The NDNF-like factor Nord is a Hedgehog-induced extracellular BMP modulator that regulates Drosophila wing patterning and growth
Figures
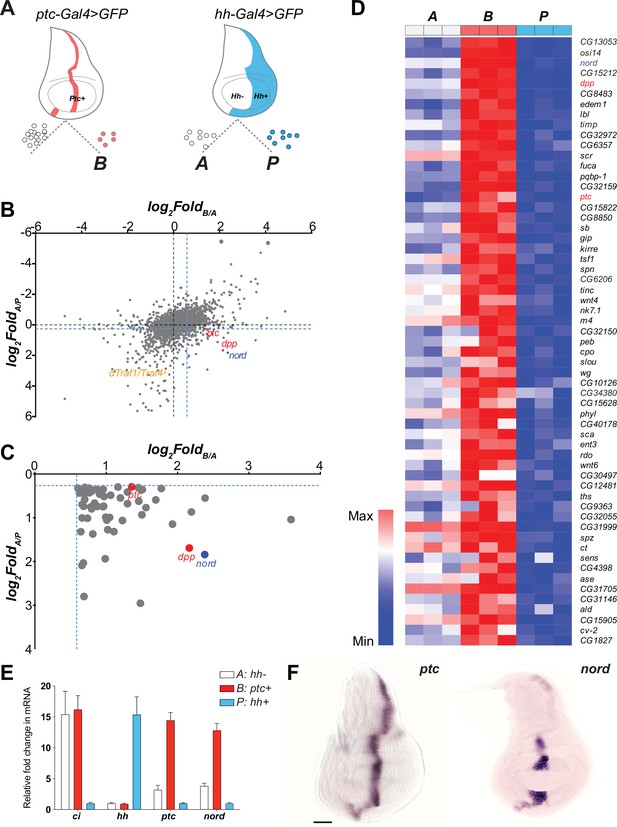
nord is a novel target gene of the Drosophila Hedgehog (Hh) signaling pathway.
(A) Schematic diagram of Drosophila wing imaginal disc: posterior compartment (P: hh+), anterior compartment (A: hh-), and anterior compartment cells adjacent to the A/P boundary (B: ptc+). A, P, and B cells from wing imaginal discs of third instar larvae carrying hh-Gal4 or ptc-Gal4-driven UAS-mCD8-GFP were dissociated and sorted by fluorescence-activated cell sorting (FACS). RNA was isolated and hybridized to microarrays. Differentially expressed genes were identified. (B) Dot plot shows all 14,448 annotated probe sets. Each dot indicates one probe set. The x-axis represents the log2 fold change of each gene in B vs. A cells, and the y-axis represents the log2 fold change of each gene in A vs. P cells. The blue dash lines indicated the threshold used to select differentially expressed genes in each group. Colored dots indicate known or novel Hh pathway targets dTraf1/Traf4 (orange), dpp (red), ptc (red), and nord (blue) in the graph. (C) Zoomed view of the bottom-right corner in panel (B) to show 59 differentially expressed genes, whose expression is significantly increased in B (ptc+) cells but decreased in P (hh+) cells compared with A (hh-) cells. (D) Heatmap shows the expression level of the 59 top-ranking differentially expressed genes in A, B, and P cells. The fold change in B vs. A cells was used to rank the order. (E) Fold changes of ci, hh, ptc, and nord mRNA expression, measured by quantitative reverse transcription PCR and normalized by the expression of the housekeeping gene pkg, in FACS-sorted A, B, or P cells. (F) In situ hybridization of ptc and nord in the third instar larval Drosophila wing discs. Scale bar, 50 μm.
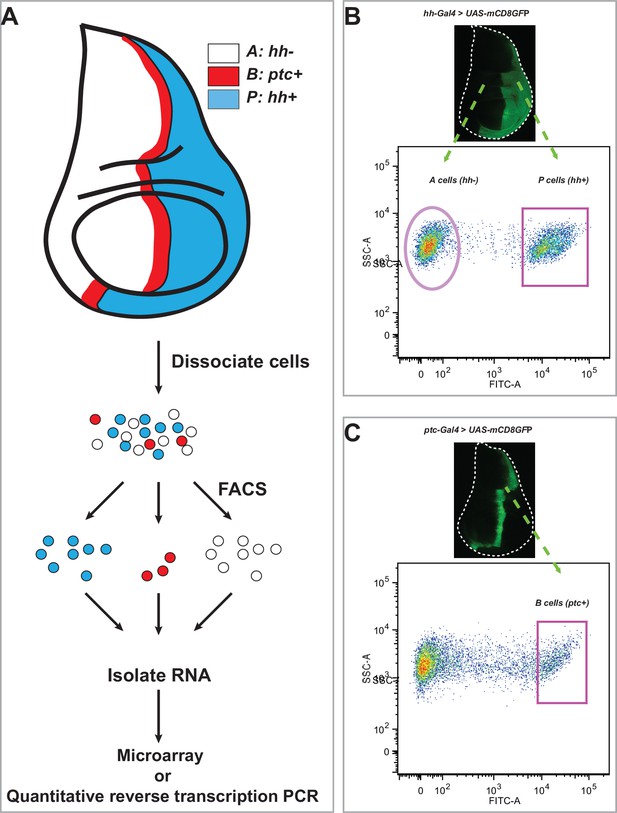
Workflow to isolate RNA from different cell populations sorted from the Drosophila wing imaginal discs.
(A) Posterior compartment cells (P: hh+; blue), anterior compartment cells (A: hh-; white), and anterior compartment cells at the A/P boundary (B: ptc+; red) from third instar Drosophila wing imaginal disc were dissociated and sorted by fluorescence-activated cell sorting (FACS). RNA from the sorted cells was isolated and subjected to either microarray or quantitative reverse transcription PCR. (B) Dissociation and FACS-based purification of P and A cells from wing imaginal discs of third instar larvae carrying hh-Gal4 and UAS-mCD8-GFP. (C) Dissociation and FACS-based purification of B cells from wing imaginal discs of third instar larvae carrying ptc-Gal4 and UAS-mCD8-GFP.
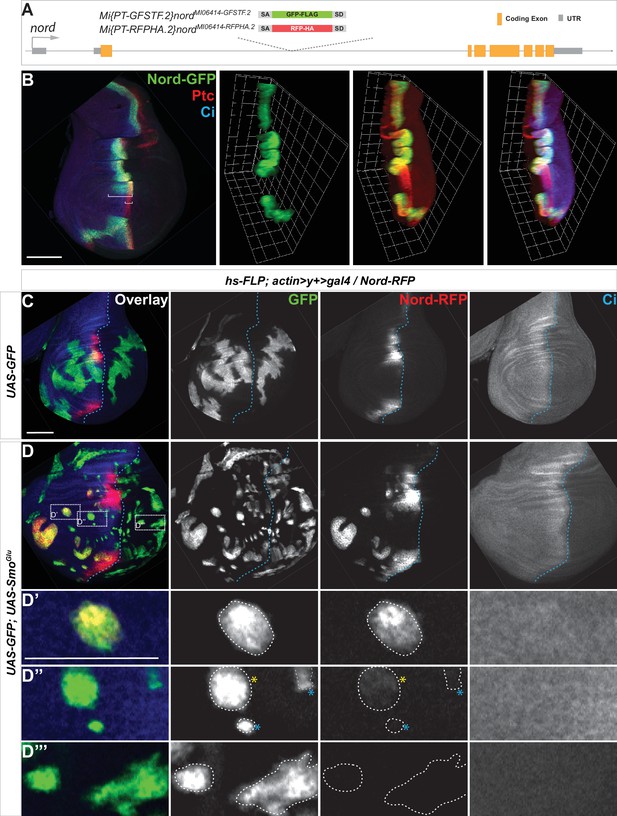
Hedgehog (Hh) signaling regulates nord expression in the Drosophila wing imaginal disc.
