Allosteric mechanism of signal transduction in the two-component system histidine kinase PhoQ
Figures
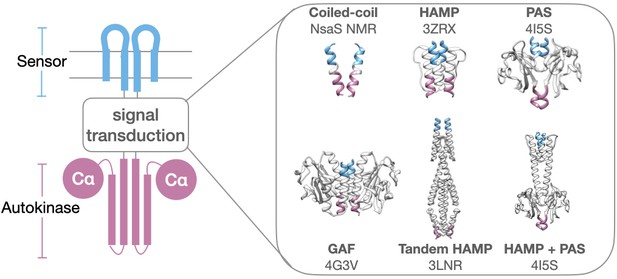
Modular architecture of histidine kinases.
Various protein folds and numbers of signal transduction domains are found inserted between sensor (blue) and autokinase (purple). Structurally elucidated examples include simple coiled-coils (NsaS), HAMP (AF1503), PAS (VicK), GAF (Nlh2), Tandem HAMP (Aer2), and HAMP/PAS domain (VicK). PDB codes are provided in figure, except for NsaS (NMR structure).
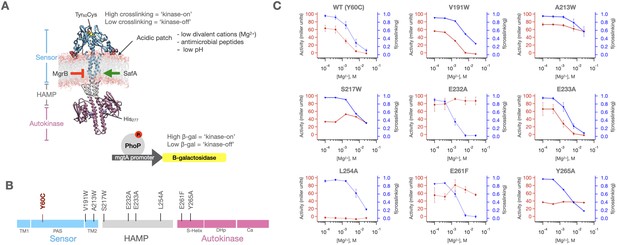
PhoQ single mutants exhibit a range of behaviors.
(A) Molecular Dynamics model of dimeric PhoQ in which the sensor (res. 1–219, blue), HAMP (res. 220–260, grey) and autokinase domains (res 261–494, purple) are annotated. The sensor contains a Y60C mutation (spheres) which shows signal state dependent crosslinking. The autokinase contains the conserved catalytic His277, which upon phosphorylation transfers a phosphoryl group to the response regulator PhoP, which then modulates a mgtA promoter-driven β-galactosidase reporter. Stimuli and regulatory proteins that modulate PhoQ activity are shown. (B) Linear topology diagram of PhoQ. The sensor, HAMP and Autokinase domains are highlighted in blue, gray, and purple, respectively. The locations of mutations in panel (C) are shown. (C) Fraction of sensor crosslinking (blue) and autokinase activity (red) determined for ‘wild type’ (Y60C) PhoQ, as well as eight mutants along the signal transduction pathway (n = 9 for WT, n = 2 for A213W, E232A, E233A, L254A, and E261F, n = 1 for V191W, S217W, and Y265A). The sensor state and autokinase activity do not show identical ligand-dependent behavior as would be predicted by a concerted signaling mechanism. Error bars correspond to± SD, where applicable.
-
Figure 2—source data 1
[Mg2+] dependent activity and sensor crosslinking of mutants in Figure 2C.
- https://cdn.elifesciences.org/articles/73336/elife-73336-fig2-data1-v2.zip
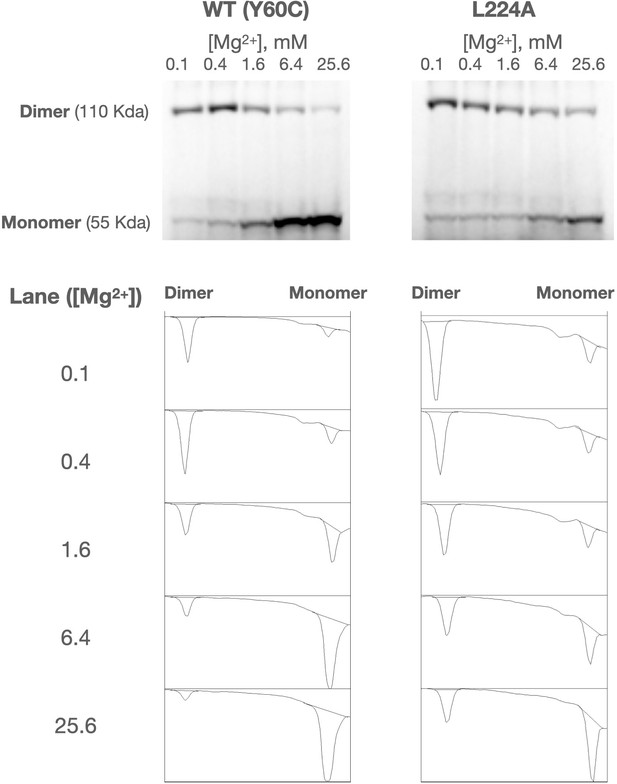
Representative PhoQ crosslinking western blot and quantification of WT (Y60C) and Y60C/L224A mutant.
