Evolution of host-microbe cell adherence by receptor domain shuffling
Figures
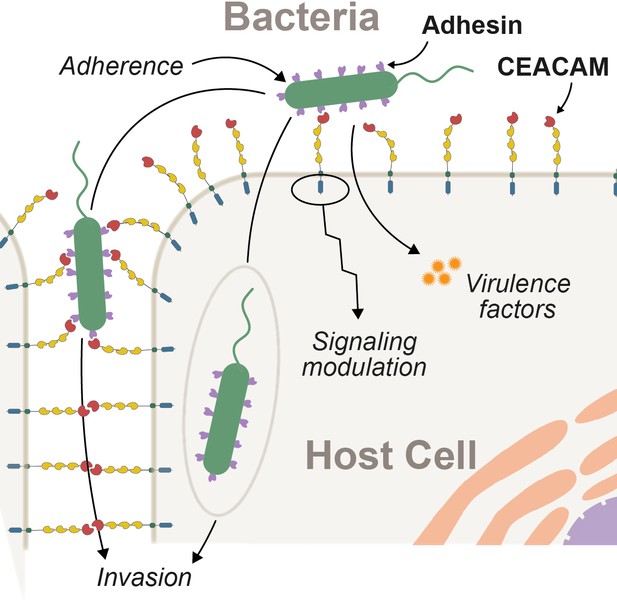
Interactions between epithelial carcinoembryonic antigen-associated cell adhesion molecules (CEACAMs) and bacterial adhesins.
Bacterial attachment to host cells via adhesin proteins (purple) facilitates epithelial adherence. Adhesins also contribute to pathogenicity by promoting invasion, modulation of host cell signaling pathways, and by promoting the delivery of virulence factors into the host cell cytoplasm.
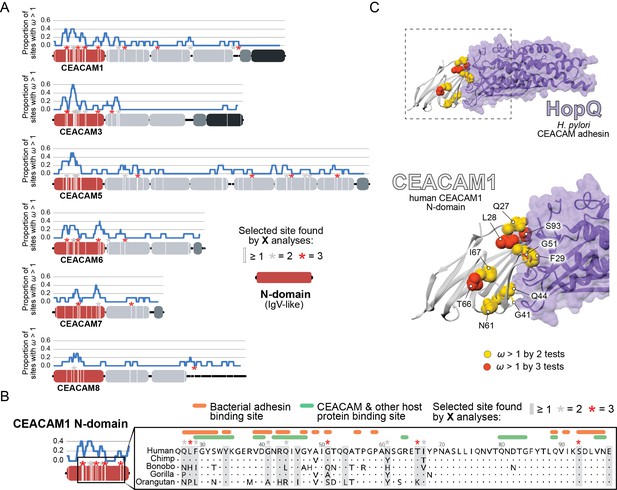
Rapid evolution of primate carcinoembryonic antigen-related cell adhesion molecule (CEACAM) N-domains.
(A) Sites in CEACAM proteins exhibiting elevated ω. Domain structure of CEACAMs outlined in red (N-domain), light gray (IgC-like domains), dark gray (transmembrane domain), and black (cytoplasmic domain). All rapidly evolving sites identified by at least one phylogenetic analysis (PAML, FUBAR, or MEME) are marked by a white line, sites identified by two or three tests signified by gray and red asterisks, respectively. Blue line shows the proportion of rapidly evolving sites identified across a 10 amino acid sliding window. (B) Multiple sequence alignment of hominid CEACAM1 residues 26–98. Sites identified as evolving under positive selection and sites known to influence adhesin and host protein binding are highlighted (Figure 2—source data 1F). (C) Protein co-crystal structure of human CEACAM1 (gray) and the HopQ adhesin (purple) from Helicobacter pylori strain G27 (PDB ID: 6GBG). CEACAM1 sites identified as evolving under positive selection by two or more tests are highlighted.
-
Figure 2—source code 1
Code to generate graphs and images for Figure 2A.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig2-code1-v3.zip
-
Figure 2—source data 1
(a) Summary of primate carcinoembryonic antigen-related cell adhesion molecule (CEACAM) sequences used in analyses.
