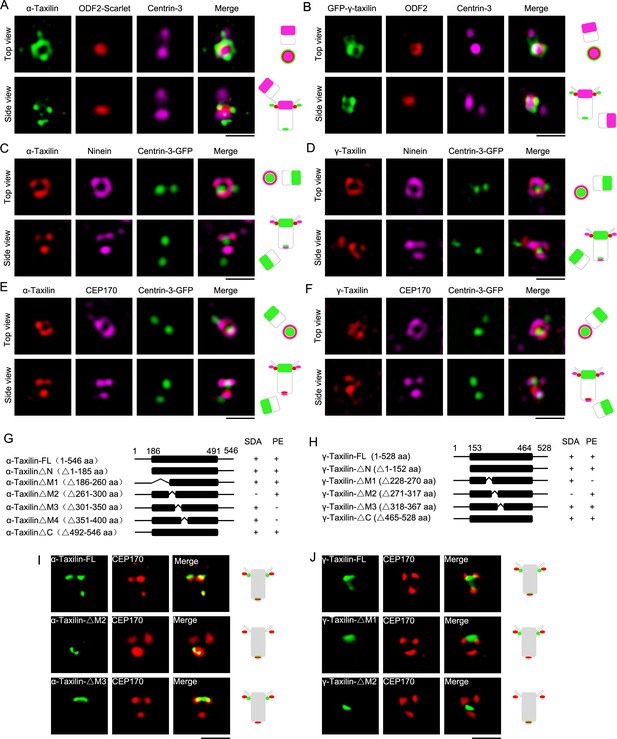
Structured illumination microscopy (SIM) images and characterizations of α-taxilin and γ-taxilin at the centrosomes.
(A) Immunostained α-taxilin (green) and centrin-3 (magenta) in RPE-1 cells transfected with ODF2-Scarlet (red). Scale bar, 1 µm. The cartoons to the right of each set of images graphically depict the merge images. (B) Immunostained ODF2 (red) and centrin-3 (magenta) in RPE-1 cells transfected with GFP-γ-taxilin (green). Scale bar, 1 µm. (C) Immunostained α-taxilin (red) and ninein (magenta) in RPE-1 cells transfected with centrin-3-GFP (green). Scale bar, 1 µm. (D) Immunostained γ-taxilin (red) and ninein (magenta) in RPE-1 cells transfected with centrin-3-GFP (green). Scale bar, 1 µm. (E) Immunostained α-taxilin (red) and CEP170 (magenta) in RPE-1 cells transfected with centrin-3-GFP (green). Scale bar, 1 µm. (F) Immunostained γ-taxilin (red) and CEP170 (magenta) in RPE-1 cells transfected with centrin-3-GFP (green). Scale bar, 1 µm. (G–H) Schematic showing full-length (FL) and deletion mutants (Δ) of α-taxilin (G) and γ-taxilin (H). N, N terminus; M, middle; C, C terminus; +, positive; −, negative; SDA, subdistal appendages; PE, proximal end. (I) Immunofluorescence images of HA-tagged FL α-taxilin and deletion mutants (green) in (G) and CEP170 (red) in RPE-1 cells. Scale bar, 1 µm. (J) Immunofluorescence images of CEP170 (red) in RPE-1 cells transfected with GFP-tagged FL γ-taxilin and deletion mutants (green) in (H). Scale bar, 1 µm. Arrowheads in (I) and (J) show α-taxilin and γ-taxilin SDA localizations, respectively.
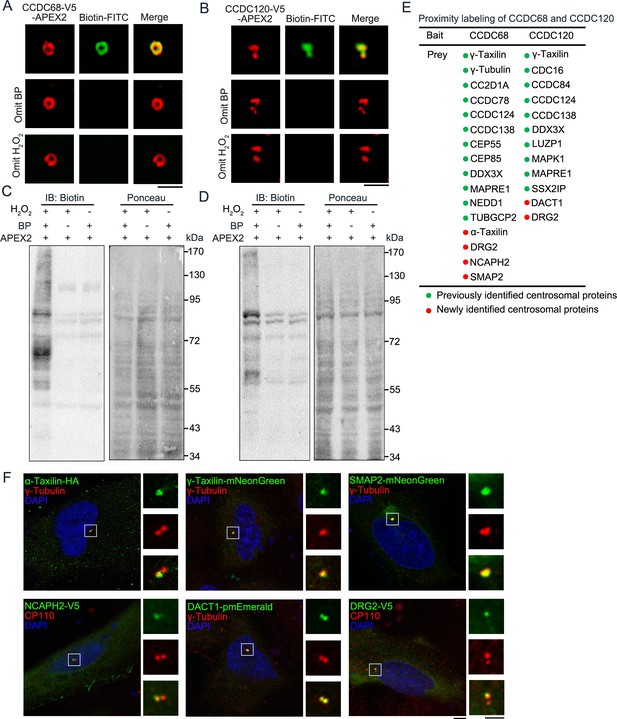
α-Taxilin and γ-taxilin proposed as subdistal appendage proteins as shown by CCDC68 and CCDC120 proximal labeling.
(A) Structured illumination microscopy (SIM) images of HeLa cells expressing CCDC68-V5-APEX2 (red) targeted to the SDA (top view), co-localized and surrounded by biotin-labeled endogenous centrosome proteins (green, line 1). Samples omitting biotin-phenol (BP) or H2O2 were set as negative controls (lines 2 and 3). Scale bar, 1 µm. (B) SIM images of HeLa cells expressing CCDC120-V5-APEX2 (red) targeted to SDA (side view), co-localized and surrounded by biotin-labeled endogenous centrosome proteins (green, line 1). Samples omitting BP or H2O2 were set as negative controls (lines 2 and 3). Scale bar, 1 µm. (C–D) Immunoblots showing CCDC68-V5-APEX2 (C) and CCDC120-V5-APEX2 (D) mediated biotinylation of endogenous proteins with anti-Biotin antibody (left panel). HEK-293T cells were transfected with CCDC68-V5-APEX2 (C) and CCDC120-V5-APEX2 (D) and labeled as in A and B. The right panels show Ponceau S staining of the same membranes. Samples omitting BP or H2O2 were set as negative controls (lines 2 and 3). (E) List of centrosomal proteins identified using mass spectrometric examination of APEX2-mediated biotin proximal labeled CCDC68-V5-APEX2 and CCDC120-V5-APEX2. (F) Confocal images of the indicated fusion proteins (green) co-localized with immunostained centrosome markers CP110 (red) or γ-tubulin (red) in U2OS cells. DNA was stained with DAPI (blue). Scale bar, 5 µm. The right-hand images are enlargement of the indicated white box. Scale bar, 2.5 µm.