LDS buffer solubilized membrane samples are separated by SDS-PAGE and western blotted using an anti-pentaHis antibody. Relative amounts of dimeric and monomeric PhoQ are measured to generate a crosslinking efficiency between 0 and 1.
-
Figure 2—figure supplement 1—source data 1
Western blot image for PhoQ sensor crosslinking of WT (Y60C) and L224A mutant.
- https://cdn.elifesciences.org/articles/73336/elife-73336-fig2-figsupp1-data1-v2.zip
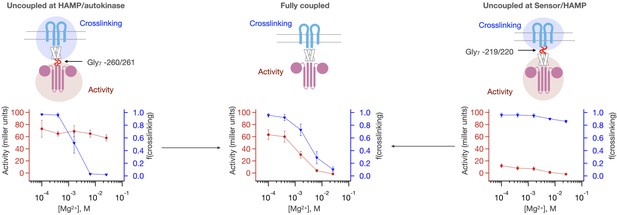
Intrinsic activities of the PhoQ sensor and autokinase domains are altered by coupling to HAMP.
Gly7 insertions are introduced either between the HAMP domain and the autokinase (Gly7 - 260/261, left, n = 3) or between the Sensor and HAMP domain (Gly7 –219/220, right, n = 2) to disrupt allosteric coupling between sensor and autokinase. Both the sensor and autokinase by themselves show high ‘kinase-on’ propensity (red trace, left; blue trace, right). The HAMP domain potentiates the ‘kinase-off’ state, resulting in a more [Mg2+] responsive sensor (blue trace, left), or a lower basal activity autokinase (red trace, right). The fully coupled protein shows correlated sensor/autokinase activity(red and blue traces, middle, n = 9).
-
Figure 3—source data 1
[Mg2+]-dependent activity and sensor crosslinking of PhoQ Gly7 insertions.
- https://cdn.elifesciences.org/articles/73336/elife-73336-fig3-data1-v2.zip
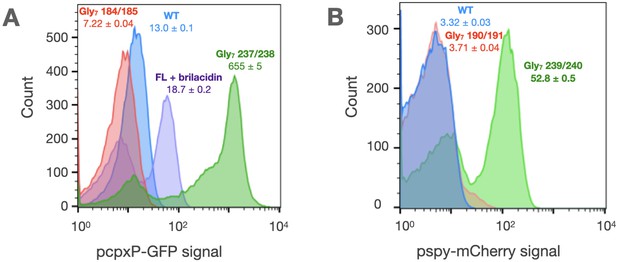
Glycine disconnections in CpxA and BaeS.
(A) The activity of CpxA constructs is measured in AFS51 strain (ΔcpxA) using a pcpxP::GFP reporter. The activity of WT CpxA (blue) is responsive to the antimicrobial mimetic, brilacidin (Scott et al., 2008; Mensa et al., 2011) (purple). The autokinase domain of CpxA when uncoupled (Gly7 237/238) shows very high kinase activity (green), which is repressed to basal levels by the addition of the HAMP domain alone (Gly7 184/185, red). (B) The activity of BaeS constructs is measured in a ΔbaeS ΔcpxA strain using a pspy::mCherry reporter. The autokinase domain of BaeS when uncoupled shows high kinase activity (Gly7 239/240, green) relative to WT (blue), which is repressed by the addition of the HAMP domain alone (Gly7 190/191, red). Median reporter fluorescence values ± STE (n = 20,000) are reported below labels for single experiment.
-
Figure 4—source data 1
Raw flow cytometry data for Gly7 insertions in CpxA and BaeS.
- https://cdn.elifesciences.org/articles/73336/elife-73336-fig4-data1-v2.zip
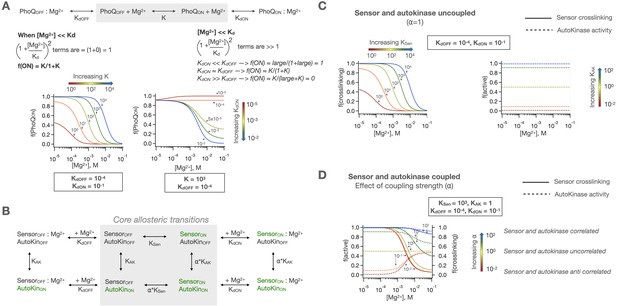
Concerted and two-domain allosteric models for PhoQ signaling.