Table summarizing primate CEACAM sequences extracted for evolutionary analyses and phylogenetic reconstructions. (b) Summary of primate CEACAM identification. Table summarizing BLAST results, genome annotation, and sequence analyses used to identify human CEACAM orthologs in primates. (c) Additional notes on primate CEACAM identification. Table of additional notes on CEACAM sequences used in analyses. (d) PAML NS sites results summary. Table of PAML NS sites tests of selection in primate CEACAMs. (e) Summary of sites identified by evolutionary analyses. Table of sites identified as evolving under positive selection by evolutionary analyses and GARD predicted recombination breakpoints. (f) References for CEACAM1 binding sites. Table of references for sites identified as contributing to CEACAM1 binding with host proteins and bacterial adhesins as well as the specific sites identified.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig2-data1-v3.xlsx
-
Figure 2—source data 2
Trimmed carcinoembryonic antigen-related cell adhesion molecule (CEACAM) sequences and primate species trees used for evolutionary analyses.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig2-data2-v3.zip
-
Figure 2—source data 3
Results files for evolutionary analyses.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig2-data3-v3.zip
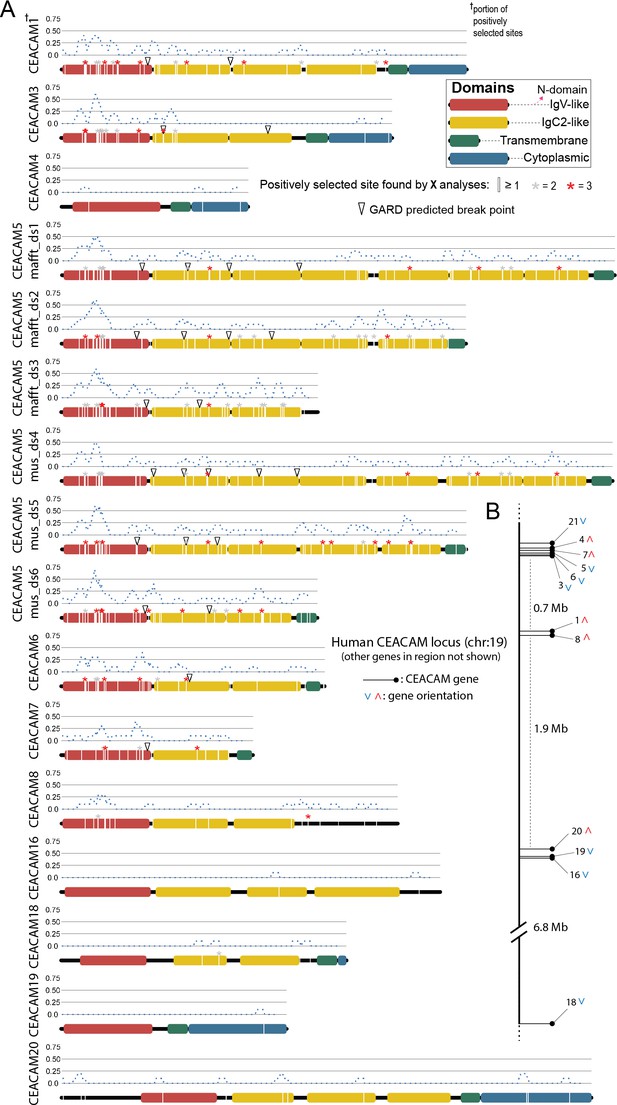
Primate carcinoembryonic antigen-related cell adhesion molecule (CEACAM) evolutionary analysis summary.
Sites with elevated dN/dS in all human CEACAM proteins. (A) Sites in CEACAM proteins identified as evolving rapidly in specific domains by one (white line), two (gray asterisks), or three (red asterisks) evolutionary analyses. Dotted blue line indicates the proportion of sites identified as evolving rapidly across a 10 amino acid sliding window. Open triangles show GARD predictions of the approximate locations of recombination breakpoints. (B) Location of human CEACAM genes along chromosome 19. Other genes on chromosome 19 not shown.
-
Figure 2—figure supplement 1—source code 1
Code to generate graphs and images for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig2-code2-v3.zip
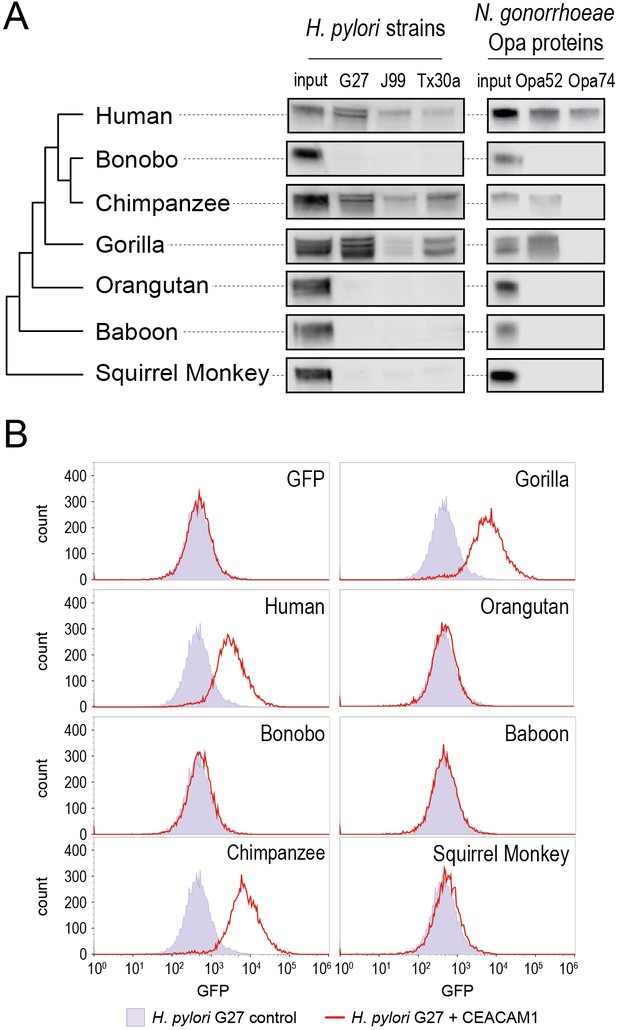
Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) divergence in great apes restricts bacterial adhesin recognition.
(A) Binding between primate GFP-tagged CEACAM1 N-domain orthologs and bacteria determined by pulldown assays and visualized by western blotting. Input is 10% CEACAM1 protein used in bacterial pulldowns. Primate species relationships indicated by phylogenetic tree. (B) Pulldown experiments of Helicobacter pylori strain G27 incubated with CEACAM1 N-domain constructs or GFP assayed by flow cytometry. Binding indicated by relative GFP fluorescence. Representative western blot and flow cytometry experiments are depicted. For flow cytometry all tests shown were performed as part of a single experiment using H. pylori strain G27 alone as a negative control.