-
Figure 1—figure supplement 1—source data 1
Full immunoblots labeled and unlabeled for Figure 1—figure supplement 1C and D.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
CCDC68 and CCDC120 proximity spectrometry data.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig1-figsupp1-data2-v1.zip
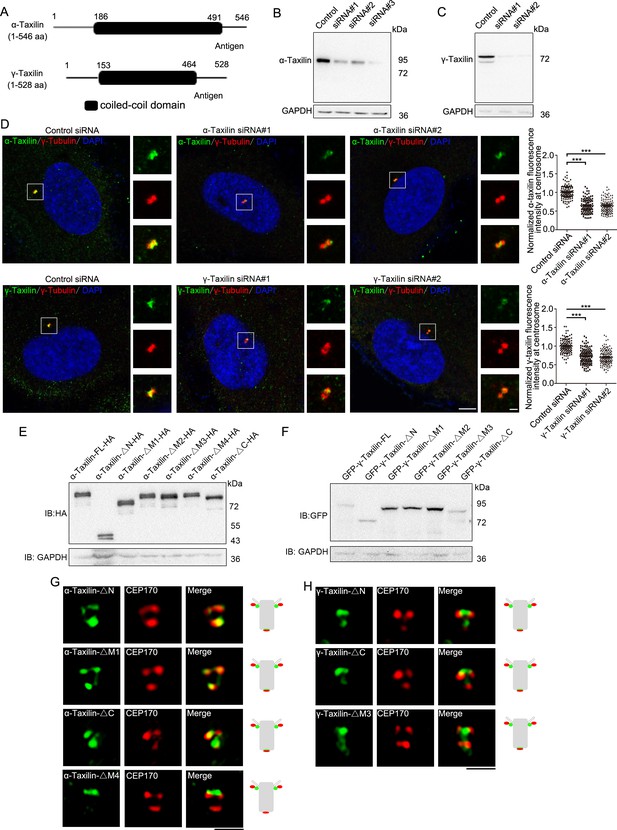
α-Taxilin and γ-taxilin antibodies’ specificities and localization characteristics.
(A) α-Taxilin and γ-taxilin schematic showing their N-terminus, M region (coiled-coil domain), and C-terminus. (B) Immunoblots showing the anti-α-taxilin antibody specificity in control and α-taxilin siRNA-treated U2OS cells. GAPDH was the loading control. (C) Immunoblots showing the anti-γ-taxilin antibody specificity in control and α-taxilin siRNA-treated U2OS cells. GAPDH was the loading control. (D) Confocal images and comparisons of the centrosomal localizations of endogenous α-taxilin or γ-taxilin (green) in control- or siRNA-treated RPE-1 cells. Immunostained γ-tubulin (red) served as the centrosome marker. DNA was stained with DAPI (blue). Scale bar, 5 µm. The right-hand images were enlargement of the indicated white box. Scale bar, 1 µm. Data are Mean ± SEM. Statistical significance was determined by one-way ANOVA from three independent experiments. n > 100. ***, p < 0.001. (E) Immunoblot showing the HA-tagged α-taxilin full-length (FL) and the deletion mutants described in Figure 1G. GAPDH was the loading control. (F) Immunoblot showing the GFP-γ-Taxilin FL and the deletion mutants described in Figure 1H. GAPDH was the loading control. (G) Structured illumination microscopy (SIM) images of immunostained HA-tagged α-taxilin deletion mutants (green) and CEP170 (red) in RPE-1 cells. Scale bar, 1 µm. (H) SIM images of immunostained CEP170 (red) in RPE-1 cells transfected with GFP-tagged γ-taxilin deletion mutants (green). Scale bar, 1 µm. The cartoons to the right of the images in (G) and (H) graphically depict the merge images.
-
Figure 1—figure supplement 2—source data 1
Full immunoblots labeled and unlabeled for Figure 1—figure supplement 2B, C, E and F.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig1-figsupp2-data1-v1.zip
-
Figure 1—figure supplement 2—source data 2
Data of normalized α-taxilin fluorescence intensity at centrosome of control- and α-taxilin-siRNA treated RPE-1 cells.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig1-figsupp2-data2-v1.docx
-
Figure 1—figure supplement 2—source data 3
Data of normalized γ-taxilin fluorescence intensity at centrosome of control- and γ-taxilin-siRNA treated RPE-1 cells.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig1-figsupp2-data3-v1.docx
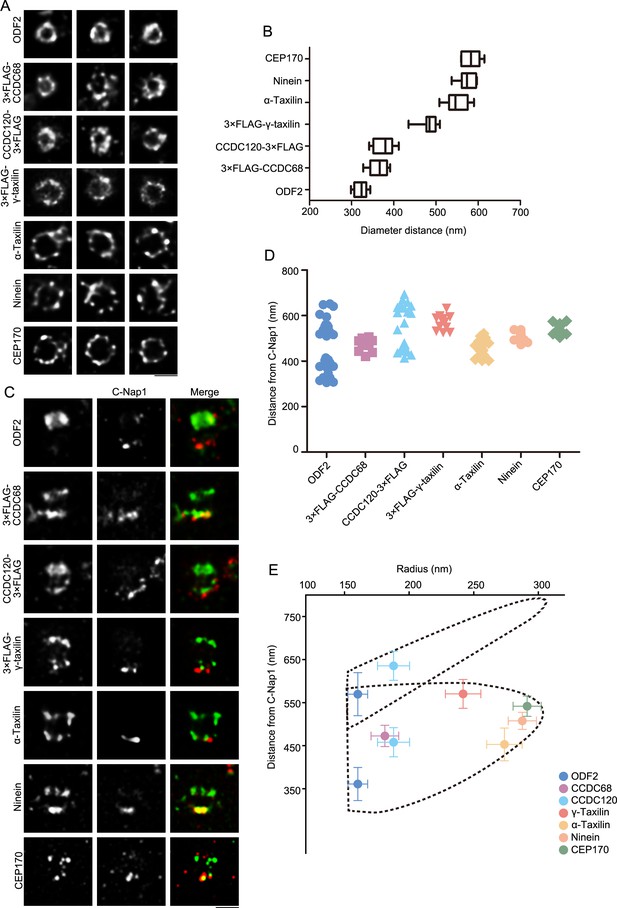
Specific localizations of subdistal appendage (SDA) proteins, including α-taxilin and γ-taxilin.
(A) Representative simulated emission depletion (STED) super-resolution images showing top views of the subdistal appendage (SDA) protein distribution patterns. For ODF2, α-taxilin, ninein and CEP170, RPE-1 cells were immunostained with appropriate antibodies. For CCDC68, CCDC120 and γ-taxilin, RPE-1 cells overexpressed with 3×FLAG-tagged CCDC68, CCDC120 or γ-taxilin full-length were immunostained with FLAG antibody. Scale bar, 500 nm. (B) Diameter analysis of SDA proteins showing ring size diameter. Data are Mean ± SD. n ≥ 7, box = 25th and 75th percentiles. (C) Representative two-color STED super-resolution images showing side view of the SDA proteins (green) and the centriole proximal end protein C-Nap1 (red). Scale bar, 500 nm. (D) A scatter plot describing the distance of SDA proteins relative to C-Nap1. Data are Mean ± SD. n ≥ 11, box = 25th and 75th percentiles. (E) Relative localization of SDA proteins in radial and lateral directions of the mother centriole. The upper dotted lines reveal the slanted arrangement of distal appendage (DA) and the lower dotted lines represent the triangular SDA structure, respectively. Data are Mean ± SD.