(A) Schematic diagram of the wild-type nord locus and the protein-trap alleles of nord. The EGFP-FlAsH-StrepII-3xFLAG (GFSTF) or TagRFP-T-3xHA (RFPHA) tag was inserted in the appropriate orientation and reading frame of nord, which permitted visualization of the Nord protein localization in vivo. (B) Wing imaginal discs from late third instar larvae carrying nord-GFP (Mi{PT-GFSTF.2}nordMI06414-GFSTF.2/+) were immunostained for GFP (green), Ptc (red), and Ci (blue). Maximum intensity z-projection and 3D reconstruction from a confocal image stack show nord expression in a representative wing imaginal disc. White brackets indicate the expression range of Ptc or Nord-GFP. (C, D) Wing imaginal discs from late third instar larvae carrying nord-RFP (Mi{PT-RFPHA.2}nordMI06414-RFPHA.2/+) and flip-out clones expressing the indicated UAS-transgenes were immunostained for HA (Nord-RFP, red), GFP (flip-out clones, green), and Ci (A compartment, blue). (D’–D”’) Zoomed view of the indicated area from panel (D). Note that ectopic nord-RFP is induced in UAS-SmoGlu-expressing clones located in the A compartment flanking the wing pouch (D’), but not in the P compartment (D”’). In the central wing pouch (D”), little (yellow star) or none (blue star) ectopic Nord-RFP was detected in SmoGlu-expressing flip-out clones. Dashed white lines indicate the clone boundary; dashed blue lines indicate the A/P compartment boundary, which is determined by the expression of endogenous Ci. Scale bar, 50 μm.
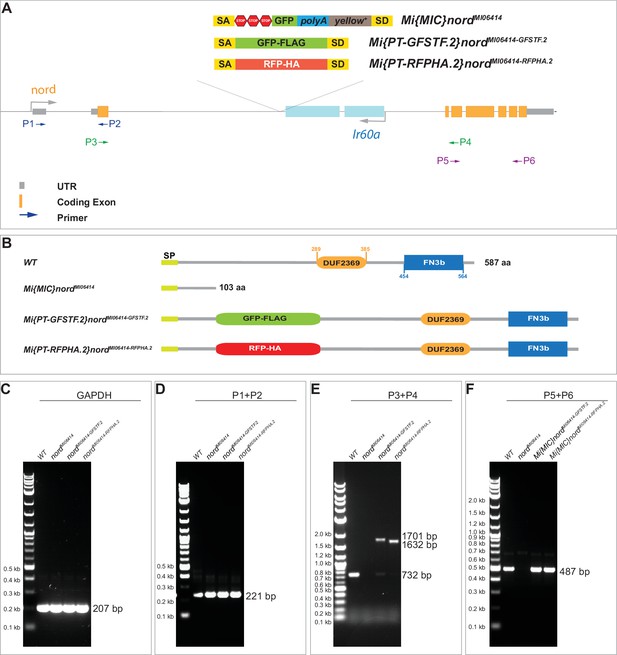
Schematic diagram of the wild-type nord locus, the gene-trap, and the protein-trap alleles of nord.
(A) Schematic diagram of the wild-type nord locus, the gene-trap, and the protein-trap alleles of nord. A Minos-Mediated Integration Cassette (MiMIC) consisting of a splice acceptor site followed by stop codons in all three reading frames was inserted into the first coding intron of nord in the nord gene-trap allele Mi{MIC}nordMI0641. The EGFP-FlAsH-StrepII-3xFLAG (GFSTF) or TagRFP-T-3xHA (RFPHA) protein-trap cassette was inserted into the first coding intron in the appropriate orientation and reading frame of nord, which resulted in the Nord-GFP or Nord-RFP fusion. Note that the gene Ir60a (light blue) is transcribed from the opposite strand. (B) Schematic diagram of the predicted protein or polypeptide products from the wild-type nord locus, the gene-trap, and the protein-trap alleles of nord. (C–F) RT-PCR confirmation of the predicted transcripts from the wild-type nord locus, the gene-trap, and the protein-trap alleles of nord. RNA is isolated and reverse-transcribed from the indicated fly lines. The location of PCR primers is marked in (A).

Alignment of Nord, Nord-GFP, and Nord-RFP encoded by transcripts from the wild-type or nord protein-trap alleles.
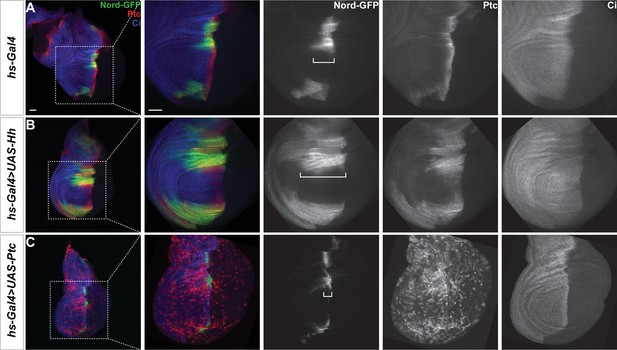
Hedgehog (Hh) signaling regulates nord expression in the Drosophila wing disc.
(A–C) Larvae carrying nord-GFP (Mi{PT-GFSTF.2}nordMI06414-GFSTF.2/+) and hs-Gal4 in combination with the indicated transgenes were heat shocked at 37°C for 30 min. 24–48 hr later, the wing imaginal discs were collected and immunostained for GFP (green), Ptc (red), and Ci (blue). Note that Nord-GFP expression domain is expanded anteriorly with ectopic expression of Hh (B), but reduced with ectopic expression of Ptc (C). White brackets indicate the expression range of Nord-GFP. Scale bar, 50 μm.
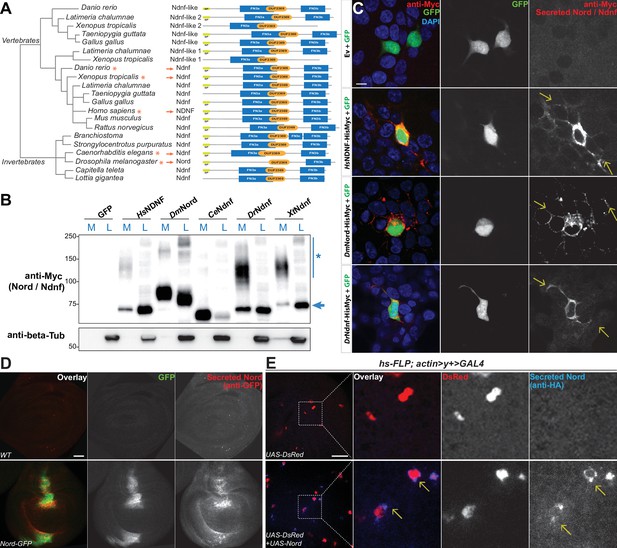
Nord belongs to a family of secreted proteins.
(A) Phylogenetic analysis of Nord homologs from different species. The phylogenetic analysis of Nord homologs was performed with eggNOG (http://eggnogdb.embl.de/#/app/home). The phylogenetic tree is shown with domains predicted using Pfam (https://www.ebi.ac.uk/interpro/). The protein diagrams were then drawn proportional in length to the number of residues. Red stars (*) and red arrows (→) indicate the Ndnf/Nord proteins selected for further analysis. SP, signal peptide; FN3, fibronectin type 3 domain; DUF2369, domain of unknown function 2369. (B) Western blot analysis of HisMyc-tagged Nord or Ndnf protein in medium (M) and cell lysate (L) from transiently expressed in HEK-293 cells. Smeared band (*) indicates a portion of slow migrating Ndnf/Nord protein that may undergo extensive post-translational modification. Blue arrow (→) indicates Ndnf/Nord proteins that migrate as their predicted size. Hs, Homo sapiens; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Dr, Danio rerio; Xt, Xenopus tropicalis. (C) HEK-293 cells co-transfected with GFP and HisMyc-tagged Nord or Ndnf followed by cell-surface staining with anti-Myc antibody to reveal secreted Nord or Ndnf protein (yellow arrows). Note that secreted Nord-Myc and Ndnf-Myc (anti-Myc, red) were also detected several cell diameters away from the expressing cells (GFP, green). Scale bar, 10 μm. (D) Wing imaginal discs from third instar WT or nord-GFP (Mi{PT-GFSTF.2}nordMI06414-GFSTF.2/+) larvae were immunostained for GFP (red) without detergent treatment to label the secreted pool of Nord-GFP proteins. The total Nord-GFP proteins were detected by fluorescence from the GFP tag (green). Scale bar, 20 μm. (E) Wing imaginal discs from third instar larvae carrying flip-out clones expressing UAS-DsRed alone or in combination with UAS-Nord. The discs were immunostained in the absence of detergent to label the secreted pool of HA-tagged UAS-Nord (anti-HA, blue). Flip-out clones were detected by fluorescence from the UAS-DsRed transgene (red). Scale bar, 20 μm.
-
Figure 3—source data 1
Uncropped Western blot for Figure 3.