(A) In a concerted model for signaling, PhoQ has an intrinsic on-off equilibrium (constant = K) which is modulated by Mg2+ binding to either states with corresponding Kds. This allows for modulation of both low and high activity asymptotes and the midpoint of transition but requires perfect correlation between sensor and autokinase signaling states. Equations for calculating population fractions are shown in Methods (Equation 1). (B) The sensor and autokinase domains of PhoQ are allowed to sample both ‘kinase-on’ and ‘kinase-off’ states with equilibrium constants KSen and KAK when the other domain is in the ‘kinase-off’ state. When the other domain is in the ‘kinase-on’ state, the equilibria are scaled by the coupling constant, ⍺. This allows for semi-independent fractions of sensor and autokinase in the ‘kinase-on’ state, which are computed as shown in Methods (Equation 2). (C) In the uncoupled case (⍺ = 1), KSen modulates the sensor identically to the previously described concerted signaling mechanism, while KAK sets the basal autokinase activity. (D) The coupling of these domains with ⍺≠one results in [Mg2+] dependent activity that is either correlated (⍺ > 1) or anticorrelated (⍺ < 1). As ⍺ gets larger, the two domains act more as one concerted protein.
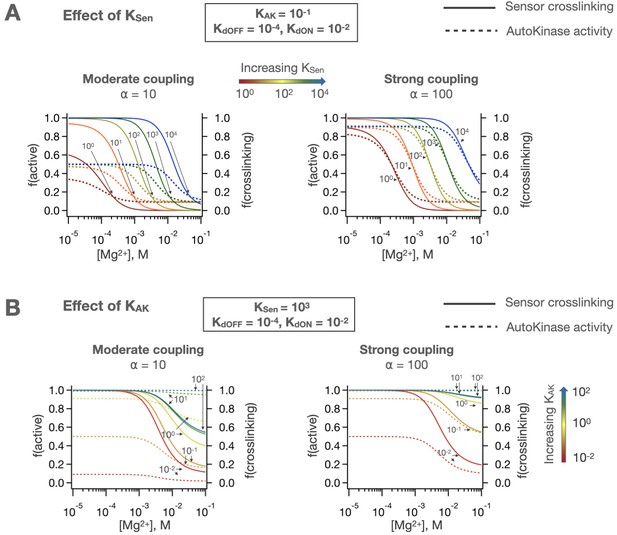
Effects of KSen and KAK on two-domain signaling.
(A) Changes in the intrinsic equilibrium of the sensor affect autokinase activity through coupling, and similarly (B) changes in the intrinsic equilibrium of the autokinase domain can alter the ligand-dependent crosslinking behavior of the sensor.
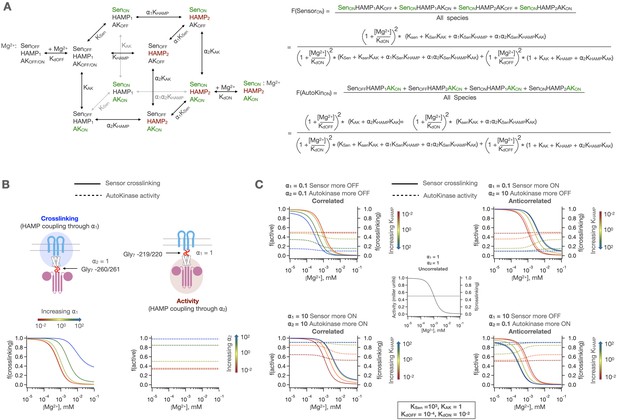
3-domain allosteric coupling model for PhoQ signaling.
(A) The HAMP domain is allowed to sample a two-state equilibrium between ‘HAMP1’ and ‘HAMP2’ states with the equilibrium constant KHAMP. The sensor and autokinase domains of PhoQ are allowed to sample both ‘kinase-on’ and ‘kinase-off’ signaling states while coupled to ‘HAMP1’ state in adjacent HAMP domain with equilibria KSen and KAK, respectively. When adjacent states are in ‘kinase-on’ or ‘HAMP2’ states, the equilibria for transition are scaled by ⍺1 (sensor-HAMP) or ⍺2 (HAMP-autokinase). Predicted fraction of sensor crosslinking or autokinase activity are computed as shown to the right. Please note that Mg2+ binding is allowed for all eight possible signaling states but are omitted except for two reference states for clarity. Similarly, three equilibria arrows and constants have been shaded grey to spatially differentiate them from nearby equilibria. (B) The HAMP domain allows for the independent modulation of the basal state of the sensor or autokinase. When ⍺2 = 1, the HAMP domain modulates the [Mg2+] dependent transition of the sensor, and when ⍺1 = 1, the HAMP domain modulates the basal activity level of the autokinase. (C) The two allosteric coupling constants allow for both correlated and anticorrelated modulation of sensor and autokinase and allow for potentiation of both the ‘kinase-on’ and ‘kinase-off’ states.