-
Figure 2—source data 1
The diameter of subdistal appendage (SDA) proteins, including α-taxilin and γ-taxilin.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig2-data1-v1.docx
-
Figure 2—source data 2
The longitudinal positions of subdistal appendage (SDA) proteins, including α-taxilin and γ-taxilin.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig2-data2-v1.docx
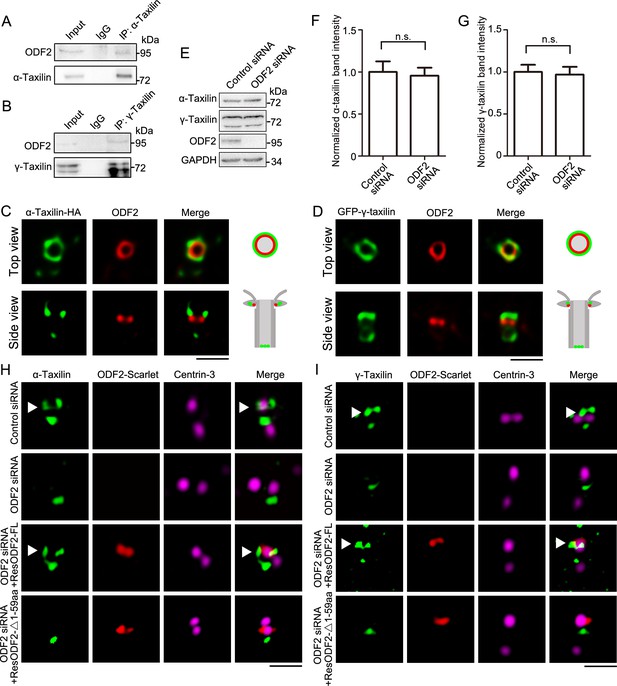
ODF2 is responsible for α-taxilin and γ-taxilin localization at the subdistal appendage (SDA).
(A) Immunoblots of the endogenous immunoprecipitation (IP) assay of ODF2 and α-taxilin using anti-α-taxilin antibody in lysates of RPE-1 cells. IgG was the control. (B) Immunoblots of the endogenous IP assay of ODF2 and γ-taxilin using anti-γ-taxilin antibody in lysates of RPE-1 cells. (C) Simulated emission depletion (STED) images of RPE-1 cells immunostained with α-taxilin-HA (green) and ODF2 (red). Scale bar, 500 nm. (D) STED images of immunostained ODF2 (red) in RPE-1 cells transfected with GFP-γ-taxilin (green). The cartoons to the right of each set of images in (C) and (D) graphically depict the merge images. Scale bar, 500 nm. (E) Immunoblots of α-taxilin and γ-taxilin protein levels in control- or ODF2-siRNA treated RPE-1 cells. GAPDH was the loading control. (F–G) Comparisons of α-taxilin (F) and γ-taxilin (G) band intensities in (E). Data are Mean ± SEM. Statistical significance was determined by two-tailed Student’s t-tests of six independent experiments. n.s., not significant. (H) Structured illumination microscopy (SIM) images of immunostained α-taxilin (green) and centrin-3 (magenta) in control- or ODF2-siRNA treated RPE-1 cells and those cells rescued by transfection with either siRNA-resistant scarlet-tagged ODF2 full-length (FL) or the 1–59 aa ODF2 deletion mutant (red). Scale bar, 1 µm. (I) SIM images of immunostained γ-taxilin (green) and centrin-3 (magenta) in control- or ODF2-siRNA treated RPE-1 cells and those cells rescued by transfection with either siRNA-resistant scarlet-tagged ODF2 FL or the 1–59 aa ODF2 deletion mutant (red). Scale bar, 1 µm. Arrowheads in (H) and (I) show α-taxilin and γ-taxilin SDA localizations, respectively.
-
Figure 3—source data 1
Data of normalized α-taxilin band intensity in control- and ODF2-siRNA treated RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig3-data1-v1.docx
-
Figure 3—source data 2
Data of normalized γ-taxilin band intensity in control- and ODF2-siRNA treated RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig3-data2-v1.docx
-
Figure 3—source data 3
Full immunoblots labeled and unlabeled for Figure 3A, B and E.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig3-data3-v1.zip
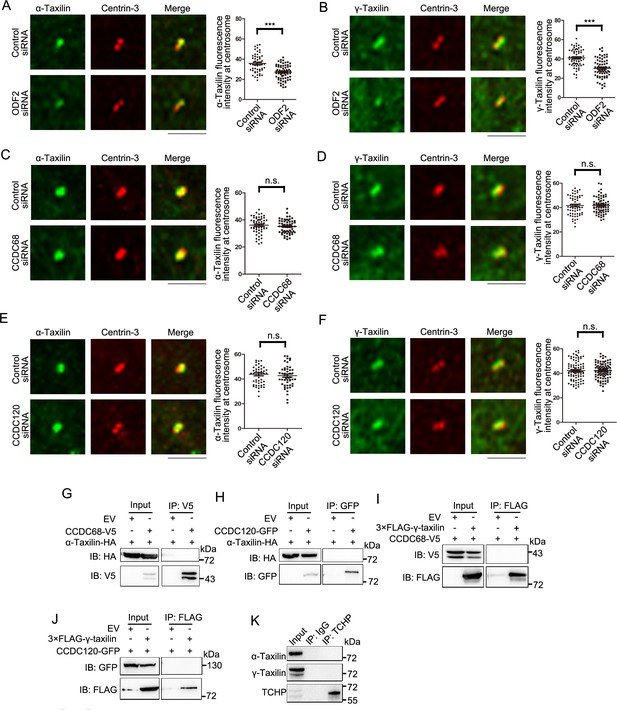
The α-taxilin and γ-taxilin assembly at the centrosome.
(A–B) Confocal images and comparisons of immunostained α-taxilin (A) and γ-taxilin (B) (green) at the centrosome in control- or ODF2-siRNA treated RPE-1 cells. Immunostained centrin-3 (red) served as the centrosome marker. (C–D) Confocal images and comparisons of α-taxilin (C) and γ-taxilin (D) (green) at the centrosome in control- or CCDC68-siRNA treated RPE-1 cells. Immunostained centrin-3 (red) served as the centrosome marker. (E–F) Confocal images and comparisons of α-taxilin (E) and γ-taxilin (F) (green) at the centrosome in control- or CCDC120-siRNA treated RPE-1 cells. Immunostained centrin-3 (red) served as the centrosome marker. For (A–F), scale bars, 5 µm. Data are Mean ± SEM. Statistical significance was determined by two-tailed Student’s t-tests. n > 50. ***, p < 0.001; n.s., not significant. (G) Lysates of HEK-293T cells co-overexpressing CCDC68-V5 and α-taxilin-HA were subjected to immunoprecipitation (IP) with anti-V5 antibody and immunoblotted (IB) with anti-V5 and anti-HA antibodies. (H) Lysates of HEK-293T cells co-overexpressing CCDC120-GFP and α-taxilin-HA were subjected to IP with anti-GFP antibody and IB with anti-GFP and anti-HA antibodies. (I) Lysates of HEK-293T cells co-overexpressing CCDC68-V5 and 3×FLAG-γ-taxilin were subjected to IP with anti-FLAG and IB with anti-V5 and anti-FLAG antibodies. EV, empty vector. (J) Lysates of HEK-293T cells co-overexpressing CCDC120-GFP and 3×FLAG-γ-taxilin were subjected to IP with anti-FLAG and IB with anti-GFP and anti-FLAG antibodies. (K) Endogenous IP assay of α-taxilin and γ-taxilin with TCHP using anti-TCHP antibody in lysates of HEK-293T cells and IB with anti-α-taxilin, anti-γ-taxilin, and anti-TCHP antibodies. IgG was the control.
