Legionella pneumophila modulates host energy metabolism by ADP-ribosylation of ADP/ATP translocases
Figures
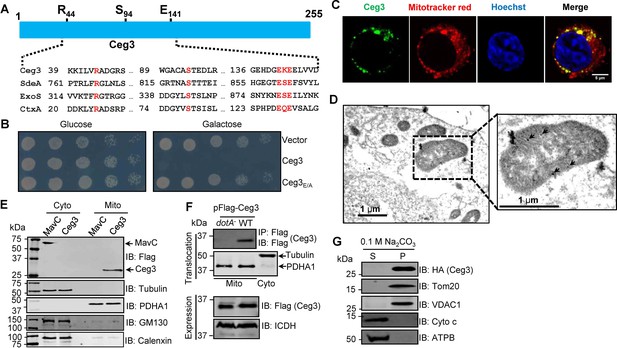
Ceg3 is a mitochondria-associated effector that inhibits yeast growth by a putative mART activity.
(A) Sequence alignment of the central region of Ceg3 with three bacterial proteins of mART activity. The strictly conserved residues essential for catalysis are in red. SdeA, ExoS, and CtxA are from Legionella pneumophila, Pseudomonas aeruginosa, and Vibrio cholerae, respectively. (B) The predicted mART motif is critical for Ceg3-mediated yeast toxicity. Serially diluted yeast cells expressing Ceg3 or Ceg3E/A from a galactose-inducible promotor were spotted on the indicated media for 3 days before image acquisition. Similar results were obtained in at least three independent experiments. (C) Ceg3 co-localizes with a mitochondrial marker. HeLa cells were transfected with plasmids that direct the expression of Flag-Ceg3 for 18 hr and then incubated with fresh cell medium containing Mitotracker Red for 30 min. Cells were fixed and immunostained with antibodies specific for Flag (green). Images were acquired with a Zeiss LSM 880 confocal microscope. Scale bar, 5 μm. The colocalization of Ceg3 with mitochondria was quantitated by Pearson correlation coefficient with ImageJ. (D) Flag-Ceg3 localized to the mitochondria detected by Immunogold labeling. HEK293T cells expressing Flag-Ceg3 were fixed, stained with a mouse anti-Flag antibody and an anti-mouse IgG conjugated with 10 nm gold particles sequentially. Images were acquired with a Tecnai T12 electron microscopy. Area highlighted by rectangles (dashed line) on the left panel is magnified in the right panels. Gold particles were indicated by white arrows. Scale bar, 1 μm. (E) Ceg3 fractionated with mitochondria. The cytosol and mitochondrial fractions of HEK293T cells transfected to express Flag-Ceg3 or Flag-MavC were probed with antibodies specific for the indicated proteins. (F) Ceg3 translocated into host cells by the Dot/Icm system is targeted to mitochondria in cells infected with L. pneumophila. HEK293T cells expressing the FcγII receptor were infected with opsonized bacteria expressing 5xFlag-Ceg3 at an MOI of 100 for 2 hr. Lysates of isolated mitochondria were subjected to immunoprecipitation with beads coated with the Flag antibody, precipitates resolved by SDS-PAGE were detected by the Flag-specific antibody (top). The quality of cell fractionation was evaluated by detecting PDHA1 and tubulin. A cytosolic fraction sample was included as an additional control (middle). The expression of Ceg3 in bacteria was detected with the Flag antibody, and ICDH was probed as a loading control (bottom). (G) Ceg3 is an integral mitochondrial membrane protein. Mitochondria isolated from HEK293T cells expressing HA-Ceg3 were subjected to extraction with 0.1 M Na2CO3 (pH 11) for 30 min. Relevant proteins in soluble (S) and pellet (P) fractions separated by centrifugation were probed by immunoblotting with the indicated antibodies. MOI, multiplicity of infection.
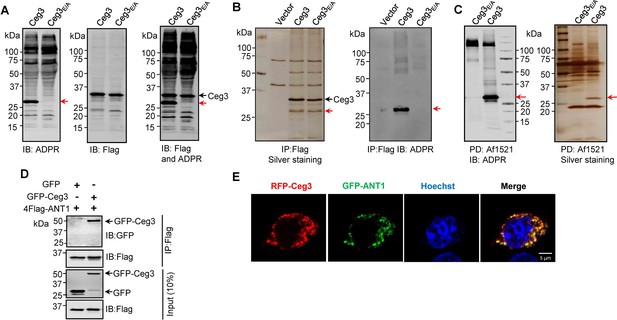
Identification of the cellular targets of Ceg3.