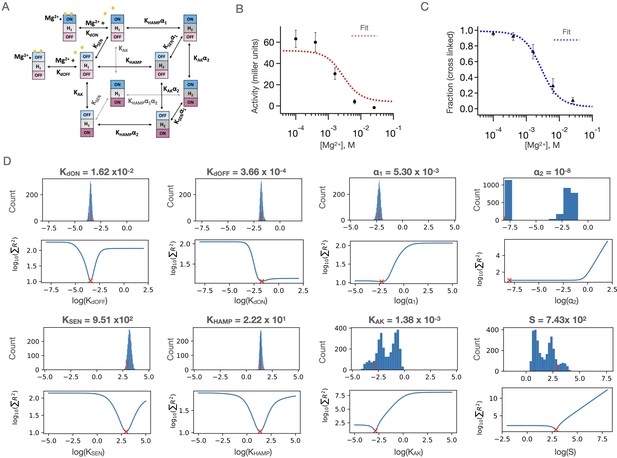
Results of three-domain two-state allosteric model fit of PhoQ activity.
(A) Three-domain two-state allosteric model used for fitting (see also Figure 6A) (B) Fits to the [Mg2+]-dependent kinase activity and (C) sensor crosslinking for ‘wild type’ Y60C PhoQ are shown. Error bars correspond to± SD for n = 9 biological replicates. (D) Bootstrapped confidence intervals (top) and residual sweep analyses (bottom) are shown for all eight global parameters. The value of the fit is indicated with red (x) and (|) marks. The confidence intervals of parameters S, KAK and ⍺2 are further parsed in Figure 7—figure supplement 2, and the confidence intervals for ⍺2 are further parsed in Figure 7—figure supplement 3.
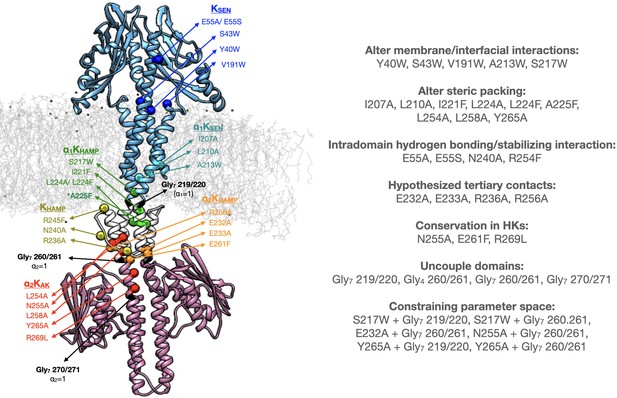
Point mutations and Gly7 insertions in PhoQ.
A molecular dynamics model of PhoQ shows the location of mutations on one monomer with colored Cβ spheres. Colors correspond to mutation labels in Figure 8.
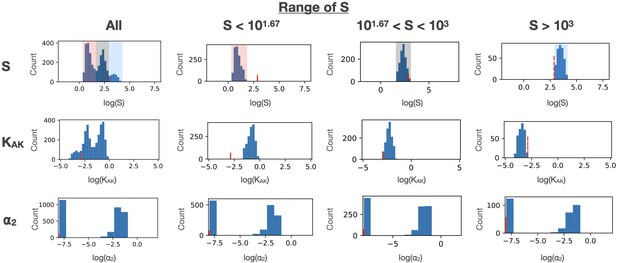
Effect of constraining S and KAK.
The confidence intervals for KAK and ⍺2 parameters are shown as a function of different ranges of S values. S and KAK show strong correlation.
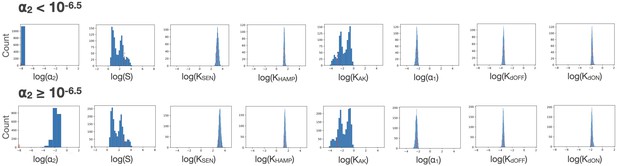
Effect of constraining ⍺2.
The confidence intervals for all global parameters are shown as a function of different ranges of ⍺2 values.
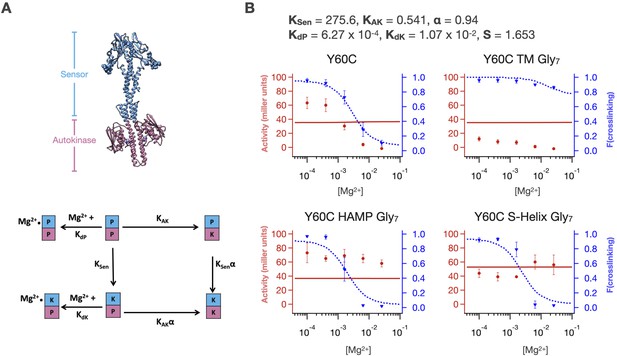
A two-domain two-state allosteric coupling model does not fit set of PhoQ mutants.
(A) In a two-domain signaling model, PhoQ is split into two domains, the ‘Sensor’ which includes the periplasmic PAS domain, TM and HAMP domains, and the ‘Autokinase’. These two domains have their own intrinsic equilibria, KSen and KAK, which are coupled in their ‘on’ states by the parameter ⍺. The population ensemble is perturbed by Mg2+ binding to both states of the Sensor. (B) Global fitting of PhoQ single point mutant and Gly7 insertion data set cannot simultaneously fit sensor crosslinking and autokinase activity data. Representative fits for WT phoQ and Gly7 disconnections are shown.