-
Figure 3—figure supplement 1—source data 1
Data of normalized α-taxilin fluorescence intensity at the centrosome of control- and ODF2-siRNA treated RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig3-figsupp1-data1-v1.docx
-
Figure 3—figure supplement 1—source data 2
Data of normalized γ-taxilin fluorescence intensity at the centrosome of control- and ODF2-siRNA treated RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig3-figsupp1-data2-v1.docx
-
Figure 3—figure supplement 1—source data 3
Data of normalized α-taxilin fluorescence intensity at the centrosome of control- and CCDC68-siRNA treated RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig3-figsupp1-data3-v1.docx
-
Figure 3—figure supplement 1—source data 4
Data of normalized γ-taxilin fluorescence intensity at the centrosome of control- and CCDC68-siRNA treated RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig3-figsupp1-data4-v1.docx
-
Figure 3—figure supplement 1—source data 5
Data of normalized α-taxilin fluorescence intensity at the centrosome of control- and CCDC120-siRNA treated RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig3-figsupp1-data5-v1.docx
-
Figure 3—figure supplement 1—source data 6
Data of normalized γ-taxilin fluorescence intensity at the centrosome of control- and CCDC120-siRNA treated RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig3-figsupp1-data6-v1.docx
-
Figure 3—figure supplement 1—source data 7
Full immunoblots labeled and unlabeled for Figure 3—figure supplement 1G-K.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig3-figsupp1-data7-v1.zip
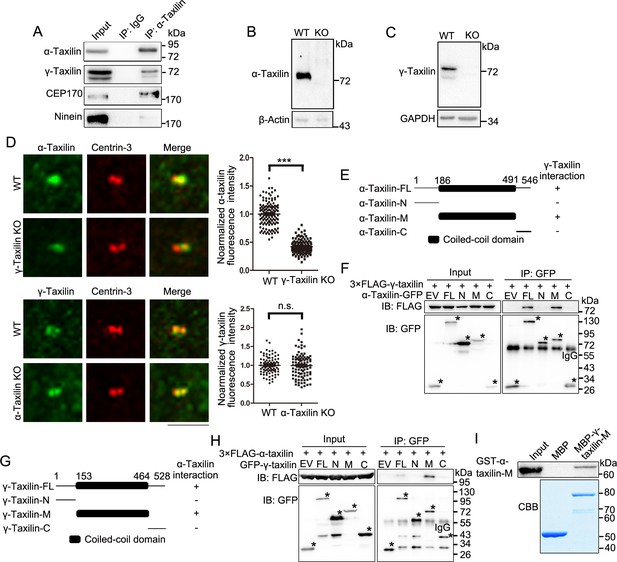
γ-Taxilin interacts with α-taxilin via their coiled-coil domains.
(A) Endogenous immunoprecipitation (IP) assays of α-taxilin with γ-taxilin, CEP170, and ninein using anti-α-taxilin antibody in lysates of HEK-293T cells and were immunoblotted with the indicated antibodies. IgG was the control. (B–C) Immunoblots showing knockout of α-taxilin (B) and γ-taxilin (C) in RPE-1 knockout (KO) cells. β-Actin or GAPDH was used as loading controls. (D) Confocal images of immunostained α-taxilin (green) in wild-type (WT) and γ-taxilin KO RPE-1 cells, as well as immunostained γ-taxilin (green) in WT and γ-taxilin KO RPE-1 cells. Immunostained Centrin-3 (red) was used as a centrosome marker. Data were analyzed by two-tailed Student’s t-tests with three experimental replicates and expressed as Mean ± SEM. n > 80; ***, p < 0.001; n.s., not significant. (E) Schematic showing the interactions of full-length (FL) α-taxilin and its truncated mutants (N terminus, [M] Middle, and C terminus) with γ-taxilin. +, positive; −, negative. (F) Lysates of HEK-293T cells co-overexpressing GFP-tagged FL α-taxilin or its truncated mutants from (E) with 3× FLAG-γ-taxilin were subjected to IP and immunoblotted (IB) with anti-GFP and anti-FLAG antibodies. EV, empty vector. The stars marked the indicated bands in (E). (G) Schematic showing FL γ-taxilin and its truncated mutants’ interactions with α-taxilin. (H) Lysates of HEK-293T cells co-overexpressing GFP-tagged FL γ-taxilin or its truncated mutants from (G) with 3×FLAG-α-taxilin were subjected to IP with anti-GFP and IB with anti-GFP and anti-FLAG antibodies. The stars marked the indicated bands in (G). (I) In vitro binding assay of MBP-γ-taxilin-M from (G) (expressed in E. coli, purified, and stained with Coomassie brilliant blue [CBB]) with GST-α-taxilin-M from (E) (expressed in E. coli and pulled down and detected by IB using the GST antibody).
-
Figure 4—source data 1
Centrosomal α-taxilin fluorescence intensities in wildtype (WT) and γ-taxilin knockout (KO) RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig4-data1-v1.docx
-
Figure 4—source data 2
Centrosomal γ-taxilin fluorescence intensities in wildtype (WT) and α-taxilin knockout (KO) RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig4-data2-v1.docx
-
Figure 4—source data 3
Full immunoblots labeled and unlabeled for Figure 4A, B, C, F and H.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig4-data3-v1.zip
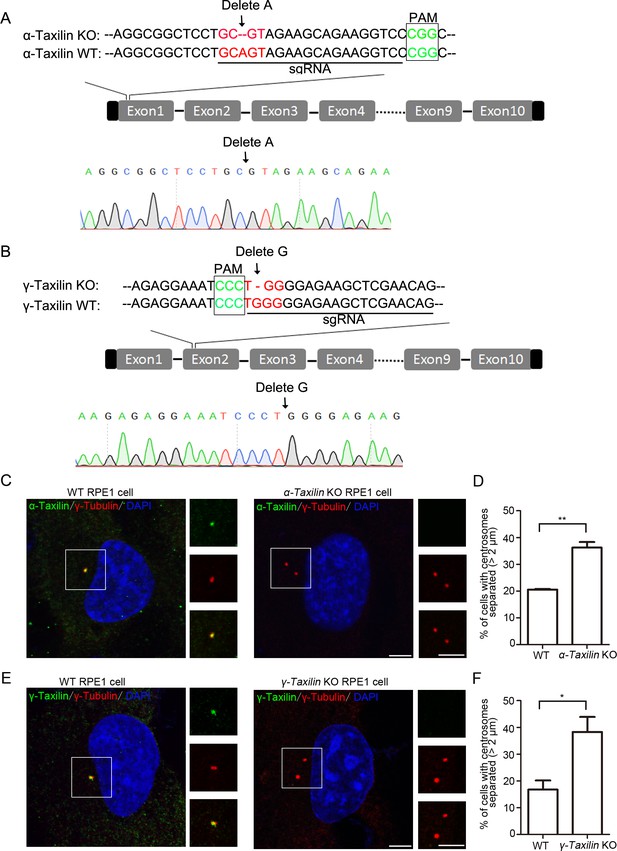
Characterizations of α-taxilin or γ-taxilin wildtype (WT) and knockout (KO) RPE-1 cells.