(A) Detection of modified proteins by the ADPR-specific antibody. Lysates of HEK293T cells expressing 4xFlag-Ceg3 or 4xFlag-Ceg3E/A were probed with antibodies specific for ADPR-modification (left), the Flag tag (middle), or both (right). Note that the band indicated by a red arrow in samples expressing Ceg3 detected by the ADPR antibody represents its potential targets. (B) Substrate probing by immunoprecipitation (IP). Lysates of HEK293T cells transfected to express 4xFlag-Ceg3 or 4xFlag-Ceg3E/A were subjected to IP with beads coated with the Flag antibody and the products resolved by SDS-PAGE were detected by silver staining (left) or probed with the ADPR-specific antibody (right). Note the presence of a band in samples expressing Ceg3 but not Ceg3E/A when detected with the ADPR antibody (red arrows). (C) Enrichment of ADP-ribosylated proteins by Af1521 from cells transfected to express Ceg3. Lysates of HEK293T cells expressing 4xFlag-Ceg3 or 4xFlag-Ceg3E/A were incubated with beads coated with recombinant Af1521 and the pulldown products resolved by SDS-PAGE were detected by immunoblotting with the ADP-ribose antibody (left) or by silver staining (right) (red arrows). (D) Interactions between ANT1 and Ceg3. Lysates of HEK293T cells co-transfected to express 4xFlag-ANT1 and GFP-Ceg3 (or GFP) were subjected to IP with beads coated with the Flag antibody and bound proteins resolved by SDS-PAGE were detected with Flag and GFP antibodies, respectively. The expression of 4xFlag-ANT1, GFP-Ceg3, and GFP were similarly probed in total cell lysates as input. (E) Colocalization of Ceg3 and ANT1. HeLa cells transfected to express RFP-Ceg3 and GFP-ANT1 were fixed and analyzed. Images were acquired using a Zeiss LSM 880 confocal microscope. Scale bar, 5 μm. The colocalization of Ceg3 with ANT1 was quantitated by Pearson correlation coefficient with ImageJ.
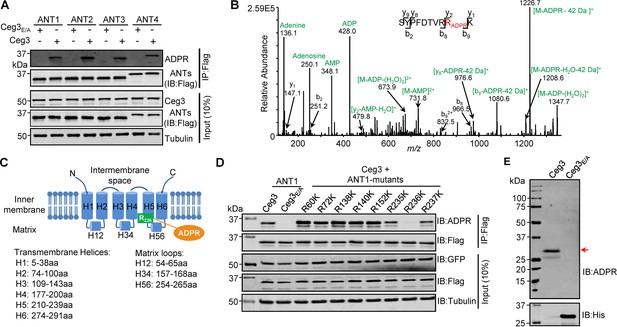
Determination of the modification sites on ADP/ATP translocases induced by Ceg3.
(A) Ceg3 attacks all four ANT isoforms by ADP-ribosylation. Flag-tagged ANTs isolated from lysates of cells co-expressing GFP-Ceg3 or GFP-Ceg3E/A by immunoprecipitation were probed with antibodies specific for ADPR and Flag (top). The expression of relevant proteins was probed in total cell lysates with antibodies specific for GFP (Ceg3), Flag (ANTs), and Tubulin (bottom), respectively. (B) Mass spectrometric analysis of ADP-ribosylated 4xFlag-ANT1V227K/R237K. Mono-ADP-ribosylation modification was detected in the peptide -S228YPFDTVRRK237-. Tandem mass (MS/MS) spectrum shows the fragmentation pattern of the modified peptide, with many ADP-ribosylation-specific marker ions and neutral loss fragments highlighted in green. (C) The schematic topology of ANT1 based on the structure of the bovine ADP/ATP carrier (PDB: 1OKC). Note the positioning of Arg236 at the end of helix 5. (D) Mutation of R236 but not neighboring arginine residues in ANT1 abolished Ceg3-induced modification. In each case, Flag-ANT1 or its mutants isolated from HEK293T cells co-expressing GFP-Ceg3 or GFP-Ceg3E/A were probed for ADPR modification (top) or for protein levels in IP products. The expression of Ceg3 and ANT1 or their mutants was probed in total lysates (input). Tubulin was detected as a loading control. (E) Ceg3 induces ADP-ribosylation modification of yeast ADP/ATP carriers. Lysates of yeast cells expressing His6-Ceg3 or His6-Ceg3E/A driven by a galactose-inducible promotor were detected for ADPR modification (top) (red arrow) or for the expression of Ceg3 (bottom). Note that the expression of wild-type Ceg3 was not detectable despite the presence of strong modification signals.
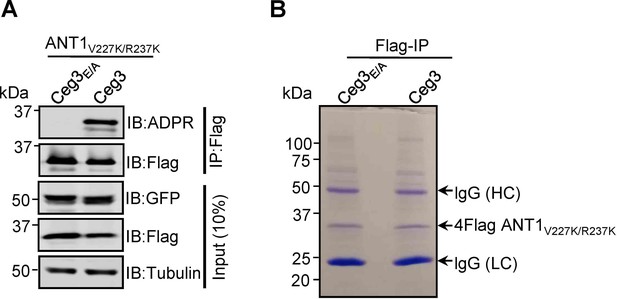
Lys substitutions of V227 and R237 in ANT1 do not affect Ceg3-induced ADP-ribosylation.