-
Figure 3—source data 1
Raw and labeled western blot images for Figure 3A and flow cytometry data for Figure 3B.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig3-data1-v3.zip

Helicobacter pylori G27 Δhopq pulldown.
Binding assay to assess interactions between H. pylori strain G27 Δhopq and GFP-tagged CEACAM1 N-domain constructs for human, chimpanzee, and gorilla, by pulldown experiments and visualization by western blot.
-
Figure 3—figure supplement 1—source data 1
Raw and labeled western blot images for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig3-figsupp1-data1-v3.zip
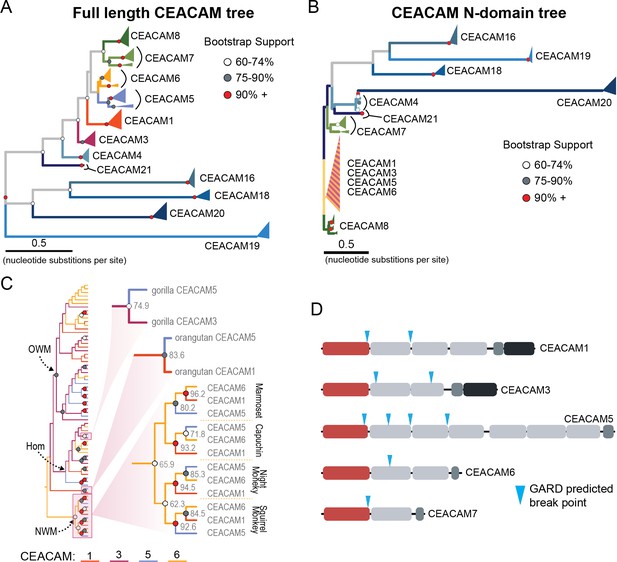
Recurrent episodes of gene conversion among adhesin-binding carcinoembryonic antigen-related cell adhesion molecules (CEACAMs).
(A) Maximum likelihood-based phylogeny of full-length primate CEACAM protein-coding sequences. (B) Phylogeny of the IgV-like (N-domain) of primate CEACAM proteins. (C) Expanded cladogram view of the clade containing the N-domains of CEACAM1, CEACAM3, CEACAM5, and CEACAM6 from panel B. Arrows indicate nodes designating clades for Old World monkeys (OWM), hominoids (Hom), and New World monkeys (NWM). Specific subclades, gorilla CEACAM3 and CEACAM5, orangutan CEACAM5 and CEACAM1, and NWM are further magnified and highlighted with bootstrap support at nodes. (D) Domain structures of CEACAM proteins predicted to have undergone recombination by GARD analysis with sites of predicted breakpoints highlighted (blue arrows). CEACAM N-domains are denoted in red.
-
Figure 4—source code 1
Code to generate images for Figure 4D.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig4-code1-v3.zip
-
Figure 4—source data 1
Sequence alignments of trimmed carcinoembryonic antigen-related cell adhesion molecule (CEACAM) sequences used for phylogenetic reconstructions.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig4-data1-v3.zip
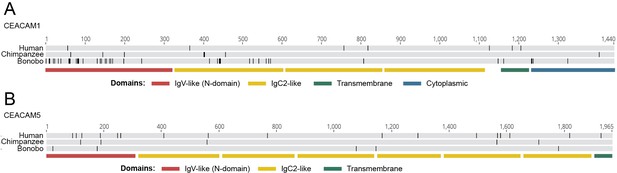
Alignment of human-pan carcinoembryonic antigen-related cell adhesion molecule (CEACAM) sequences.
Human, chimpanzee, and bonobo CEACAM1 (A) and CEACAM5 (B) alignments by MAFFT translation alignment implemented in Geneious Prime 2020.2.2. Black lines mark differences from consensus. Lower bars show location of CEACAM domains.
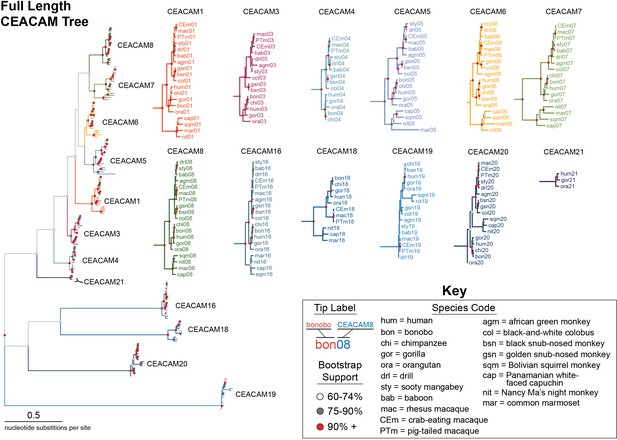
Expanded full-length carcinoembryonic antigen-related cell adhesion molecule (CEACAM) tree.
Maximum likelihood-based phylogeny of full-length CEACAM protein-coding sequences as represented in Figure 4A, with clades expanded. Clades encompassing individual CEACAM orthologs are shown isolated and expanded.