(A–B) Sequence alignments of partial α-taxilin (A) and γ-taxilin (B) coding sequences from their WT and KO RPE-1 cells. The protospacer adjacent motif (PAM) sequences are in green font, the sgRNA target sequences are underlined, and mutated sequences are in red font. (C) Confocal images of immunostained α-taxilin (green) in WT and α-taxilin KO RPE-1 cells. Immunostained γ-tubulin (red) served as the centrosome marker. DNA was stained with DAPI (blue). Scale bar, 5 µm. (D) Comparisons of the percentages of cells with centrosomes separated by >2 μm in the WT and α-taxilin KO RPE-1 cells in C. Data are Mean ± SEM. Statistical significance was determined by Student’s t-test of three independent experiments. n > 100. **, p < 0.01. (E) Confocal images of immunostained γ-taxilin (green) in WT or γ-taxilin KO RPE-1 cells. Immunostained γ-tubulin (red) served as the centrosome marker. DNA was stained with DAPI (blue). Scale bar, 5 µm. (F) Comparison of the percentages of cells with centrosomes separated by >2 μm in the WT and γ-taxilin KO RPE-1 cells in E. Data are Mean ± SEM. Statistical significance was determined by Student’s t-test of three independent experiments. n > 100. *, p < 0.05.
-
Figure 4—figure supplement 1—source data 1
Percentage of cells with centrosome separated over 2 µm in WT and α-taxilin KO RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig4-figsupp1-data1-v1.docx
-
Figure 4—figure supplement 1—source data 2
Percentage of cells with centrosome separated over 2 µm in WT and γ-taxilin KO RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig4-figsupp1-data2-v1.docx
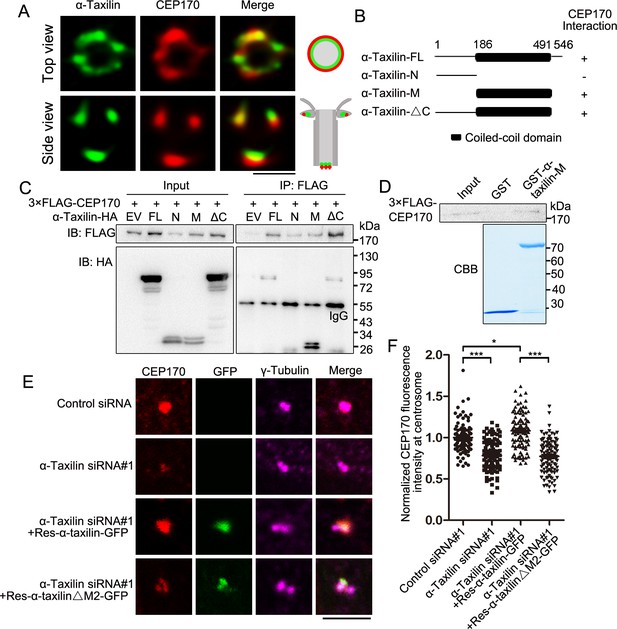
α-Taxilin recruits CEP170 to the subdistal appendage (SDA).
(A) Simulated emission depletion (STED) images of RPE-1 cells immunostained with α-taxilin (green) and CEP170 (red). Scale bar, 500 nm. The cartoons to the right of the images graphically depict the merge images. (B) Schematic showing the full-length (FL) α-taxilin and the truncated mutants (N terminus, [M] Middle, deleted C terminus). +, positive; −, negative. (C) Lysates of HEK-293T cells co-overexpressing HA-tagged α-taxilin-FL or the indicated truncated mutants in (B) with 3×FLAG-CEP170 were immunoprecipitated (IP) with anti-FLAG and immunoblotted (IB) with anti-HA and anti-FLAG antibodies. EV, empty vector. (D) In vitro binding of GST-α-taxilin-M from (B) (expressed in E. coli and purified) with 3×FLAG-CEP170 (expressed in HEK-293T cells and purified). The GST-α-taxilin-M was stained with Coomassie brilliant blue (CBB) and the 3×FLAG-CEP170 was pulled down and IB using FLAG antibody. (E) Confocal images of immunostained CEP170 (red) and γ-tubulin (magenta) in control- or α-taxilin-siRNA treated RPE-1 cells, and those cells rescued with siRNA-resistant GFP-tagged α-taxilin-FL or the α-taxilin M2 deletion mutant (△261–300 aa) (green). Scale bar, 5 µm. (F) Comparisons of CEP170 fluorescence intensities at the centrosomes in (E). Statistical significance was determined with one-way ANOVA with three replicates. Data are Mean ± SEM. n > 100; *, p < 0.05; ***, p < 0.001; n.s., not significant.
-
Figure 5—source data 1
Data of centrosomal CEP170 fluorescence intensity in control and α-taxilin siRNA treated RPE-1 cells, and rescued by overexpressed full-length α-taxilin or the α-taxilin M2 deletion mutant (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig5-data1-v1.docx
-
Figure 5—source data 2
Full immunoblots labeled and unlabeled for Figure 5C and D.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig5-data2-v1.zip
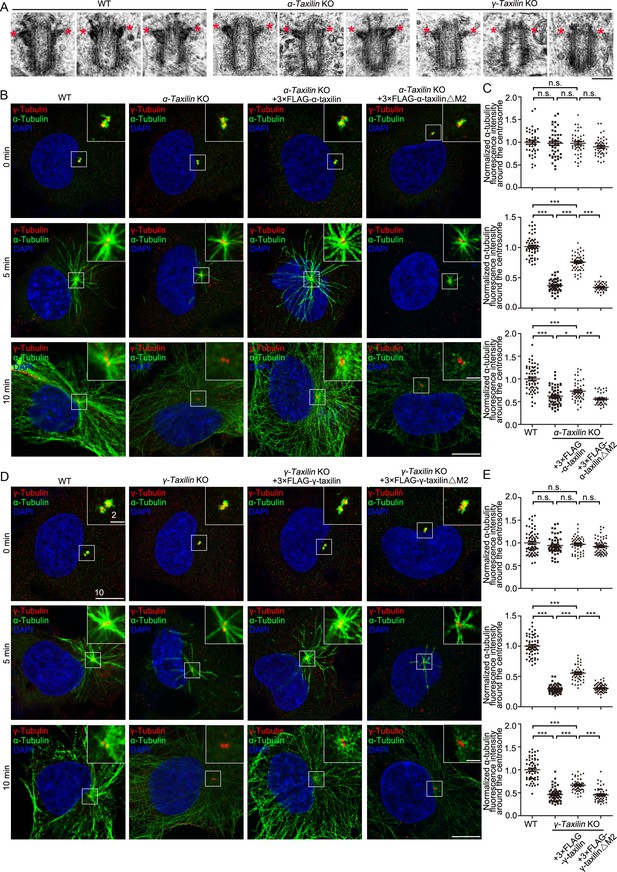
α-Taxilin and γ-taxilin knockout (KO) inhibits microtubule reformation in RPE-1 cells after cold depolymerization.