-
Figure 7—figure supplement 4—source data 1
Two-domain two-state model fitting parameter values for PhoQ mutant dataset.
- https://cdn.elifesciences.org/articles/73336/elife-73336-fig7-figsupp4-data1-v2.xlsx
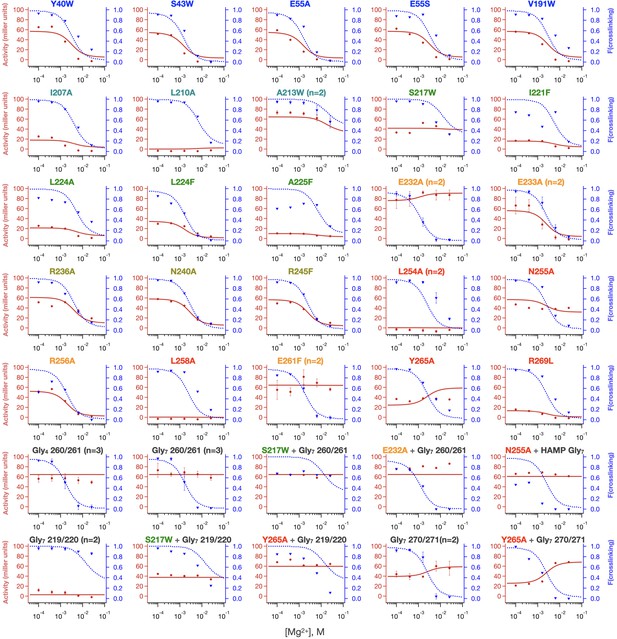
Local fits of sensor crosslinking and kinase activity for 35 PhoQ mutations.
Fits to activity (red line, closed circles) and sensor crosslinking (blue dashed line, triangles) are shown for the entire PhoQ dataset. The color of mutations matches the color scheme in Figure 7—figure supplement 1 to indicate locally varied parameters, and these parameters are listed in Table 1 and Table 2. Confidence intervals and residual sweep analyses are presented in Figure 8—figure supplement 1. Poor fits are highlighted in Figure 8—figure supplement 2.
-
Figure 8—source data 1
[Mg2+]-dependent activity and sensor crosslinking of PhoQ mutants.
- https://cdn.elifesciences.org/articles/73336/elife-73336-fig8-data1-v2.zip
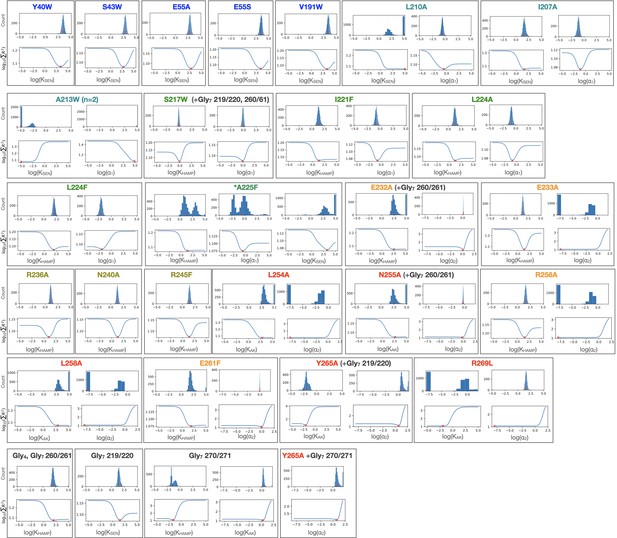
Bootstrapped confidence intervals and residual sweep analyses for PhoQ mutant fits.
Histograms from 3,061 convergent fits of simulated datasets for each local variable are shown in top panels. Residual sweeps in which the sum of residuals of the global fit is plotted as a function of indicated parameter being varied locally is shown in the bottom panels. Values of parameter fits are shown with red (x) and (|) marks.
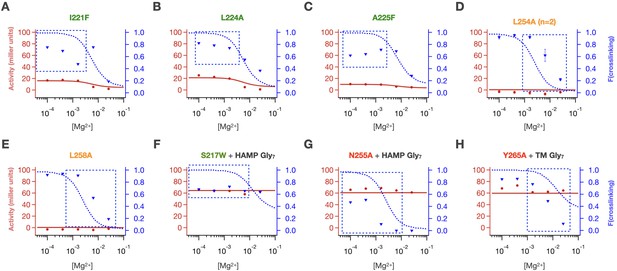
Poor fits were obtained for crosslinking at low [Mg2+] for (A) I221F, (B) L224A and (C) A225F.
Poor midpoints of crosslinking transitions were fit for (D) L254A and (E) L258A. Some combinations of mutations had poor crosslinking fits (F) S217W + HAMP Gly7, (G) N255A + HAMP Gly7 and activity fits (H) Y265A + sensor Gly7.