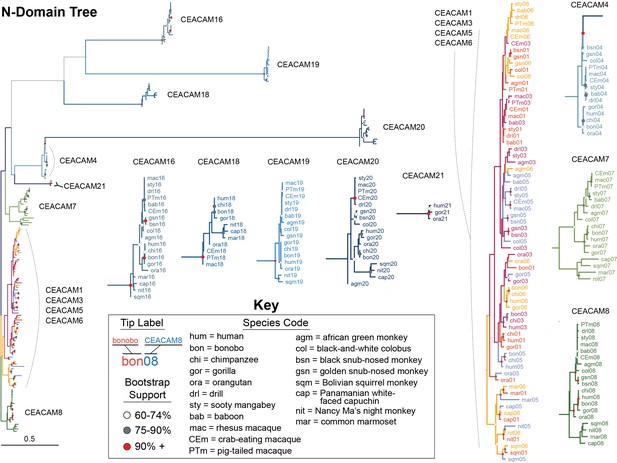
Expanded carcinoembryonic antigen-related cell adhesion molecule (CEACAM) N-domain tree.
Maximum likelihood-based phylogeny of CEACAM IgV-like (N-domain) sequences as represented in Figure 4B, with clades expanded. Clades encompassing individual CEACAM orthologs along with the CEACAM1, CEACAM3, CEACAM5, and CEACAM6 clade are shown isolated and expanded.
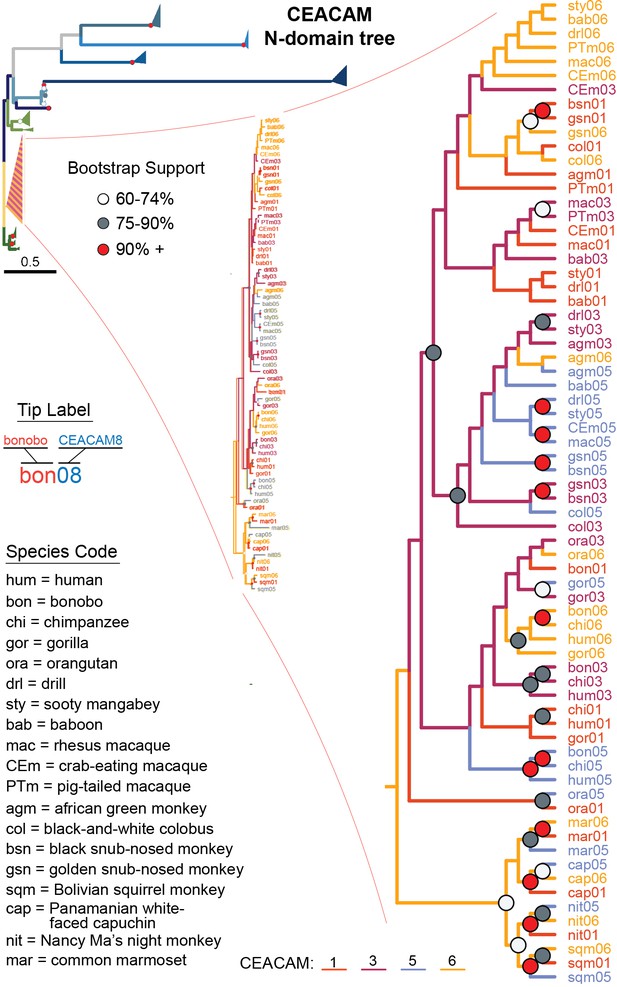
Expanded view of carcinoembryonic antigen-related cell adhesion molecule (CEACAM)1,3,5,6 N-domain clade.
Expanded view of CEACAM1, CEACAM3, CEACAM5, and CEACAM6 clade from Figure 4B.
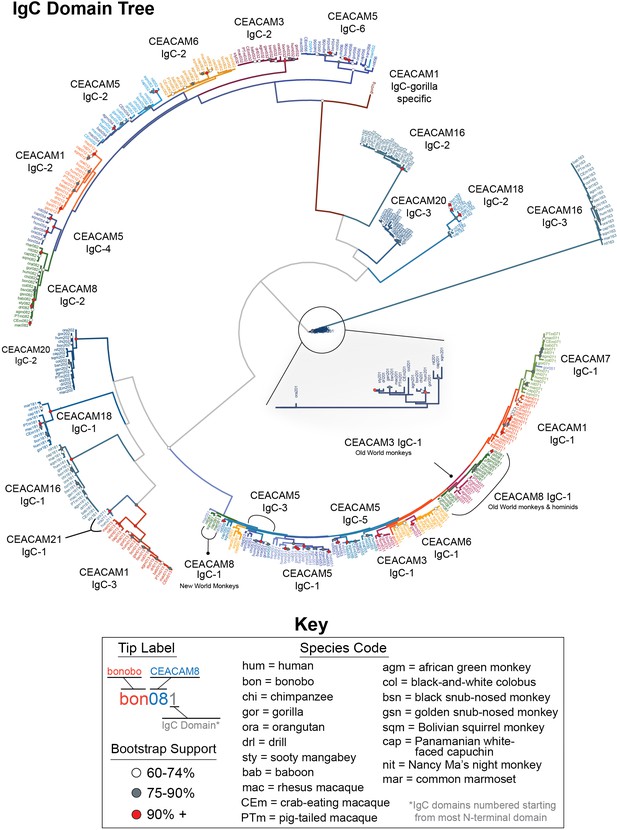
Expanded carcinoembryonic antigen-related cell adhesion molecule (CEACAM) IgC domains tree.
Maximum likelihood-based phylogeny of CEACAM IgC-like domain sequences. Expanded view of CEACAM20 clade shown.