(A) Transmission electron microscope (TEM) images of the mother centriole of wild-type (WT), α-taxilin and γ-taxilin KO RPE-1 cells. Red stars marked subdistal appendages. Scale bar, 200 nm. (B) Confocal images of microtubule reformation at 0, 5, and 10 min after rewarming. Wild-type (WT), α-taxilin KO RPE-1 cells, and those cells rescued with 3×FLAG-tagged full-length (FL) α-taxilin and the α-taxilin M2 deletion mutant (△261–300 aa, shown in Figure 1G) were immunostained with antibodies against α-tubulin (green) and centrosome marker γ-tubulin (red). DNA was stained with DAPI (blue). Scale bars, 5 µm. (C) Comparisons of α-tubulin fluorescence intensities around the centrosome in (B). Statistical significance was determined by one-way ANOVA. n > 30. Data are mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant. (D) Confocal images of microtubule reformation at 0, 5, and 10 min after rewarming. WT, γ-taxilin KO RPE-1 cells, and those cells rescued with 3×FLAG-tagged γ-taxilin-FL and the γ-taxilin M2 deletion mutant (△271–317 aa, shown in Figure 1H) were immunostained with antibodies against α-tubulin (green) and centrosome marker γ-tubulin (red). DNA was stained with DAPI (blue). Scale bars, 5 µm. (E) Statistical analyses of α-tubulin fluorescence intensity around the centrosome in (D). Statistical significance of α-tubulin fluorescence intensity at 0, 5, and 10 min was determined by one-way ANOVA. n > 40. Data are Mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
-
Figure 6—source data 1
Data of normalized centrosomal α-tubulin fluorescence intensity in wild-type (WT), α-Taxilin knockout (KO) RPE-1 cells, and cells rescued by overexpressed 3×FLAG-α-taxilin or 3× FLAG-α-taxilin△M2 (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig6-data1-v1.docx
-
Figure 6—source data 2
Data of normalized centrosomal α-tubulin fluorescence intensity in wild-type (WT), γ-Taxilin knockout (KO) RPE-1 cells, and rescued by overexpression of 3×FLAG-γ-Taxilin or 3×FLAG-γ-Taxilin△M2 (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig6-data2-v1.docx
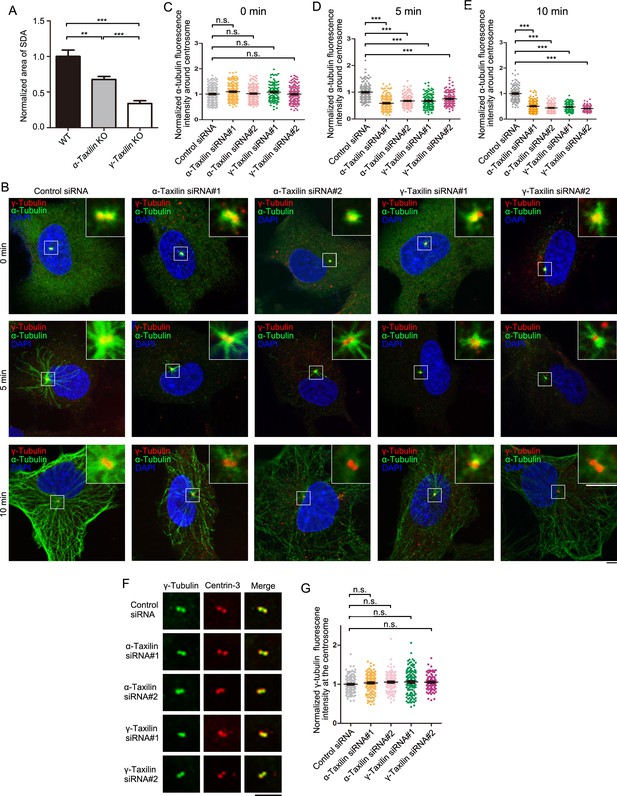
α-Taxilin and γ-taxilin depletion inhibits microtubule reformation in RPE-1 cells after cold depolymerization.
(A) Comparisons of subdistal appendage (SDA) areas from Figure 6A. Statistical significance was determined by one-way ANOVA. Data are Mean ± SEM. n = 8. **, p < 0.01; ***, p < 0.001. (B) Confocal images of microtubule reformation with immunostained α-tubulin (green) in control-, α-taxilin- or γ-taxilin-siRNA treated RPE-1 cells. Immunostained γ-tubulin (red) was used as the centrosome marker. DNA was stained with DAPI (blue). Scale bars, 10 µm. (C–E) Comparisons of α-tubulin fluorescence intensity at 0 (C), 5 (D), and 10 min (E) post-warming was determined by one-way ANOVAs of three independent experiments per time period. n > 100. Data are Mean ± SEM. ***, p < 0.001; n.s., not significant. (F) Confocal images of immunostained γ-tubulin (green) and centrin-3 (red) in control-, α-taxilin-, or γ-taxilin-siRNA treated RPE-1 cells. Scale bar, 5 μm. (G) Comparisons of γ-tubulin fluorescence intensities at the centrosomes in (F). Statistical significance was determined by one-way ANOVA. n > 90. Data are Mean ± SEM. n.s., not significant.
-
Figure 6—figure supplement 1—source data 1
Data of normalized areas of SDAs in wild-type (WT), α-taxilin and γ-taxilin KO RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig6-figsupp1-data1-v1.docx
-
Figure 6—figure supplement 1—source data 2
Data of normalized centrosomal α-tubulin fluorescence intensity in control-, α-taxilin-, or γ-taxilin-siRNA treated RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig6-figsupp1-data2-v1.docx
-
Figure 6—figure supplement 1—source data 3
Data of normalized centrosomal γ-tubulin fluorescence intensity in control-, α-taxilin-, or γ-taxilin-siRNA treated RPE-1 cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig6-figsupp1-data3-v1.docx
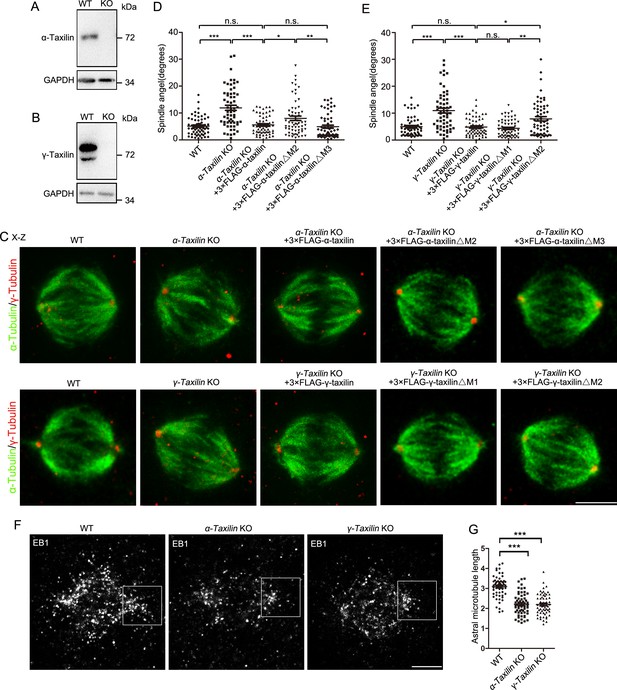
α-Taxilin and γ-taxilin knockout (KO) increases spindle angle orientation during mitosis.