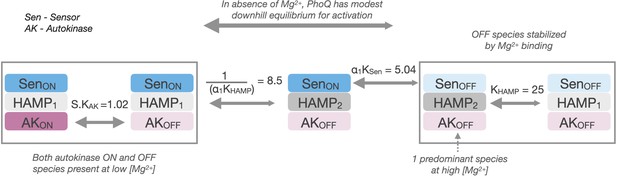
Allosteric pathway for PhoQ activation.
In the absence of Mg2+, PhoQ has a moderate downhill equilibrium to a mixture of active states. Mg2+ binding is sufficient for overpowering this equilibrium and stabilizing the ‘kinase-off’ state, resulting in a predominantly Sensor-off/HAMP2/Autokinase-off population.
Tables
List of mutant parameter fits.
Local parameters whose values remained within 10% of the global fit value are highlighted in bold font and green background. Parameters whose value drifted to one end of the explored parameter range are highlighted in italicized font and orange background. Key: ‘TM7’ → Gly7 insertion at 219/220; ‘HAMP 4’ → Gly4 insertion at 260/261; ‘HAMP 7’ / ‘H7’ → Gly7 insertion at 260/261; ‘SH7’ → Gly7 insertion at 270/271.
| Mutation | KSen | KHAMP | KAK | α1 | α2 | S | KdOFF | KdON |
|---|---|---|---|---|---|---|---|---|
| Y60C | 9.5 E + 02 | 2.2 E + 01 | 1.4 E –03 | 5.3 E –03 | 1.0 E –08 | 7.4 E + 02 | 3.7 E –04 | 1.6 E –02 |
| Y60C HAMP 4 | 4.5 E + 01 | 1.0 E + 00 | ||||||
| Y60C HAMP 7 | 1.0 E + 00 | |||||||
| Y60C SH7 | 2.1 E + 01 | 7.7 E –04 | 1.7 E + 00 | |||||
| Y60C TM7 | 1.0 E + 00 | |||||||
| Y40W | 1.4 E + 03 | |||||||
| S43W | 3.8 E + 02 | |||||||
| E55A | 4.1 E + 02 | |||||||
| E55S | 1.5 E + 03 | |||||||
| V191W | 1.2 E + 03 | |||||||
| I207A | 6.9 E + 02 | 1.1 E –01 | ||||||
| L210A | 3.1 E –03 | 9.1 E + 04 | ||||||
| A213W | 4.0 E + 04 | 1.0 E –05 | ||||||
| S217W | 7.1 E –01 | 7.6 E –01 | ||||||
| S217W + H7 | 1.0 E + 00 | |||||||
| S217W + TM7 | 1.0 E + 00 | |||||||
| I221F | 2.0 E + 01 | 1.5 E –01 | ||||||
| L224A | 1.6 E + 01 | 1.3 E –01 | ||||||
| L224F | 6.8 E + 01 | 1.2 E –02 | ||||||
| A225F | 1.0 E + 03 | 2.4 E + 01 | 2.3 E –01 | |||||
| E232A | 1.1 E + 02 | 1.4 E + 00 | ||||||
| E232A + H7 | 1.0 E + 00 | |||||||
| E233A | 2.3 E + 01 | 1.0 E –08 | ||||||
| R236A | 8.6 E + 00 | |||||||
| N240A | 1.7 E + 01 | |||||||
| R245F | 2.2 E + 01 | |||||||
| L254A | 1.0 E –05 | 1.0 E –08 | ||||||
| N255A | 1.3 E –03 | 4.9 E –01 | ||||||
| N255A + H7 | 1.0 E + 00 | |||||||
| R256A | 3.6 E + 01 | 1.0 E –08 | ||||||
| L258A | 1.0 E –05 | 1.0 E –08 | ||||||
| E261F | 3.9 E + 01 | 9.9 E –01 | ||||||
| Y265A | 4.1 E –04 | 3.2 E + 00 | ||||||
| Y265A + TM7 | 1.0 E + 00 | |||||||
| Y265A + SH7 | 3.8 E + 00 | |||||||
| R269L | 3.3 E –04 | 1.0 E –08 |
parameters used in fitting.