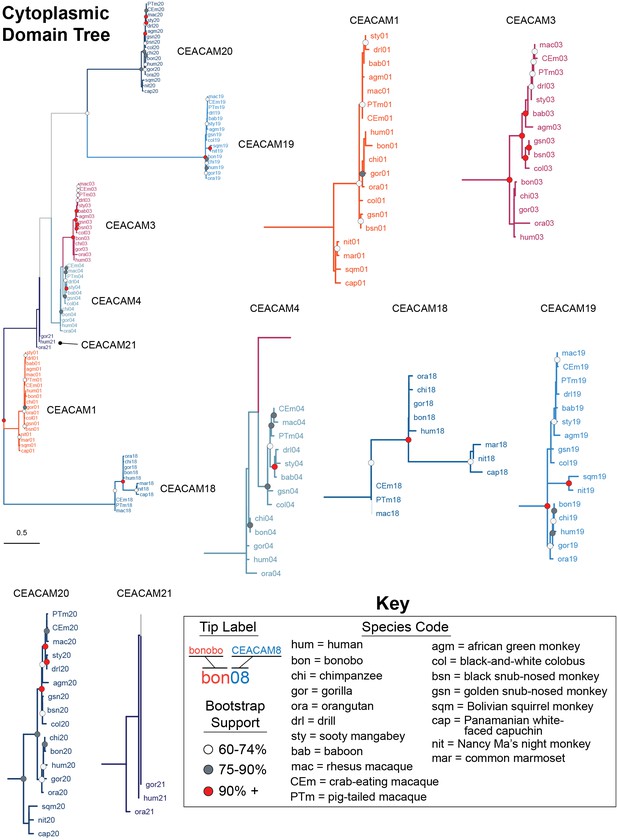
Expanded carcinoembryonic antigen-related cell adhesion molecule (CEACAM) cytoplasmic domain tree.
Maximum likelihood-based phylogeny of CEACAM cytoplasmic domain sequences. Clades encompassing individual CEACAM orthologs are shown isolated and expanded.
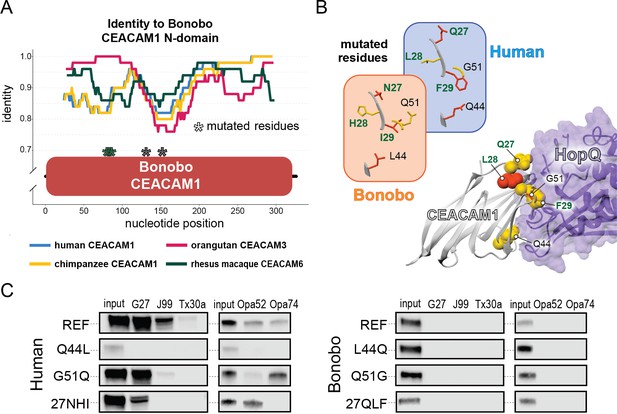
Rapid divergence of the bonobo carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) N-domain impairs bacterial adhesin recognition.
(A) Graph shows a fifty base pair sliding window plotting identity between bonobo CEACAM1 N-domain sequence and other CEACAM sequences. Asterisks mark locations of residues mutated for adhesin-binding assays. (B) Windows show amino acids and their structures at sites selected for mutational analysis in humans and bonobos. Lower right depicts a protein co-crystal structure of human CEACAM1 and Helicobacter pylori G27 HopQ with sites selected for mutagenesis highlighted. (C) Representative western blots of pulldown experiments assaying binding between chimeric human and bonobo CEACAM1 N-domain constructs and bacterial strains.
-
Figure 5—source data 1
Raw and labeled western blot images for Figure 5C.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig5-data1-v3.zip
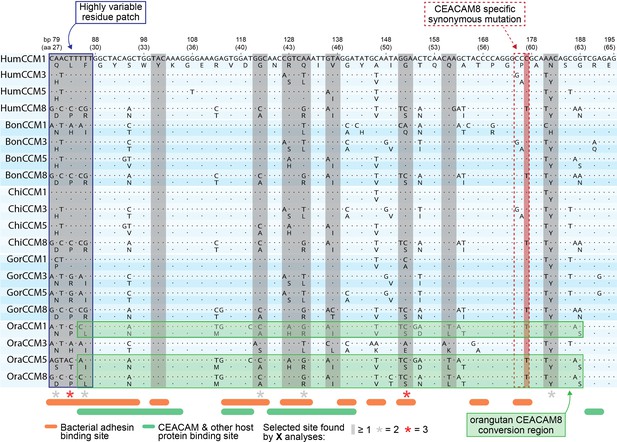
Alignment of rapidly evolving N-domain region in hominids.
Multiple sequence alignment of carcinoembryonic antigen-related cell adhesion molecule (CEACAM)1, CEACAM3, CEACAM5, and CEACAM8 orthologs for human, bonobo, chimpanzee, gorilla, and orangutan. Translation of each nucleotide sequence is positioned on the line below. Sites known to influence adhesin and host protein binding (Figure 2—source data 1F) are indicated as are sites identified as evolving under positive selection.