- https://cdn.elifesciences.org/articles/73357/elife-73357-fig3-data1-v2.pdf
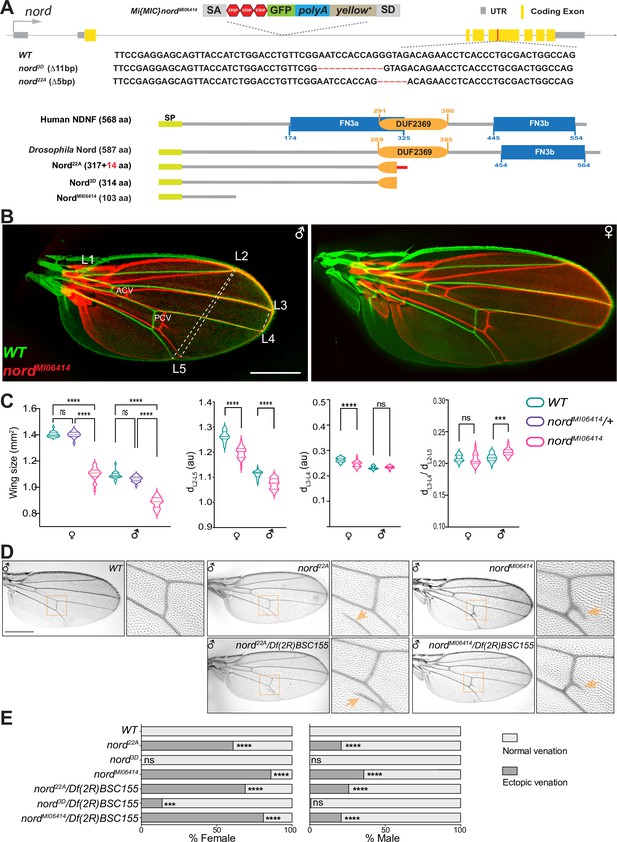
Nord is required for proper crossvein patterning and growth of the Drosophila wing.
(A) Upper panel: schematic diagram of the wild-type nord locus, the gene-trap, and the CRISPR alleles of nord. A Minos-Mediated Integration Cassette (MiMIC) cassette consisting of a splice acceptor site followed by stop codons in all three reading frames was inserted into the first coding intron of nord in the nord gene-trap allele nordMI0641. Detailed view of the deleted regions in the nord mutant alleles generated by the CRISPR/Cas9 system. Lower panel: schematic diagram of the human Neuron-Derived Neurotrophic Factor (NDNF) protein, Drosophila Nord protein, and the predicted polypeptide products from the indicated nord mutant alleles. (B) Adult wings obtained from male or female flies with indicated genotypes were pseudo-colored and overlapped to show the size difference. ACV, anterior crossvein; PCV, posterior crossvein; LV, longitudinal veins (L1–L5). (C) Quantification of wing size, distance between distal ends of LVs L2 and L5 (dL2–L5), L3 and L4 (dL3–L4), and the ratio of dL3–L4 /dL2–L5. Each bar shows the mean ± SD from n = 20 wings. All flies were grown at 25°C. One-way ANOVA followed by Sidak’s multiple comparison test or unpaired two-tailed t-test was used for statistical analysis. ***p<0.001, ****p<0.0001, ns, not significant; au, arbitrary units. (D) Adult wings of flies with the indicated genotypes. Yellow arrowhead indicates ectopic vein near posterior crossvein (PCV) or L5. (E) Quantification of the ectopic venation phenotype in adult wings from flies with the indicated genotypes. n > 50. Two-sided Fisher’s exact tests were used for statistical analysis. ***p<0.001, ****p<0.0001, ns, not significant. Scale bar, 500 μm.

Alignment of wild-type and truncated Nord proteins from the wild-type and nord mutant alleles.
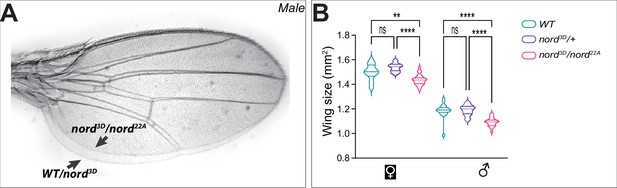
Nord is required for proper growth of the Drosophila wing.
(A) Overlapped adult wings obtained from male flies with indicated genotypes. Scale bar, 500 μm. (B) Quantification of the size of adult wing from flies with indicated genotypes. Each bar shows the mean ± SD from n > 14 adults wings, and representative images are shown in panel (A). All flies were grown at 25°C. One-way ANOVA followed by Sidak’s multiple comparison test was used for statistical analysis. ****p<0.0001, ns, not significant.
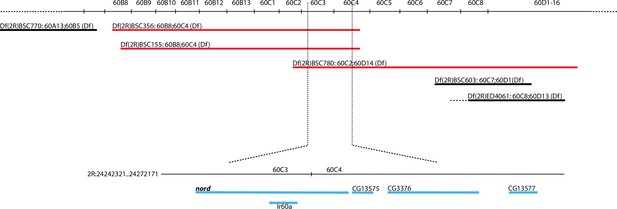
Location of deficiency lines used in the complementation test.
Mi{MIC}nordMI06414 allele failed to complement deficiency lines (red) harboring nord deletion for ectopic crossvein formation. The deficiency lines that complemented the Mi{MIC}nordMI06414 allele for ectopic crossvein formation are indicated in black.
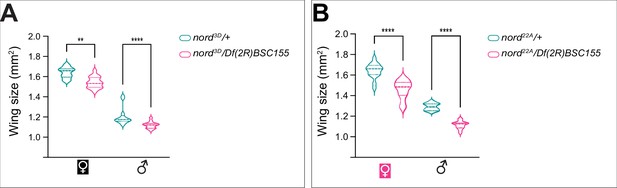
nord22A and nord3D in trans to Df(2R)BSC155 produced smaller wings compared to controls.
(A, B) Quantification of the size of adult wing from male and female flies with indicated genotypes. Each bar shows the mean ± SD from n > 10 adults wings. All flies were grown at 25°C. The unpaired two-tailed t-test was used for statistical analysis. **p<0.01, ****p<0.0001, ns, not significant.
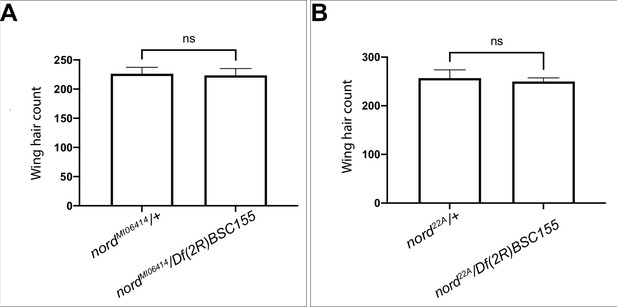
Effects of loss of nord on wing trichome density.
(A, B) Quantitation of wing hair cell density in a fixed area in the posterior wing between the L4 and L5 wing veins (see Materials and methods) for nord MI06414/ Df(2R)BSC155 or nord22A/ Df(2R)BSC155 compared to heterozygous (nordMI06414/+ or nord22A/+) controls. Each bar shows the mean ± SD, n = 10. The unpaired two-tailed t-test was used for statistical analysis. ns, not significant (p>0.05).
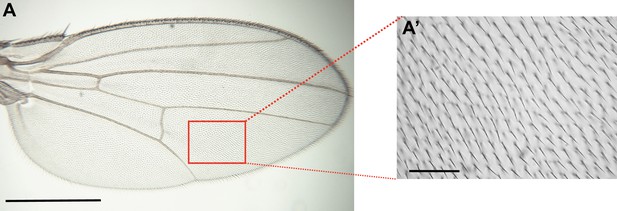
Illustration of wing trichome measurements.
(A) Adult wings were dissected, mounted, and imaged using a ×4 objective. The box indicates the regions in which wing hairs/trichome were counted for each wing. Scale bar, 500 μm. (A’) Wing trichomes from the dorsal wing surface were imaged with a ×40 objective in the region between veins 4 and 5 just distal to the posterior crossvein (PCV). Trichomes were counted manually within the imaged area (37,500 µm2). Scale bar, 50 μm.