Values in red font indicate parameters fixed to one to account for Gly7 insertion.
| Par. | Fit value | Lower bound | Upper bound | Fit datasets affected |
|---|---|---|---|---|
| KdON | 1.6 E-02 | 1.0 E-08 | 1.0 E + 02 | ALL |
| KdOFF | 3.7 E-04 | 1.0 E-08 | 1.0 E + 02 | ALL |
| α1 | 5.3 E-03 | 1.0 E-05 | 1.0 E + 05 | Y60C_Gly7 270/271, Y60C_Gly7 260/261, Y60C_Gly4 260/261, Y60C, Y40W, Y265A_Gly7 270/271, Y265A, V191W, S43W, R269L, R256A, R245F, R236A, N255A Gly7 260/261, N255A, N240A, L258A, L254A, E55S, E55A, E261F, E233A, E232A_Gly7 260/261, E232A |
| α2 | 1.0 E-08 | 1.0 E-08 | 1.0 E + 02 | Y60C_Gly7 219/220, Y60C, Y40W, V191W, S43W, S217_Gly7 219/220, S217W, R245F, R236A, N240A, L224F, L224A, L210A, I221F, I207A, E55S, E55A, Α225F, Α213W |
| KSen | 9.5 E + 02 | 1.0 E-05 | 1.0 E + 05 | Y60C_Gly7 219/220, Y60C_Gly7 270/271, Y60C_Gly7 260/261, Y60C_Gly4 260/261, Y60C, Y265A_Gly7 270/271, Y265A, S217W_Gly7 219/220, S217W_Gly7 260/261, S217W, R269L, R256A, R245F, R236A, N255A_Gly7 260/261, N255A, N240A, L258A, L254A, L224F, L224A, L210A, I221F, I207A, E261F, E233A, E232A_Gly7 260/261, E232A |
| KHAMP | 2.2 E + 01 | 1.0 E-05 | 1.0 E + 05 | Y60C_Gly7 219/220, Y60C, Y40W, Y265A_Gly7 219/220, Y265A_Gly7 270/271, Y265A, V191W, S43W, R269L, E55S, E55A, Α213W |
| KAK | 1.4 E-03 | 1.0 E-05 | 1.0 E + 05 | Y60C_Gly7 219/220, Y60C_Gly7 260/261, Y60C_Gly4 260/261, Y60C, Y40W, V191W, S43W, S217W_Gly7 219/220, S217W_Gly7 260/261, S217W, R256A, R245F, R236A, N255A_Gly7 260/261, N255A, N240A, L258A, L254A, L224F, L224A, L210A, I221F, I207A, E55S, E55A, E261F, E233A, E232A_Gly7 260/261, E232A, Α225F, Α213W |
| S | 7.4 E + 02 | 1.0 E-05 | 1.0 E + 05 | ALL |
| KSen | 1.4 E + 03 | 1.0 E-05 | 1.0 E + 05 | Y40W |
| KSen | 3.8 E + 02 | 1.0 E-05 | 1.0 E + 05 | S43W |
| KSen | 4.1 E + 02 | 1.0 E-05 | 1.0 E + 05 | E55A |
| KSen | 1.5 E + 03 | 1.0 E-05 | 1.0 E + 05 | E55S |
| KSen | 1.2 E + 03 | 1.0 E-05 | 1.0 E + 05 | V191W |
| KSen | 6.9 E + 02 | 1.0 E-05 | 1.0 E + 05 | I207A |
| α1 | 1.1 E-01 | 1.0 E-05 | 1.0 E + 05 | I207A |
| KSen | 3.1 E-03 | 1.0 E-05 | 1.0 E + 05 | L210A |
| α1 | 9.1 E + 04 | 1.0 E-05 | 1.0 E + 05 | L210A |
| α1 | 1.0 E-05 | 1.0 E-05 | 1.0 E + 05 | Α213W |
| KSen | 4.0 E + 04 | 1.0 E-05 | 1.0 E + 05 | Α213W |
| KHAMP | 7.1 E-01 | 1.0 E-05 | 1.0 E + 05 | S217W_Gly7 219/220, S217W_Gly7 260/261, S217W |
| α1 | 7.6 E-01 | 1.0 E-05 | 1.0 E + 05 | S217W_Gly7 260/261, S217W |
| KHAMP | 2.0 E + 01 | 1.0 E-05 | 1.0 E + 05 | I221F |
| α1 | 1.5 E-01 | 1.0 E-05 | 1.0 E + 05 | I221F |
| KHAMP | 1.6 E + 01 | 1.0 E-05 | 1.0 E + 05 | L224A |
| α1 | 1.3 E-01 | 1.0 E-05 | 1.0 E + 05 | L224A |
| Par. | Fit value | lower bound | Upper bound | Fit datasets affected |
| KHAMP | 6.8 E + 01 | 1.0 E-05 | 1.0 E + 05 | L224F |
| α1 | 1.2 E-02 | 1.0 E-05 | 1.0 E + 05 | L224F |
| KHAMP | 2.4 E + 01 | 1.0 E-05 | 1.0 E + 05 | Α225F |
| α1 | 2.3 E-01 | 1.0 E-05 | 1.0 E + 05 | Α225F |
| KSen | 1.0 E + 03 | 1.0 E-05 | 1.0 E + 05 | Α225F |
| KHAMP | 1.1 E + 02 | 1.0 E-05 | 1.0 E + 05 | E232A_Gly7 260/261, E232A |