(A–B) Immunoblots showing wild-type (WT) and KO α-taxilin (A) and γ-taxilin (B) in HeLa cells. GAPDH was the loading control. (C) Representative orthogonal views (x–z) of metaphase HeLa cells stained for microtubules (α-tubulin, green) and spindle poles (γ-tubulin, red). Views showed WT, α-taxilin, or γ-taxilin KO cells, and cells overexpressed with 3×FLAG-tagged full-length α-taxilin or γ-taxilin and the indicated deletion mutants (Δ) described in Figure 1G and H, respectively. Scale bars, 10 µm. (D) Comparisons of raw spindle angles in WT, α-taxilin KO HeLa cells, and cells overexpressed with 3×FLAG-tagged full-length α-taxilin or the indicated deletion mutants (Δ). Data are represented as Mean ± SEM. Statistical significance was determined by one-way ANOVA of three individual experiments. n ≥ 60. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant. (E) Comparisons of raw spindle angles in WT, γ-taxilin KO HeLa cells, and cells overexpressed with 3×FLAG-tagged full-length γ-taxilin or the indicated deletion mutants (Δ). Data are represented as Mean ± SEM. Statistical significance was determined by one-way ANOVA of three individual experiments. n ≥ 60. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant. (F) Confocal images of aster microtubules immunostained with EB1 in WT, α-taxilin, or γ-taxilin KO HeLa cells. Scale bars, 5 µm. (G) Comparisons of the aster microtubule lengths in (F). Data are mean (SEM). Statistical significance was determined by one-way ANOVA of three replicates. n ≥ 60. ***, p < 0.001.
-
Figure 7—source data 1
Spindle angles of wild-type (WT), α-taxilin knockout (KO) HeLa cells, and cells overexpressed with indicated α-taxilin full-length and deletion mutants (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig7-data1-v1.docx
-
Figure 7—source data 2
Spindle angles of wild-type (WT), γ-taxilin knockout (KO) HeLa cells, and cells overexpressed with indicated γ-taxilin full-length and deletion mutants (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig7-data2-v1.docx
-
Figure 7—source data 3
Astral microtubule length in wild-type (WT), α-taxilin, and γ-taxilin knockout (KO) HeLa cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig7-data3-v1.docx
-
Figure 7—source data 4
Full immunoblots labeled and unlabeled for Figure 7A, B.
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig7-data4-v1.zip
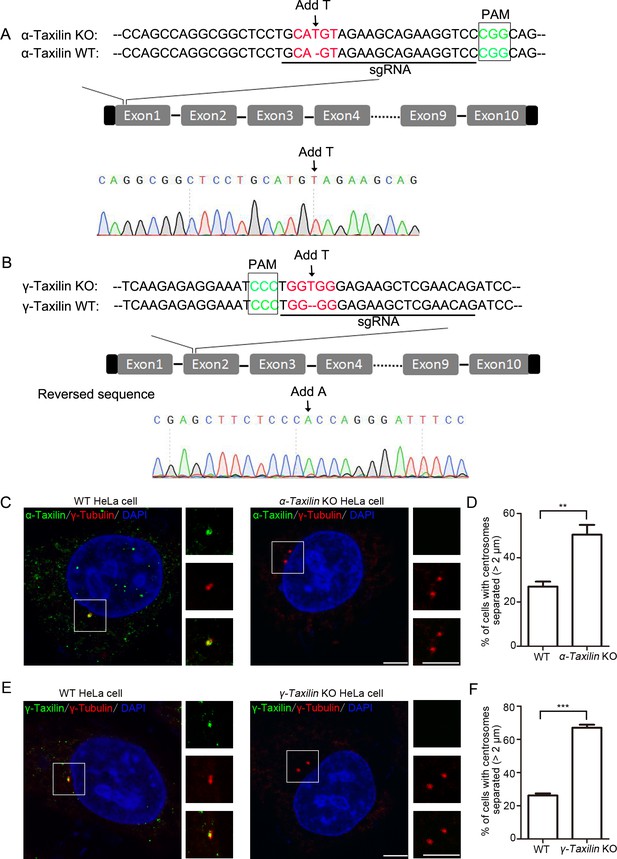
Characterizations of α-taxilin and γ-taxilin WT and KO HeLa cells.
(A–B) Sequence alignments of partial α-taxilin (A) and γ-taxilin (B) coding sequences from their wild-type (WT) and KO HeLa cells. The protospacer adjacent motif (PAM) sequences are in green font, the sgRNA target sequences are underlined, and mutated sequences are in red font. (C) Confocal images of immunostained α-taxilin (green) in WT and α-taxilin KO HeLa cells. Immunostained γ-tubulin (red) served as the centrosome marker. DNA was stained with DAPI (blue). Scale bar, 5 µm. (D) Comparisons of the percentages of cells with centrosomes separated by >2 μm in WT and α-taxilin KO HeLa cells in C. Data are Mean ± SEM. Statistical significance was determined by Student’s t-test of three independent experiments. n > 100. **, p < 0.01. (E) Confocal images of immunostained γ-taxilin (green) in WT or α-taxilin KO HeLa cells. Immunostained γ-tubulin (red) served as the centrosome marker. DNA was stained with DAPI (blue). Scale bar, 5 µm. (F) Comparisons of the percentages of cells with centrosomes separated by >2 μm in WT and γ-taxilin KO HeLa cells in (E). Data are Mean ± SEM. Statistical significance was determined by Student’s t-test of three independent experiments. n > 100. ***, p < 0.001.
-
Figure 7—figure supplement 1—source data 1
Percentage of cells with centrosome separated over 2 µm in WT and α-taxilin KO HeLa cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig7-figsupp1-data1-v1.docx
-
Figure 7—figure supplement 1—source data 2
Percentage of cells with centrosome separated over 2 µm in WT and γ-taxilin KO HeLa cells (Data provided as Mean ± SEM).