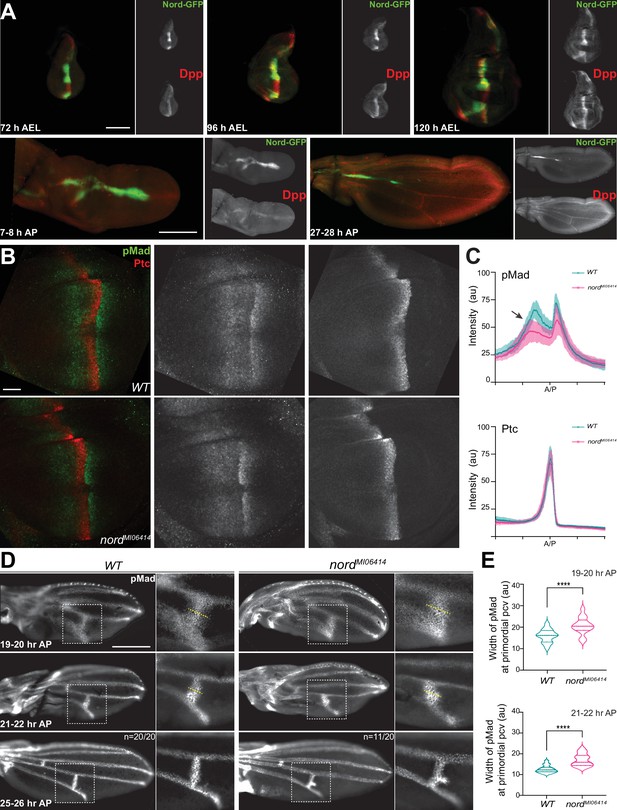
Nord modulates Bone Morphogenetic Protein (BMP) signaling at or above the level of Mad phosphorylation.
(A) Expression of Nord in the wing discs through the third instar larval and early pupal stage. Upper panel: the third instar wing discs from nord-GFP (Mi{PT-GFSTF.2}nordMI06414-GFSTF.2/+) larvae were collected at indicated time points after egg laying (AEL) and were immunostained for GFP (Nord, green) and the pro-domain of Dpp (red). Bottom panel: pupal wings from pupae carrying both nord-GFP were collected at indicated time points after pupation (AP) and were immunostained for GFP (Nord, green) and the pro-domain of Dpp (red). Scale bar, 100 μm. (B) Wing imaginal discs from third instar wild-type and nord mutant (nordMI06414) larvae were immunostained for pMad (green) and Ptc (red). Scale bar, 50 μm. (C) Plotted pixel intensity of pMad or Ptc as a function of A/P position. Each point shows the mean ± SD. n = 15. au, arbitrary units. (D) Anti-pMad staining in wild-type and nordMI06414 pupal wings at indicated hours AP. Scale bar, 100 μm. (E) Quantification of the width (yellow dashed line in panel D) of pMad signal at primordial posterior crossvein (PCV) at indicated time points. Each bar shows the mean ± SD from n > 10 pupal wings. The unpaired two-tailed t-test was used for statistical analysis. ****p<0.0001.
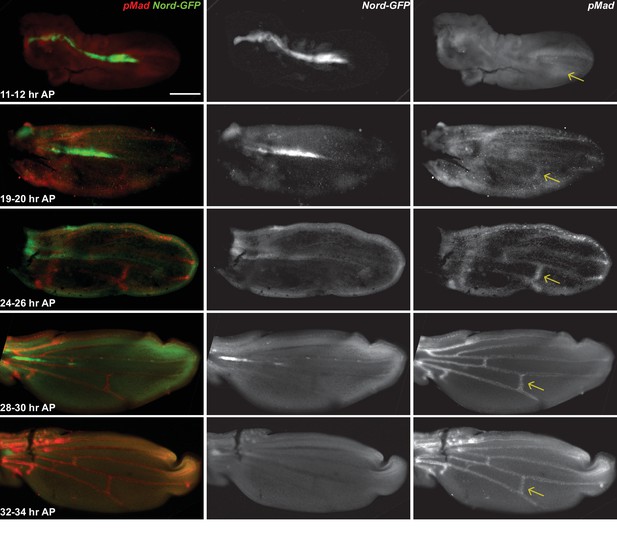
Expression of Nord in the wing discs during the early pupal stage.
(A–C) The early pupal wing discs from nord-GFP (Mi{PT-GFSTF.2}nordMI06414-GFSTF.2/+) pupae were collected at indicated time points after pupation (AP) and were immunostained for GFP (Nord, green) and pMad (red). Note that Nord is expressed in the early pupal wing during the pMad refinement at the posterior crossvein (PCV) primordia. The yellow arrows indicate the PCV primordia. Scale bar, 100 μm.
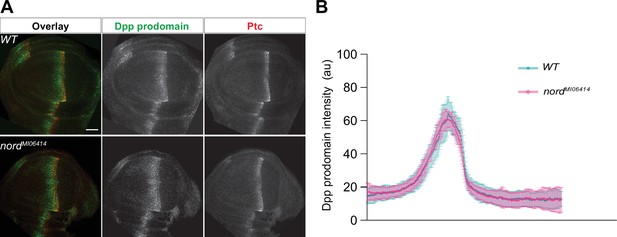
Loss of nord has little effect on the expression of Dpp in wing imaginal discs.
(A) Wing imaginal discs from late third instar wild-type and nord mutant (nordMI06414) larvae were immunostained for the pro-domain of Dpp (green) and Ptc (red). Scale bar, 20 μm. (B) Plotted pixel intensity of Dpp as a function of anteroposterior (A/P) position. The A/P position is determined by the expression of Ptc. Each point shows the mean ± SD. n = 10 wing discs. au, arbitrary units. Scale bar, 50 μm.
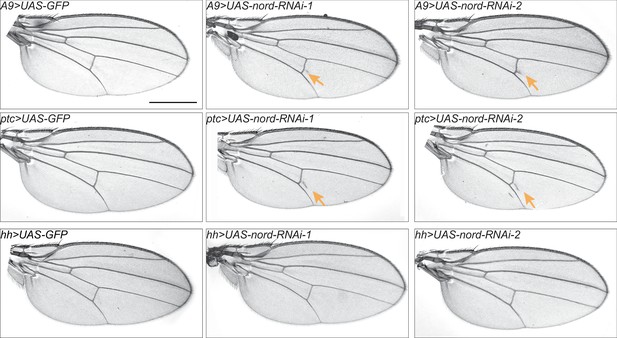
Venation phenotype in adult wings associated with nord knocking down.
Representative images of the ectopic venation phenotype in adult wings from female flies with the indicated genotypes. Scale bar, 500 μm.
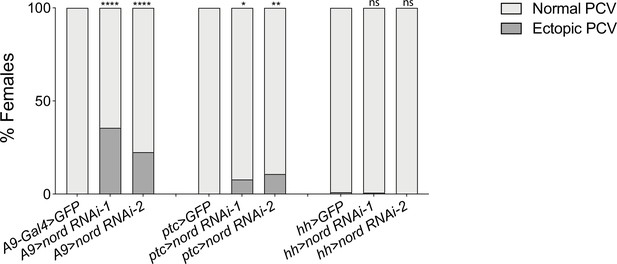
Quantification of the ectopic venation phenotype in adult wings associated with nord knocking down.
Quantification of the ectopic venation phenotype in adult wings from female flies with the indicated genotypes. n > 50. Two-sided Fisher’s exact tests were used for statistical analysis. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05, ns, not significant.
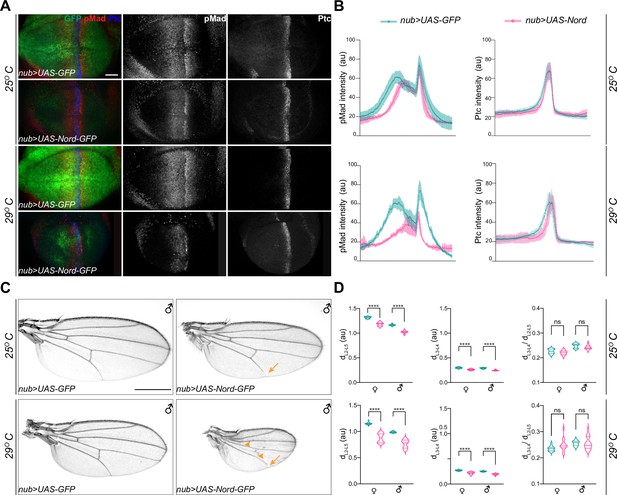
Ectopic expression of Nord attenuates Bone Morphogenetic Protein (BMP) signaling in vivo.