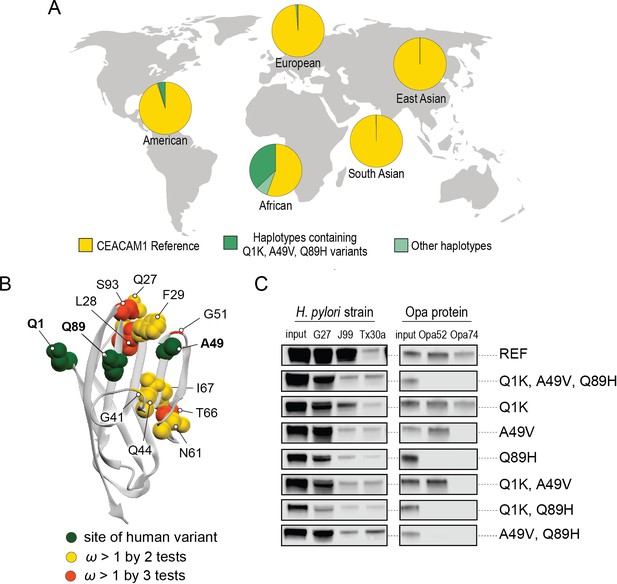
Abundant human carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) variants restrict pathogen binding.
(A) Frequency of haplotypes containing variants Q1K, A49V, and Q89H across human populations (map from BioRender.com). (B) CEACAM1 crystal structure highlighting high-frequency human variants and sites found to be evolving under positive selection across simian primates. (C) Representative western blots of pulldown experiments testing binding between combinations of high-frequency human variants in the human CEACAM1 reference background and bacterial strains.
-
Figure 6—source code 1
Code for analyzing carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) haplotypes and generating graphs for Figure 6A.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig6-code1-v3.zip
-
Figure 6—source data 1
Data files for carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) haplotypes for Figure 6A and Figure 6—figure supplements 1 and 2.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig6-data1-v3.zip
-
Figure 6—source data 2
Raw and labeled western blot images for Figure 6C.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig6-data2-v3.zip
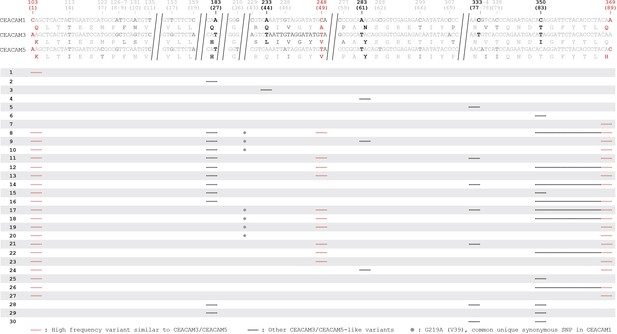
Human carcinoembryonic antigen-related cell adhesion molecule (CEACAM)-like CEACAM1 haplotypes.
Other CEACAM-like human CEACAM1 haplotypes. Alignment of human CEACAM1, CEACAM3, and CEACAM5 N-domain reference nucleotide sequences with amino acid translations below. Long invariable alignment regions are removed. Sites that differ in CEACAM3 or CEACAM5 relative to CEACAM1 are bolded. Sites found in variant CEACAM1 haplotypes are in black. Changes that encode the high-frequency variants Q1K, A49V, and Q89H are in red. Below alignment each row is a unique human CEACAM1 N-domain haplotype. Lines indicate variant regions in CEACAM1. Only haplotypes that increase similarity to CEACAM3 or CEACAM5 are shown.
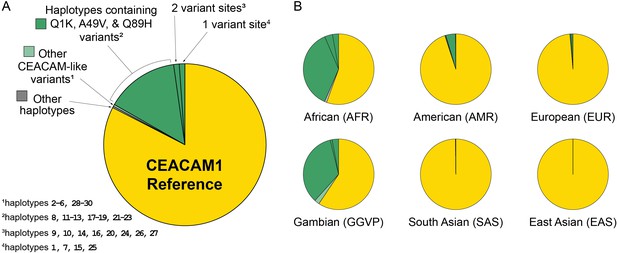
Human carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) haplotype frequencies.
Frequency of variant human CEACAM1 haplotypes. (A) Overall frequency of CEACAM1 variants Q1K, 449V, Q89H, and other variant haplotypes in humans. The indicated CEACAM-like haplotypes are enumerated in Figure 6—figure supplement 1. (B) Frequency of CEACAM1 variants across different human populations.
-
Figure 6—figure supplement 2—source code 1
Code for analyzing carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) haplotypes and generating graphs for Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig5-code5-v3.zip
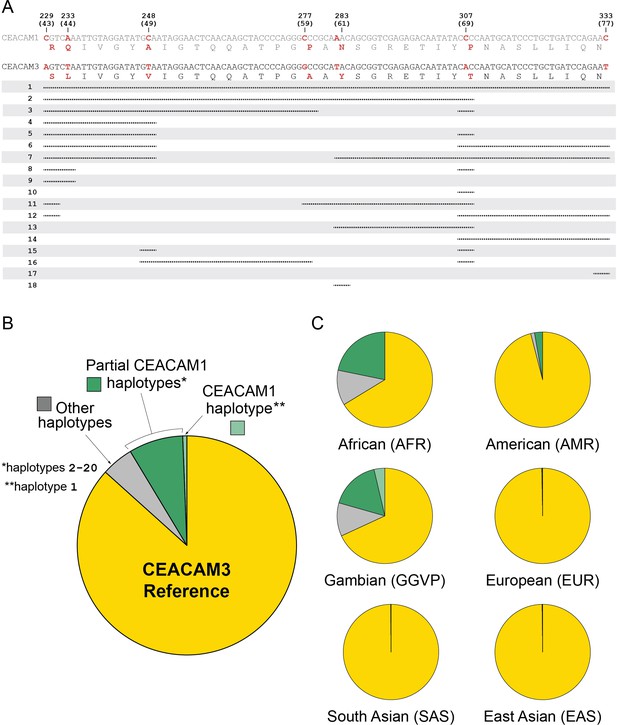
Human carcinoembryonic antigen-related cell adhesion molecule 3 (CEACAM3) variation.