| α2 | 1.4 E + 00 | 1.0 E-08 | 1.0 E + 02 | E232A |
| KHAMP | 2.4 E + 01 | 1.0 E-05 | 1.0 E + 05 | E233A |
| α2 | 1.0 E-08 | 1.0 E-08 | 1.0 E + 02 | E233A |
| KHAMP | 8.6 E + 00 | 1.0 E-05 | 1.0 E + 05 | R236A |
| KHAMP | 1.7 E + 01 | 1.0 E-05 | 1.0 E + 05 | N240A |
| KHAMP | 2.2 E + 01 | 1.0 E-05 | 1.0 E + 05 | R245F |
| KAK | 1.0 E-05 | 1.0 E-05 | 1.0 E + 05 | L254A |
| α2 | 1.0 E-08 | 1.0 E-08 | 1.0 E + 02 | L254A |
| KAK | 1.3 E-03 | 1.0 E-05 | 1.0 E + 05 | N255A_Gly7 260/261, N255A |
| α2 | 4.9 E-01 | 1.0 E-08 | 1.0 E + 02 | N255A |
| KHAMP | 3.6 E + 01 | 1.0 E-05 | 1.0 E + 05 | R256A |
| α2 | 1.0 E-08 | 1.0 E-08 | 1.0 E + 02 | R256A |
| KAK | 1.0 E-05 | 1.0 E-05 | 1.0 E + 05 | L258A |
| α2 | 1.0 E-08 | 1.0 E-08 | 1.0 E + 02 | L258A |
| KHAMP | 3.9 E + 01 | 1.0 E-05 | 1.0 E + 05 | E261F |
| α2 | 9.9 E-01 | 1.0 E-08 | 1.0 E + 02 | E261F |
| KAK | 4.1 E-04 | 1.0 E-05 | 1.0 E + 05 | Y265A_Gly7 219/220, Y265A_Gly7 270/271, Y265A |
| α2 | 3.2 E + 00 | 1.0 E-08 | 1.0 E + 02 | Y265A_Gly7 219/220, Y265A |
| KAK | 3.3 E-03 | 1.0 E-05 | 1.0 E + 05 | R269L |
| α2 | 1.0 E-08 | 1.0 E-08 | 1.0 E + 02 | R269L |
| KHAMP | 4.5 E + 01 | 1.0 E-05 | 1.0 E + 05 | Y60C_Gly7 260/261, Y60C_Gly4 260/261 |
| α2 | 1.0 E + 00 | Y60C_Gly7 260;261, Y60C_Gly4 260/261 | ||
| α2 | 1.0 E + 00 | S217W_Gly7 260/261 | ||
| α2 | 1.0 E + 00 | E232A_Gly7 260/261 | ||
| α2 | 1.0 E + 00 | N255A_Gly7 260/261 | ||
| α1 | 1.0 E + 00 | Y60C_Gly7 219/220 | ||
| α1 | 1.0 E + 00 | S217W_Gly7 219/220 | ||
| α1 | 1.0 E + 00 | Y265A_Gly7 219/220 | ||
| KHAMP | 2.1 E + 01 | 1.0 E-05 | 1.0 E + 05 | Y60C_Gly7 270/271 |
| KAK | 7.8 E-04 | 1.0 E-05 | 1.0 E + 05 | Y60C_Gly7 270/271 |
| α2 | 1.7 E + 00 | 1.0 E-08 | 1.0 E + 02 | Y60C_Gly7 270/271 |
| α2 | 3.8 E + 00 | 1.0 E-08 | 1.0 E + 02 | Y265A_Gly7 270/271 |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/73336/elife-73336-transrepform1-v2.docx
-
Source code 1
HPC job submission file to run global fitting of PhoQ activity/crosslinking data on UCSF's wynton compute cluster.
- https://cdn.elifesciences.org/articles/73336/elife-73336-supp1-v2.zip
-
Source code 2
HPC job submission file to run bootstrap fitting for confidence interval analysis of the global PhoQ fit.
- https://cdn.elifesciences.org/articles/73336/elife-73336-supp2-v2.zip
-
Source code 3
Python script slightly changed from 'phoq_fit_local_global.py' to load properly in an iPython session.
- https://cdn.elifesciences.org/articles/73336/elife-73336-supp3-v2.zip
-
Source code 4
Python script containing code for global and local fitting of multi-state models to PhoQ activity/crosslinking data.
- https://cdn.elifesciences.org/articles/73336/elife-73336-supp4-v2.zip
-
Source code 5
Python script containing code fto run bootstrap fitting for confidence intervals of the global PhoQ fit.
- https://cdn.elifesciences.org/articles/73336/elife-73336-supp5-v2.zip
-
Source code 6
Three-domain, two-state model parameter fit values for PhoQ mutant dataset.
- https://cdn.elifesciences.org/articles/73336/elife-73336-supp6-v2.zip