(A) Larvae expressing the indicated transgenes driven by nub-Gal4 were raised at 25 or 29°C. Wing imaginal discs were collected from these larvae in the late third instar stage, and then immunostained for anti-pMad (red), anti-Ptc (blue), and anti-GFP (green). Scale bar, 20 μm. (B) Plotted pixel intensity of pMad or Ptc as a function of anteroposterior (A/P) position. Each point shows the mean ± SD. 25°C: n = 12. 29°C: n = 7. (C) Adult wings from male flies, which expressed the indicated transgenes driven by nub-Gal4 and were raised at 25 and 29°C. Scale bar, 500 μm. Arrows indicate reduced L5, and arrowheads indicate reduced crossveins. (D) Quantification of distance between distal ends of longitudinal veins L2 and L5 (dL2–L5), L3 and L4 (dL3–L4), and the ratio of dL3–L4 /dL2–L5. Each bar shows the mean ± SD from n > 14 wings. au, arbitrary units. The unpaired two-tailed t-test was used for statistical analysis. ****p<0.0001, ns, not significant.
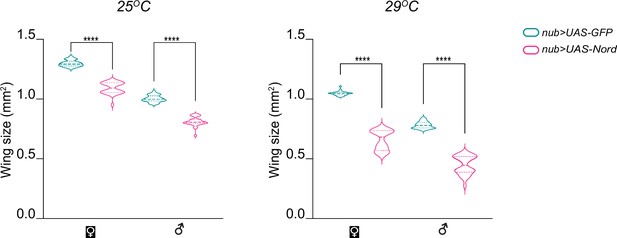
Ectopic Nord expression leads to reduction of the wing size.
Quantification of the adult wing size reduction associated with ectopic Nord expression driven by nub-Gal4. The comparisons were made separately in males and females at indicated temperatures. Each box shows the mean ± SD from n = 30 adults wings, and representative images are shown in Figure 7C. Two-tailed unpaired t-test was used for statistical analysis. ****p<0.0001.
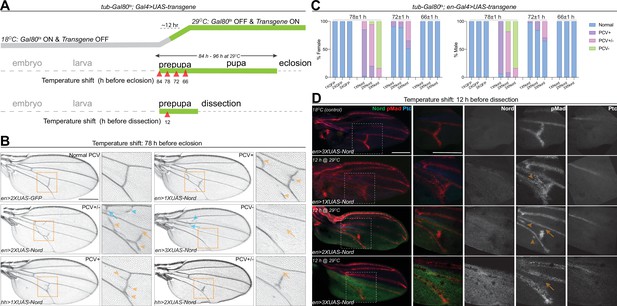
Nord is a dosage-dependent modulator of the Bone Morphogenetic Protein (BMP) signaling.
(A) Upper row: schematic diagram of temporal UAS-Nord expression in the posterior compartment of wing discs using the temperature-sensitive Gal80 (Gal80ts) system. At the permissive temperature (18°C), Gal4 activity is blocked by Gal80ts. At the restrictive temperature (29°C), Gal80ts is unable to repress Gal4, which then induces expression of the UAS-transgenes. Middle and lower rows: timing of temperature shift. Embryos and larvae are grown at 18°C, and prepupae are transferred to 29°C at the indicated time points before eclosion (middle row) or before dissection (lower row). (B) Representative adult wings from flies that carry the indicated transgenes under the control of tub-Gal80ts together with hh-Gal4 or en-Gal4. The animals were grown at 18°C till prepupa stage, and then transferred to 29°C at 78 hr before eclosion. Yellow arrowhead indicates ectopic posterior crossvein (PCV); yellow arrow indicates reduced PCV; blue arrowhead indicates ectopic anterior crossvein (ACV); blue arrow indicates reduced ACV. Scale bar, 500 μm. (C) Quantification of PCV phenotypes in adult wings of female and male flies with indicated genotypes. The animals were grown at 18°C till prepupa stage, and then transferred to 29°C around 78, 72, or 66 (±1) hr before eclosion. n > 30 wings for each genotype at a given temperature shift time point. (D) Representative pupal wing from larvae that carry the indicated transgenes under the control of tub-Gal80ts together with en-Gal4. The animals were grown at 18°C till prepupa stage, and then transferred to 29°C at 12 hr before dissection. The collected pupal wings were immunostained for anti-pMad (red), anti-Ptc (blue), and anti-GFP (Nord-GFP, green). Yellow arrowhead indicates ectopic pMad around the primordial PCV; yellow arrow indicates reduced pMad signal around the primordial PCV. Scale bar, 100 μm.
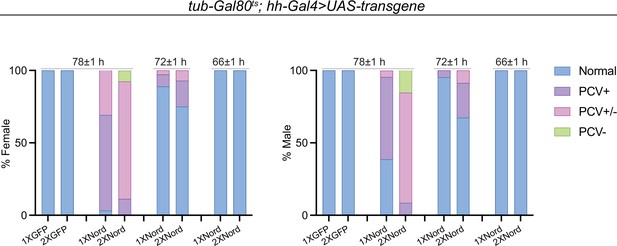
Spaciotemporal expression of exogenous Nord in the pupal wing leads to abnormal posterior crossvein (PCV) patterning.
Quantification of PCV phenotypes in adult wings from female and male flies carrying tub-Gal80ts together with hh-Gal4-driven UAS-GFP or UAS-Nord. The animals were grown at 18°C till prepupa stage, and then transferred to 29°C around 78, 72, or 66 (±1) hr before eclosion. n > 30 wings for each genotype at a given temperature shift time point. Representative adult wings are shown in Figure 8D.
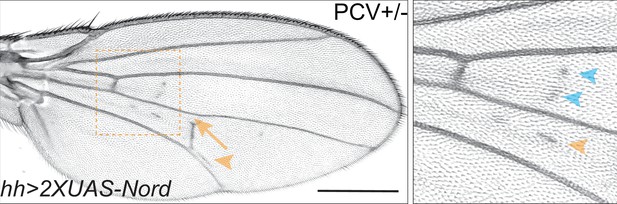
Ectopic Nord secreted from the posterior compartment induces ectopic crossvein growth in the anterior compartment.
Adult wings from flies that carry 2XUAS-Nord under the control of tub-Gal80ts together with hh-Gal4. The animals were grown at 18°C till prepupa stage, and then transferred to 29°C at 78 hr before eclosion. Yellow arrowhead indicates ectopic posterior crossvein (PCV); yellow arrow indicates reduced PCV; blue arrowhead indicates ectopic crossvein in the anterior compartment. Scale bar, 500 μm.
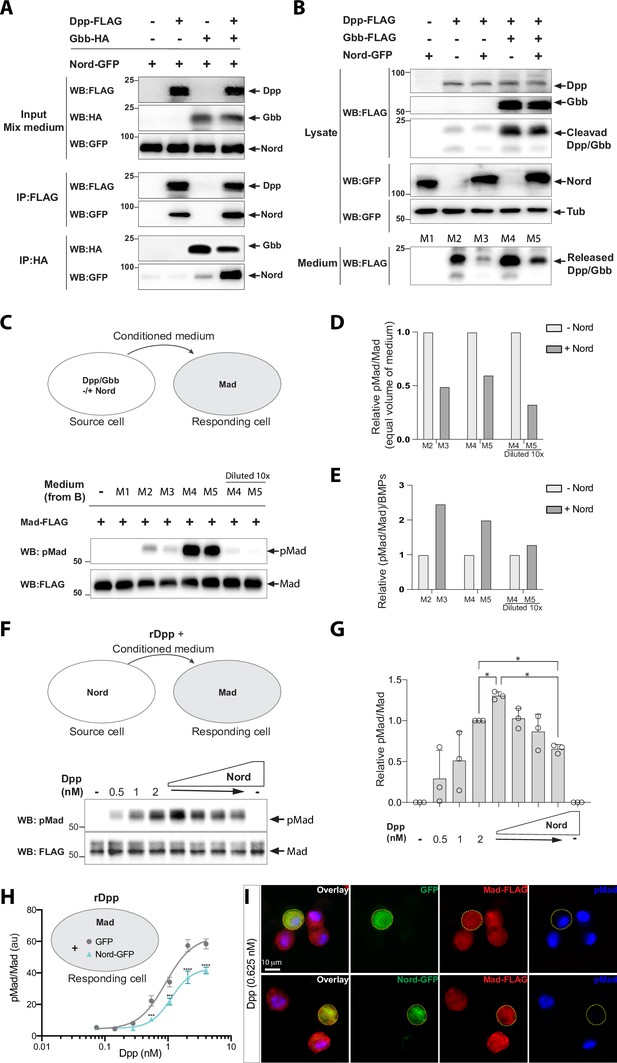
Nord binds to Decapentaplegic (Dpp) and attenuates Bone Morphogenetic Protein (BMP) signaling in vitro.