Human CEACAM1-like CEACAM3 haplotypes. (A) Alignment of human CEACAM1 and CEACAM3 reference sequences. Disagreements are bolded in red with the amino acid translation below each sequence. Below alignment each row represents a unique human CEACAM3 haplotype. Lines indicate variant regions that match the human CEACAM1 reference sequence. Only haplotypes that increase similarity to the human CEACAM1 reference sequence are shown. (B) Overall frequency of variant CEACAM3 haplotypes in humans. The CEACAM1-like haplotypes indicated are enumerated in panel A. (C) Frequency of CEACAM3 variants across different human populations.
-
Figure 6—figure supplement 3—source code 1
Code for analyzing carcinoembryonic antigen-related cell adhesion molecule 3 (CEACAM3) haplotypes and generating graphs for Figure 6—figure supplement 3.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig6-code6-v3.zip
-
Figure 6—figure supplement 3—source data 1
Data files for carcinoembryonic antigen-related cell adhesion molecule 3 (CEACAM3) haplotypes for Figure 6—figure supplement 3.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig6-figsupp3-data1-v3.zip
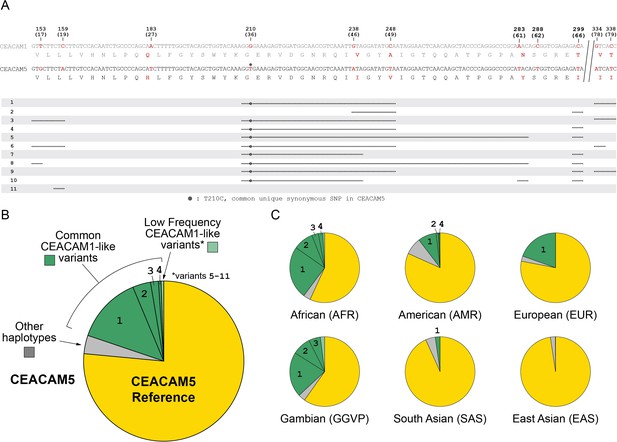
Human carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) variation.
Human CEACAM1-like CEACAM5 haplotypes. (A) Alignment of human CEACAM1 and CEACAM5 reference sequences. Disagreements are bolded in red with the amino acid translation below each sequence. Below alignment each row represents a unique human CEACAM5 haplotype. Lines indicate variant regions that match the human CEACAM1 reference sequence. Only haplotypes that increase similarity to the human CEACAM1 reference sequence are shown. (B) Overall frequency of variant CEACAM5 haplotypes in humans. The CEACAM1-like haplotypes indicated are enumerated in panel A. (C) Frequency of CEACAM5 variants across different human populations.
-
Figure 6—figure supplement 4—source code 1
Code for analyzing and generating graphs for carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) haplotypes for Figure 6—figure supplement 4.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig7-code7-v3.zip
-
Figure 6—figure supplement 4—source data 1
Data files for carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) haplotypes for Figure 6—figure supplement 4.
- https://cdn.elifesciences.org/articles/73330/elife-73330-fig6-figsupp4-data1-v3.zip
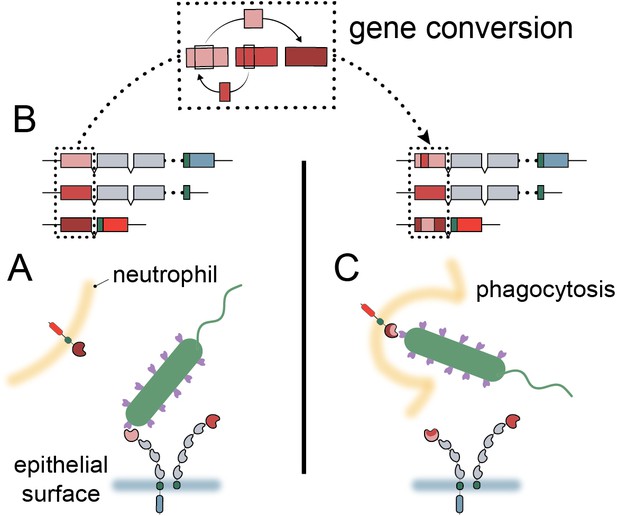
Model of carcinoembryonic antigen-related cell adhesion molecule (CEACAM) evolution in primates.