- https://cdn.elifesciences.org/articles/73252/elife-73252-fig7-figsupp1-data2-v1.docx
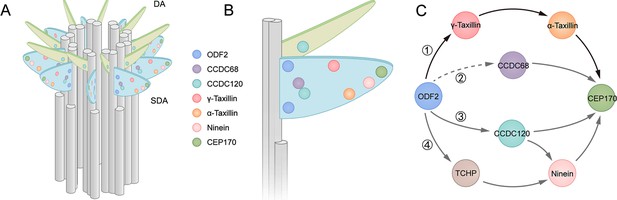
Models showing α-taxilin and γ-taxilin localization and assembly in the subdistal appendage (SDA).
(A) A 3D model of a mother centriole, illustrating the localization of various SDA proteins, including ODF2, CCDC68, CCDC120, γ-taxilin, α-taxilin, ninein, and CEP170 in order of increasing diameter size. (B) Close view of the SDA structure from (A). (C) Model showing the hierarchical relationships of SDA proteins with ODF2 at the SDA root: (1) ODF2 recruits γ-taxilin to the SDA, which in turn hierarchically recruits α-taxilin and then CEP170; (2) CCDC68 is recruited to the SDA, which further recruits CEP170 (Huang et al., 2017); (3) ODF2 recruits ninein and CEP170 via CCDC120 (Huang et al., 2017); (4) ODF2 recruits ninein via TCHP (Ibi et al., 2011). DA, distal appendage.
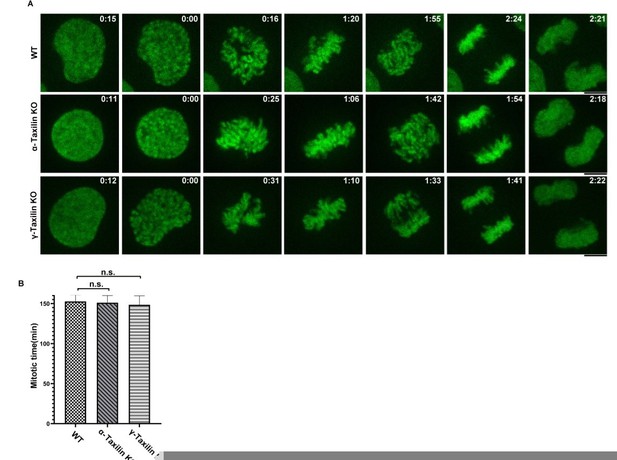
A.
Effects of taxilin loss on mitotic times. WT HeLa cells and α-taxilin or γ-taxilin knockout (KO) HeLa cells stably expressing mNeonGreen-H2B were photographed every 3 min for 24 h. Representative still frames from time-lapse experiments are shown with elapsed times shown in each frame. Scale bar, 5 µm. B. Quantification of the time intervals from nuclear envelope breakdown to anaphase onset. Data were mean ± SEM of three independent experiments. n>100. One-way ANOVA was used for statistical analysis. n.s., not significant.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo-sapiens) | CCDC68 | GenBank | NM_025214.3 | |
| Gene (Homo-sapiens) | CCDC120 | GenBank | NM_001163321.4 | |
| Gene (Homo-sapiens) | α-taxilin | GenBank | NM_175852.4 | |
| Gene (Homo-sapiens) | γ-taxilin | GenBank | XM_024452398.1 | |
| Gene (Homo-sapiens) | SWAP2 | GenBank | AY358944.1 | |
| Gene (Homo-sapiens) | DACT1 | GenBank | NM_016651.6 | |
| Gene (Homo-sapiens) | ODF2 | GenBank | NM_001242352.1 | |
| Cell line (Homo-sapiens) | HeLa | ATCC | CCL-2 | Cell line authenticated by ATCC |
| Cell line (Homo-sapiens) | U2OS | ATCC | HTB-96 | Cell line authenticated by ATCC |
| Cell line (Homo-sapiens) | HEK-293T | ATCC | CRL-3216 | Cell line authenticated by ATCC |
| Cell line (Homo-sapiens) | RPE-1 | ATCC | CRL-4000 | Cell line authenticated by ATCC |
| Antibody | α-taxilin (Rabbit polyclonal) | Proteintech | 27558–1-AP | IF(1:100), IB (1:1000) |
| Antibody | α-tubulin(Mouse monoclonal) | Sigma-Aldrich | T9026 | IF (1:1000) |
| Antibody | β-actin(Mouse monoclonal) | Cell Signaling Technology | 8H10D10 | IB (1: 10000) |
| Antibody | γ-taxilin(Rabbit polyclonal) | Proteintech | 22357–1-AP | IF(1:100), IB (1:1000) |
| Antibody | γ-tubulin(Rabbit polyclonal) | Sigma-Aldrich | T3559 | IF (1:200) |
| Antibody | Biotin(Mouse monoclonal) | Sigma-Aldrich | BN-34 | IB (1:500) |
| Antibody | Biotin-FITC(Goat polyclonal) | Sigma-Aldrich | F6762 | IF (1:100) |
| Antibody | Centrin-3(Mouse monoclonal) | Abnova | ABN-H0000 1070-M01 | IF (1:200) |
| Antibody | CEP170(Rabbit polyclonal) | Abcam | ab72505 | IF (1:100); IB (1:1000) |
| Antibody | CEP170(Rabbit polyclonal) | Proteintech | 27325–1-AP | IF (1:100) |
| Antibody | CEP170(Mouse monoclonal) | Invitrogen | 72-413-1 | IF (1:200) |
| Antibody | EB1(Rabbit polyclonal) | Proteintech | 17717–1-AP | IF (1:100) |
| Antibody | FLAG(Mouse monoclonal) | Sigma-Aldrich | F3165 | IB (1:10000) |
| Antibody | GAPDH(Mouse monoclonal) | ABclonal | AC002 | IB (1:20000) |
| Antibody | GFP(Rabbit polyclonal) | Laboratory produced | IB (1:10000) | |
| Antibody | GST(Rabbit polyclonal) | Laboratory produced | IB (1:500) | |
| Antibody | HA(Mouse monoclonal) | Sigma-Aldrich | H9658 | IF (1:1000); IB (1:10000) |
| Antibody | ninein(Rabbit polyclonal) | Abcam | ab231181 | IF (1:100); IB (1:1000) |
| Antibody | ninein(Mouse monoclonal) | Invitrogen | 79.160–7 | IF (1:100) |
| Antibody | ODF2(Rabbit polyclonal) | Proteintech | 12058–1-AP | IF (1:100); IB (1:1000) |
| Antibody | TCHP(Rabbit polyclonal) | ATLAS antibodies | HPA038638 | IB (1:1000) |
| Antibody | V5(Mouse monoclonal) | Innovative Research | R960CUS | IF (1:1000); IB (1:10000) |
| Recombinant DNA reagent | α-taxilin gRNA (plasmid) | This paper | gRNA (5’-GCAGTAGAAGCAGAAGGTCC-3’) | |
| Recombinant DNA reagent | γ-taxilin gRNA (plasmid) | This paper | gRNA (5’-CTGTTCGAGCTTCTCCCCCA-3’) | |
| Software, algorithm | Image J | NIH (Schneider et al., 2012) | ||
| Software, algorithm | Prism 5 | GraphPad |