(A) Co-immunoprecipitation of Nord with the BMP ligands Dpp and Glass-bottom boat (Gbb). Medium from S2 cells transfected for expression of GFP-tagged Nord were mixed with medium from cells expressing FLAG-tagged Dpp and HA-tagged Gbb alone or in combination, followed by incubation with anti-FLAG or anti-HA antibody-coupled beads overnight at 4°C. Precipitated proteins were analyzed by Western blotting with indicated antibodies. Nord was immunoprecipitated with Dpp and, to a lesser extent, with Gbb. The amount of immunoprecipitated Nord was increased when Dpp and Gbb were co-transfected. (B) S2 cells were transfected for expression of FLAG-tagged Dpp or Gbb with or without GFP-tagged Nord (source cell). Both cell lysate and conditioned medium from the source cells were collected and followed by Western blot analysis. Loading was controlled by probing the blot for tubulin. The amount of Dpp or Dpp-Gbb ligands released into the medium was reduced when Nord was co-expressed in the source cells. (C) Comparison of BMP signaling activities of conditioned media in a cell-based signaling assay. After incubating the conditioned media collected in (B) with S2 cells stably expressing the FLAG-Mad transgene (Mad-S2, responding cell) for 1 hr at room temperature, the responding cells were washed and lysed. The lysates were probed with anti-pMad and anti-FLAG antibodies to detect both the phosphorylated Mad and total Mad protein, respectively. (D, E) Quantification of the Western blot data in panels (B) and (C). The levels of the secreted BMP ligands from the medium (anti-FLAG in panel B), phosphorylated Mad (anti-pMad in panel C), and total Mad (anti-FLAG in panel C) were measured based on the band intensity. The signaling activity from an equal volume of conditioned medium was determined by the ratio of pMad and the corresponding total Mad (pMad/Mad) in panel (C), which was then normalized to the condition without the exogenous Nord-GFP to calculate a relative signaling activity (D), or normalized to the secreted ligand amount to calculate the relative ligand activity ([pMad/Mad]/BMPs) (E). (F) Mad-S2 cells were treated with a recombinant Dpp peptide (rDpp) in the absence or presence of conditioned medium containing raising levels of Nord for 1 hr at room temperature, the responding cells were washed and lysed. The lysates were probed with anti-pMad and anti-FLAG antibodies to detect both the phosphorylated Mad and total Mad protein, respectively. (G) Quantification of the Western blot data in panel (F). The phosphorylated Mad (anti-pMad) and total Mad (anti-FLAG) levels were measured based on the band intensity. The signaling activity from each conditioned medium was determined by the ratio of pMad and the corresponding total Mad (pMad/Mad), which was then normalized to the condition with 2 nM Dpp but no additional Nord. Panel (F) is representative of n = 3 independent experiments. Each point shows the mean ± SD. One-way ANOVA test with Tukey’s multiple comparison was used for statistical analysis, and a significant difference was considered by *p<0.05. (H) Mad-S2 cells were transiently transfected for expression of GFP or Nord-GFP. 48 hr after transfection, the cells were treated with recombinant Dpp peptides for 1 hr. Upon treatment, the cells were washed, fixed, and stained by anti-FLAG to detect total Mad and anti-pMad to detect phosphorylated Mad. The average pMad levels were measured and normalized to the total Mad levels, and then plotted against different Dpp concentrations (0–5 nM). Each point shows the mean ± SD, n > 10. au, arbitrary units. The unpaired two-tailed t-test was used for statistical analysis. ***p<0.001, ****p<0.0001. (I) Representative images of Mad-S2 cells treated by 0.625 nM Dpp. Scale bar, 10 μm.
-
Figure 8—source data 1
Uncropped Western blot for Figure 8.
- https://cdn.elifesciences.org/articles/73357/elife-73357-fig8-data1-v2.pdf
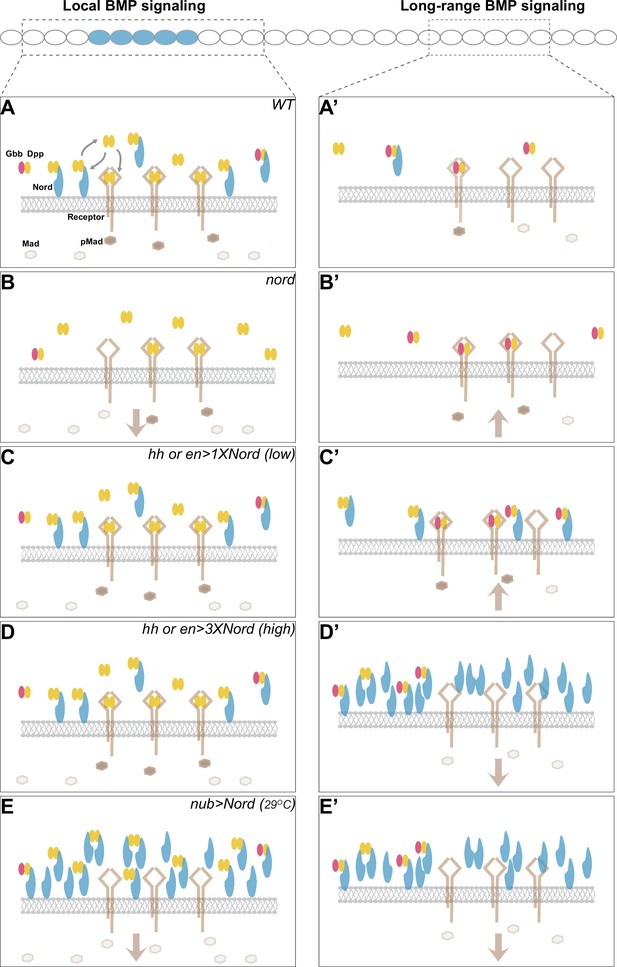
A model for dosage-dependent modulation of Bone Morphogenetic Protein (BMP) signaling by the Hedgehog (Hh) target gene nord.
Illustration of potential concentration- and location-dependent mechanism of Nord as a dosage-dependent BMP signaling modulator. The source cells of Nord (and Decapentaplegic [Dpp]) are labeled in blue, and the responding cells in focus are indicated by the dashed line. Note that Nord could be either diffusible or membrane/matrix-associated. (A, A’) In the wild-type wing discs, nord is induced by the Hh signal along the anteroposterior (A/P) compartment boundary flanking the central wing pouch, and thus is expressed in a subset of Dpp-secreting cells. We propose that membrane/matrix-associated Nord-mediated binding of Dpp and Dpp-Gbb both enhances the local BMP signaling activity by augmenting ligand concentration near the Nord/Dpp-secreting cells and impedes the mobilization of Dpp, especially the long-range BMP signaling mediator Dpp-Gbb heterodimer. The diffusible pool of Nord is more likely to interfere with BMP reception. (B, B’) Loss of nord simultaneously leads to reduced local BMP and increased long-range BMP activities, and therefore gives rise to the seemingly opposite phenotypes of reduced wing size and ectopic posterior crossvein (PCV), both of which are attributable to alteration of BMP activity at the level of Mad phosphorylation (pMad). (C–E, C’-E’) Nord misexpression experiments. Low levels of ectopic Nord in the P compartment increase BMP signaling activity (C, C’), whereas high levels of Nord, either in the P compartment (D, D’) or throughout the wing pouch (E, E’), inhibit BMP signaling activity. The altered BMP signaling activities are reflected by the pMad levels.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Drosophila melanogaster) | hs-FLP | Golic and Lindquist, 1989 | FBti0000785 | Chr X |
| Genetic reagent (D. melanogaster) | Actin>y+>Gal4 | Ito et al., 1997 | FBti0009983 | Chr 2 |
| Genetic reagent (D. melanogaster) | ptc-Gal4 | Hinz et al., 1994 | FBal0287777 | Chr 2 |
| Genetic reagent (D. melanogaster) | hh-Gal4 | BDSC | RRID:BDSC_67046 | Chr 3 |
| Genetic reagent (D. melanogaster) | en-Gal4 | BDSC | RRID:BDSC_30564 | Chr 2 |
| Genetic reagent (D. melanogaster) | nub-Gal4 | BDSC | RRID:BDSC_25754 | Chr 2 |
| Genetic reagent (D. melanogaster) | MS1096-Gal4 | BDSC | RRID:BDSC_8860 | Chr X |
| Genetic reagent (D. melanogaster) | MS1096-Gal4; UAS-Dcr-2 | BDSC | RRID:BDSC_25706 | Chr X; Chr 2 |
| Genetic reagent (D. melanogaster) | A9-Gal4 | BDSC | RRID:BDSC_8761 | Chr X |