(A) Bacterial adhesins recognize a subset of epithelial CEACAM proteins and avoid binding with decoy CEACAM receptors present on neutrophils. (B) Gene conversion facilitates the shuffling of regions of the CEACAM N-domain that alter binding to bacterial adhesins. (C) Through gene conversion outlined in B, epithelial CEACAM proteins avoid binding by bacterial adhesins while the CEACAM decoy receptor gains binding, triggering bacterial clearance through phagocytosis.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Helicobacter pylori) | G27 | Baltrus et al., 2009 | ||
| Strain, strain background (Helicobacter pylori) | J99 | Alm et al., 1999 | ||
| Strain, strain background (Helicobacter pylori) | Tx30a | ATCC | 51932 | |
| Strain, strain background (Helicobacter pylori) | omp27::cat-sacB in NSH57 | Yang et al., 2019 | H. pylori strain G27 with HopQ deletion | |
| Strain, strain background (Escherichia coli) | Rosetta (DE3) pLyS | Lab collection | E. coli strain for outer membrane IPTG inducible expression of Neisserial Opa proteins | |
| Strain, strain background (Escherichia coli) | DH5α | Lab collection | E. coli strain for maintenance and propagation of pET-28a plasmid constructs | |
| Strain, strain background (Escherichia coli) | One Shot Top10 Chemically Competent cells | Thermo Fisher Scientific | C404010 | E. coli strain for cloning, maintenance and propagation of pcDNA3 GFP LIC plasmid constructs |
| Cell line (Homo sapiens) | HEK293T | ATCC | RRID:CVCL_0063; CRL-3216 | |
| Recombinant DNA reagent | pET-28a (plasmid) | Genscript | Plasmid backbone for expression of Neisserial Opa proteins | |
| Recombinant DNA reagent | pcDNA3 GFP LIC (plasmid) | Addgene | RRID:Addgene_30127; #30,127 | Plasmid backbone for expression of primate CEACAM1 N-domain constructs in HEK293T cells |
| Antibody | Mouse monoclonal antibody mixture;Mouse α-GFP clones 7.1 and 13.1 | Sigma-Aldrich | RRID:AB_390913; 11814460001 | 1:103 dilution; Primary antibody for visualization of GFP labeled CEACAM1 N-domain constructs |
| Antibody | Goat polyclonal antibody; goat α-mouse conjugated to horseradish peroxidase | Jackson ImmunoResearch | RRID:AB_10015289; 115-035-003 | 1:104 dilution; Secondary antibody for visualization of GFP labeled CEACAM1 N-domain constructs |
| Other | Advansta WesternBright ECL HRP Substrate | Thomas Scientific | K-12049-D50 | Reagent to visualize proteins bound by secondary antibody in a western blot |
| Software, algorithm | PAML4.9h | http://abacus.gene.ucl.ac.uk/software/paml.html Yang, 2007 | RRID:SCR_014932 | |
| Software, algorithm | FUBAR | https://www.datamonkey.orgMurrell et al., 2013 | RRID:SCR_010278 | |
| Software, algorithm | MEME | classic.datamonkey.orgMurrell et al., 2012 | RRID:SCR_010278 | |
| Software, algorithm | GARD | classic.datamonkey.org Kosakovsky Pond et al., 2006 | RRID:SCR_010278 | |
| Sequence-based reagent | bon_gCCM1N_F3 | This paper | PCR primer | Primer for initial amplification of bonobo CEACAM1 N-domain from genomic DNA [TTCACAGAGTGCGTGTACCC] |
| Sequence-based reagent | bon_gCCM1N_R2 | This paper | PCR primer | Primer for initial amplification of bonobo CEACAM1 N-domain from genomic DNA [CCTCCCAGGTTCAAGCGATT] |
| Sequence-based reagent | bon_gCCM1N_F1 | This paper | PCR primer | Primer for secondary amplification of bonobo CEACAM1 N-domain from genomic DNA [CAGTGGAGGGGTGAAGACAC] |
| Sequence-based reagent | bon_gCCM1N_R1 | This paper | PCR primer | Primer for secondary amplification of bonobo CEACAM1 N-domain from genomic DNA [CATGTTGGTCAGGCTGGTCT] |
| Sequence-based reagent | bon_gCCM1N_seqF1 | This paper | Sequencing primer | Primer to sequence bonobo CEACAM1 N-domain amplified from genomic DNA [CCCGTTTTTCCACCCTAATGC] |
| Sequence-based reagent | bon_gCCM1N_seqF4 | This paper | Sequencing primer | Primer to sequence bonobo CEACAM1 N-domain amplified from genomic DNA [GGGGAAAGAGTGGATGGCAA] |
| Sequence-based reagent | bon_gCCM1N_seqR2 | This paper | Sequencing primer | Primer to sequence bonobo CEACAM1 N-domain amplified from genomic DNA [TGGGGGAATCACTCACGGTA] |
| Biological sample (pan paniscus) | AG05253 | Nels Elde | RRID:CVCL_1G37 | Bonobo genomic DNA sample |
| Software, algorithm | R v4.1.2 | https://cran.r-project.org/ | RRID:SCR_003005 | |
| Software, algorithm | Python 3.7 | Python Software Foundation https://www.python.org/ | RRID:SCR_008394 | |
| Software, algorithm | JupyterNotebook 5.7.4 | Project Jupyter https://jupyter.org/ | RRID:SCR_018315 | |
| Software, algorithm | AnacondaNavigator 1.9.12 | Anaconda, Inc https://www.anaconda.com/ |
Additional files
-
Supplementary file 1
A. Oligomers and DNA templates.
Table of oligomers, DNA templates, and their order in assembly reactions used to assemble carcinoembryonic antigen-associated cell adhesion molecule 1 (CEACAM1) N-domain expression plasmids. B. Sources templates for plasmid components. Table listing sources of template sequences for CEACAM1 and other plasmid components used for expression plasmid construction.
- https://cdn.elifesciences.org/articles/73330/elife-73330-supp1-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/73330/elife-73330-transrepform1-v3.docx