| Genetic reagent (D. melanogaster) | hs-Gal4 | Halfon et al., 1997 | FBtp0065595 | Chr 2 |
| Genetic reagent (D. melanogaster) | tub-Gal80ts | BDSC | RRID:BDSC_7019 | Chr 2 |
| Genetic reagent (D. melanogaster) | tub-Gal80ts | BDSC | RRID:BDSC_7018 | Chr 3 |
| Genetic reagent (D. melanogaster) | UAS-Dicer-2 | BDSC | RRID:BDSC_24650 | Chr 2 |
| Genetic reagent (D. melanogaster) | UAS-Dicer-2 | BDSC | RRID:BDSC_24651 | Chr 3 |
| Genetic reagent (D. melanogaster) | UAS-GFP | BDSC | RRID:BDSC_1521 | Chr 2 |
| Genetic reagent (D. melanogaster) | UAS-GFP | BDSC | RRID:BDSC_1522 | Chr 3 |
| Genetic reagent (D. melanogaster) | UAS-mCD8-GFP | BDSC | RRID:BDSC_5137 | Chr 2 |
| Genetic reagent (D. melanogaster) | UAS-DsRed | BDSC | RRID:BDSC_6282 | Chr 3 |
| Genetic reagent (D. melanogaster) | UAS-Ptc | Johnson et al., 1995 | Chr 3 | |
| Genetic reagent (D. melanogaster) | UAS-Hh | Lee et al., 1992 | Chr 3 | |
| Genetic reagent (D. melanogaster) | UAS-SmoGlu | Zhang et al., 2004 | Chr 3 | |
| Genetic reagent (D. melanogaster) | UAS-nord-RNAi-1 | VDRC | RRID:VDRC_v38151 | Chr 2 |
| Genetic reagent (D. melanogaster) | UAS-nord-RNAi-2 | VDRC | RRID:VDRC_v38152 | Chr 2 |
| Genetic reagent (D. melanogaster) | w[1,118]; Df(2R)BSC770/SM6a | BDSC | RRID:BDSC_26867 | Chr 2 |
| Genetic reagent (D. melanogaster) | [1,118]; Df(2 R)BSC356/SM6a | BDSC | RRID:BDSC_24380 | Chr 2 |
| Genetic reagent (D. melanogaster) | y(1) w[1,118]; Df(2R)BSC155/CyO-Df(2R)B80, y[+] | BDSC | RRID:BDSC_9691 | Chr 2 |
| Genetic reagent (D. melanogaster) | w[1,118]; Df(2R)BSC780/SM6a | BDSC | RRID:BDSC_27352 | Chr 2 |
| Genetic reagent (D. melanogaster) | w[1,118]; Df(2R)BSC603/SM6a | BDSC | RRID:BDSC_25436 | Chr 2 |
| Genetic reagent (D. melanogaster) | w[1,118]; Df(2R)ED4061, P{w[+ mW.Scer\FRT.hs3] = 3'. RS5 + 3.3'}ED4061/SM6a | BDSC | RRID:BDSC_9068 | Chr 2 |
| Genetic reagent (D. melanogaster) | w1118; PBac{y[+ mDint2]=vas-Cas9}VK00027 | BDSC | RRID:BDSC_51324 | Chr 3 |
| Genetic reagent (D. melanogaster) | y w | BDSC | RRID:BDSC_1495 | Chr X |
| Genetic reagent (D. melanogaster) | y w; Mi{MIC}nordMI06414 | BDSC | RRID:BDSC_42389 | Chr 2 |
| Genetic reagent (D. melanogaster) | y w; Mi{PT-GFSTF.2}nordMI06414-GFSTF.2/CyO | BDSC | RRID:BDSC_60250 | Chr 2 |
| Genetic reagent (D. melanogaster) | w1118; PBac{y[+ mDint2]=vas-Cas9}VK00027 | BDSC | RRID:BDSC_51324 | Chr 3 |
| Genetic reagent (D. melanogaster) | y w; Mi{PT-RFPHA.2}nordMI06414-RFPHA.2 | This paper | Chr 2 | |
| Genetic reagent (D. melanogaster) | UAS-Nord-HA-GFP (Chr.2) | This paper | Chr 2 | |
| Genetic reagent (D. melanogaster) | UAS-Nord-HA-GFP (Chr.3) | This paper | Chr 3 | |
| Genetic reagent (D. melanogaster) | w; nord22A | This paper | Chr 2 | |
| Genetic reagent (D. melanogaster) | w; nord3D | This paper | Chr 2 | |
| Antibody | Anti-Ci (rat monoclonal) | DSHB | Cat# 2A1; RRID:AB_2109711 | IF: (1:50) |
| Antibody | Anti-Ptc (mouse monoclonal) | DSHB | Cat# Apa 1; RRID:AB_528441 | IF: (1:50) |
| Antibody | Anti-Dpp prodomain (rabbit polyclonal) | Akiyama and Gibson, 2015 | A gift from M. Gibson | IF: (1:100) |
| Antibody | Anti-GFP (rabbit polyclonal) | Molecular Probes | Cat# A-11122, RRID:AB_221569 | IF: (1:2000) |
| Antibody | Anti-GFP (chicken polyclonal) | Abcam | Cat# ab13970, RRID:AB_300798 | IF: (1:2000) |
| Antibody | Anti-beta tubulin (mouse monoclonal) | DSHB | Cat#E7; RRID:AB_2315513 | WB: (1:5000) |
| Antibody | Anti-HA.11 (mouse monoclonal, 16B12) | Covance | Cat# MMS-101P-1000; RRID: AB_291259 | IF: (1:1000) |
| Antibody | Anti-HA (rabbit polyclonal, SG77) | Thermo Fisher Scientific | Cat# 71-5500; RRID:AB_2533988 | WB: (1:1000) |
| Antibody | Anti-FLAG (mouse monoclonal, M2) | Sigma-Aldrich | Cat# F3165; RRID:AB_259529 | WB: (1:200) |
| Antibody | Anti-FLAG (rabbit polyclonal) | Sigma-Aldrich | Cat# F7425; RRID:AB_439687 | WB: (1:200) |
| Antibody | Anti-Myc (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-40; RRID:AB_627268 | WB: (1:200) |
| Antibody | Anti-anti-phospho-Smad1/5 (rabbit polyclonal, Ser463/465) | Cell Signaling Technology | Cat# 9516; RRID:AB_491015 | IF: (1:100) |
| Antibody | Anti-FLAG (mouse monoclonal, M2) Magnetic Beads | Sigma-Aldrich | Cat# M8823; RRID:AB_2637089 | IP |
| Antibody | Anti-HA (rat monoclonal, 3F10) Affinity Matrix | Roche | Cat# 11815016001;RRID:AB_390914 | IP |
| Antibody | Fluorophore-conjugated secondary antibodies | Jackson ImmunoResearch Lab | NA | IF: (1:500) |
| Antibody | HRP-conjugated secondary antibodies | Jackson ImmunoResearch Lab | NA | WB: (1:10,000) |
| Other | DAPI | MilliporeSigma | Cat# D9542 | (1 µg/mL) |
| Other | Fetal bovine serum | Omega Scientific | Cat# FB-02 | |
| Other | Schneider’s Drosophila Media | Invitrogen | Cat# 21720 | |
| Other | Dulbecco’s Modification of Eagle’s Medium (DMEM) | Corning | Cat# 10-013CM | |
| Other | Blasticidin S HCl | Thermo Fisher Scientific | Cat# R21001 | |
| Other | Penicillin-Streptomycin-Glutamine (100×) | Thermo Fisher Scientific | Cat# 10378016 | |
| Other | Antifade mounting media | VECTASHIELD | Cat# H-1000 | |
| Other | FuGENE HD transfection reagent | Promega | Cat# E2311 | |
| Other | 16% paraformaldehyde aqueous solution | Electron Microscopy Sciences | Cat# 15710 | |
| Other | Dissociation buffer | Sigma | Cat# C-1544 | |
| Other | Recombinant Dpp | R&D Systems | Cat# 59-DP-020 | |
| Other | Elastase | Sigma | Cat# E-0258 | |
| Other | Propidium iodide | Invitrogen | Cat# P3566 | |
| Other | DNAse I | Thermo Fisher Scientific | Cat# AM2222 | |
| Other | Maxima Reverse Transcriptase | Thermo Fisher Scientific | Cat# EP0742 | |
| Other | SYBR Green Supermix | Bio-Rad | Cat# 1708880 | |
| Other | RNeasy Mini | QIAGEN | Cat# 74104 | |
| Recombinant DNA reagent | pAcSV-Nord-GFP | Supplementary file 3 | Nord coding sequence (NM_138056) was fused in frame with C-terminal GFP tag and cloned into pAcSV vector | |
| Recombinant DNA reagent | pcDNA3.1-Nord-HisMyc | Supplementary file 3 | Nord coding sequence (NM_138056) was fused in frame with C-terminal HisMyc tag and cloned into pcDNA3.1 vector | |
| Recombinant DNA reagent | pcDNA3.1-HsNDNF-HisMyc | Supplementary file 3 | HsNDNF coding sequence (NM_024574) was fused in frame with C-terminal HisMyc tag and cloned into pcDNA3.1 vector | |
| Recombinant DNA reagent | pcDNA3.1-CeNdnf-HisMyc | Supplementary file 3 | CeNdnf coding sequence (NM_067881) was fused in frame with C-terminal HisMyc tag and cloned into pcDNA3.1 vector | |
| Recombinant DNA reagent | pcDNA3.1-DrNdnf-HisMyc | Supplementary file 3 | DrNdnf coding sequence (XM_684842) was fused in frame with C-terminal HisMyc tag and cloned into pcDNA3.1 vector | |
| Recombinant DNA reagent | pcDNA3.1-XtNdnf-HisMyc | Supplementary file 3 | XtNdnf coding sequence (NM_001122800) was fused in frame with C-terminal HisMyc tag and cloned into pcDNA3.1 vector | |
| Recombinant DNA reagent | pBRAcpA- Dpp-FLAG | Supplementary file 3 | ||
| Recombinant DNA reagent | pBRAcpA- Gbb-FLAG | Supplementary file 3 | ||
| Recombinant DNA reagent | pBRAcpA-Gbb-HA | Supplementary file 3 | ||
| Recombinant DNA reagent | pBRAcpA-FLAG-Mad | Supplementary file 3 | ||
| Sequence-based reagent | Primer: pkgForward: GTCCCAAGACCCGTGAGCTCTTCGC | This paper | Primers used in Figure 1 for quantitative reverse transcription PCR | |
| Sequence-based reagent | Primer: pkgReverse: TCCGTGTTCCACTTGGCGCAGCAAG | This paper | Primers used in Figure 1 for quantitative reverse transcription PCR | |
| Sequence-based reagent | Primer: ciForward: CGACCACCAGGAGGAAGTAT | This paper | Primers used in Figure 1 for quantitative reverse transcription PCR | |
| Sequence-based reagent | Primer: ciReverse: AATCGGAATAAGGCGATGAC | This paper | Primers used in Figure 1 for quantitative reverse transcription PCR | |
| Sequence-based reagent | Primer: hhForward: GGATTCGATTGGGTCTCCTA | This paper | Primers used in Figure 1 for quantitative reverse transcription PCR | |
| Sequence-based reagent | Primer: hhReverse: GAATCTGACTTGACGGAGCA | This paper | Primers used in Figure 1 for quantitative reverse transcription PCR | |
| Sequence-based reagent | Primer: ptcForward: CGATGTGGTCGATGAGAAAT | This paper | Primers used in Figure 1 for quantitative reverse transcription PCR | |
| Sequence-based reagent | Primer: ptcReverse: CGAGGTGGGACTGGAATACT | This paper | Primers used in Figure 1 for quantitative reverse transcription PCR | |
| Sequence-based reagent | Primer: nordForward: CACCGCAAAAGTGTCCTTCG | This paper | Primers used in Figure 1 for quantitative reverse transcription PCR | |
| Sequence-based reagent | Primer: nordReverse: CAGGTTCAGCACAAATCGCT | This paper | Primers used in Figure 1 for quantitative reverse transcription PCR | |
| Sequence-based reagent | Primer: ptcForward: atggaccgcgacagcctccca | Hsia et al., 2017 | Anti-sense probe (600 bp) for ptc | Primers used in Figure 1 for generating in situ probes |
| Sequence-based reagent | Primer: ptcReverse: TAATACGACTCACTATAGGG gaggtggcgcaggatctgctc | Hsia et al., 2017 | Anti-sense probe (600 bp) for ptc | Primers used in Figure 1 for generating in situ probes |
| Sequence-based reagent | Primer: nordForward: gaaatccgggtgaagctgctacg | This paper | Anti-sense probe (666 bp) for nord | Primers used in Figure 1 for generating in situ probes |
| Sequence-based reagent | Primer: nordReverse: TAATACGACTCACTATAGGG atgcagcgaagctttgggtatgg | This paper | Anti-sense probe (666 bp) for nord | Primers used in Figure 1 for generating in situ probes |
| Sequence-based reagent | P1: nord exon 1Forward: GCAAGTGGCAAGAGCTGAAC | This paper | Primers used in Figure 2—figure supplement 1 for confirming the predicted transcripts from different Nord alleles | |
| Sequence-based reagent | P2: nord exon 1/2Reverse: GTGTTCTGCGGTTTTGCCTG | This paper | Primers used in Figure 2—figure supplement 1 for confirming the predicted transcripts from different Nord alleles | |
| Sequence-based reagent | P3: nord exon 2Forward: CGCACTCAGAGGTTGTTTCA | This paper | Primers used in Figure 2—figure supplement 1 for confirming the predicted transcripts from different Nord alleles | |
| Sequence-based reagent | P4: nord exon 4/5Reverse: GCTCCTTTCCCACTTGACGA | This paper | Primers used in Figure 2—figure supplement 1 for confirming the predicted transcripts from different Nord alleles | |
| Sequence-based reagent | P5: nord exon 6Forward: AGGCTCTGTTCCGGGATTTG | This paper | Primers used in Figure 2—figure supplement 1 for confirming the predicted transcripts from different Nord alleles | |
| Sequence-based reagent | P6: nord exon 8Reverse: AAATGCAGCGAAGCTTTGGG | This paper | Primers used in Figure 2—figure supplement 1 for confirming the predicted transcripts from different Nord alleles | |
| Sequence-based reagent | GapdhForward: GCCACCTATGACGAAATCAAGGCTA | This paper | Primers used in Figure 2—figure supplement 1 for confirming the predicted transcripts from different Nord alleles | |
| Sequence-based reagent | GapdhReverse: GGAGTAACCGAACTCGTTGTCGTAC | This paper | Primers used in Figure 2—figure supplement 1 for confirming the predicted transcripts from different Nord alleles | |
| Sequence-based reagent | 5′-GGACCTGTTCGGAATCCACC-3′ | This paper | Guide RNA sequence used to generate different Nord alleles using CRISPR/Cas9 system | |
| Sequence-based reagent | 5′-GGGTGAGGTTCTGTCTACCC-3 | This paper | Guide RNA sequence used to generate different Nord alleles using CRISPR/Cas9 system | |
| Cell line (D. melanogaster) | S2 | DGRC | S2-DGRC | |
| Cell line (Homo sapiens) | embryonic kidney cell line HEK 293 | ATCC | CRL-1573 | |
| Software, algorithm | Fiji | NIH | RRID:SCR_002285 | |
| Software, algorithm | GraphPad Prism | GraphPad software | RRID:SCR_002798 |
Additional files
-
Supplementary file 1
Genes that are differentially expressed in A, B, and P cells.
Genes (probe sets) whose expression is not only higher in A cells than P cells (FoldA/P > 1.2), but also higher in the A/P boundary adjacent B cells than general A cells (FoldB/P > 1.5) were selected as potential Hedgehog (Hh)-induced target genes. A total of 61 probe sets (59 unique genes) were identified as potential Hh signaling target genes.
- https://cdn.elifesciences.org/articles/73357/elife-73357-supp1-v2.xlsx
-
Supplementary file 2
Quantification of the posterior crossvein (PCV) phenotypes associated with trans-heterozygotic combinations of Mi{MIC}nordMI06414 and tested deficiency lines.
- https://cdn.elifesciences.org/articles/73357/elife-73357-supp2-v2.pdf
-
Supplementary file 3
The genotype of larvae, pupae, or adult flies from where wing discs, pupal, or adult wings were collected and imaged in each figure.
- https://cdn.elifesciences.org/articles/73357/elife-73357-supp3-v2.docx
-
Supplementary file 4
The nucleotide sequences used to express Nord or various Neuron-Derived Neurotrophic Factor (NDNF) fusion proteins.
- https://cdn.elifesciences.org/articles/73357/elife-73357-supp4-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/73357/elife-73357-transrepform1-v2.pdf