(A) Ceg3-induced ADPR modification of ANT1V227K/R237K. Lysates of HEK293T cells co-transfected to express 4Flag-ANT1V227K/R237K and GFP-Ceg3 or GFP-Ceg3E/A were subjected to immunoprecipitation with Flag beads and the products were detected for ADPR modification or for protein levels. The expression of ANT1V227K/R237K and GFP-Ceg3 was also detected in total cell lysates. (B) Isolation of ADP-ribosylated ANT1V227K/R237K for mass spectrometric analysis. Lysates of HEK293T cells co-expressing 4Flag-ANT1V227K/R237K and GFP-Ceg3 or GFP-Ceg3E/A were immunoprecipitated with Flag beads, and products resolved by SDS-PAGE were detected by Coomassie staining. lgG (HC) and lgG (LC) indicate IgG heavy and light chains.

Sequence alignment of the four human ADP/ATP isoforms.
The alignment was generated with the Clustal Omega software. The conserved Arg sites used for mutational analysis were colored in red. The modified sites of ADP-ribosylation mediated by Ceg3 were indicated by a red arrow.
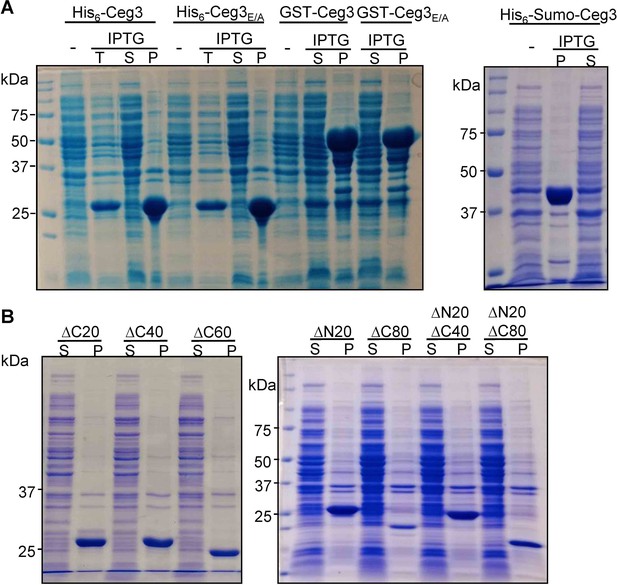
Recombinant Ceg3 expressed in Escherichia coli is insoluble.
(A) Solubility of His6-, GST-, or His6-Sumo-tagged Ceg3 in E. coli. E. coli strains harboring the appropriate plasmids were induced to express the differently tagged Ceg3 proteins with 0.2 mM IPTG. Soluble fractions obtained by centrifugation were resolved by SDS-PAGE and detected by Coomassie staining. -, no IPTG treatment; T, total lysates; S, supernatant fraction; P, pellet fraction. (B) Truncation mutagenesis did not yield a soluble version of Ceg3 in E. coli. Lysates of cells expressing His6-tagged truncations were centrifugated to obtain soluble fractions and the presence of the proteins of interest was detected by Coomassie staining. S, supernatant fraction; P, pellet fraction.
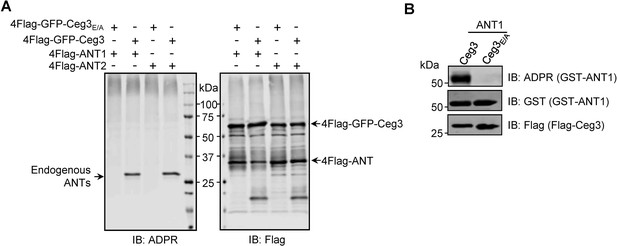
ANT1 is ADP-ribosylated by Ceg3 in Escherichia coli.
(A) Proteins purified from mammalian cells do not detectably modify ADP/ATP translocases in a cell-free assay. 4xFlag-GFP-Ceg3 and 4xFlag-ANTs proteins purified from HEK293T cells were incubated with 1 mM NAD at 37°C for 1 hr. Samples resolved by SDS-PAGE were probed for ADPR modification or for the relevant proteins. Note the detection of ADPR signals for modified endogenous ANTs. (B) E. coli strains harboring the appropriate plasmids were induced to express Flag-Ceg3 and GST-ANT1 proteins with 0.2 mM IPTG. Bacterial lysates resolved by SDS-PAGE were probed for ADPR modification and the expression of Flag-Ceg3 and GST-ANT1.
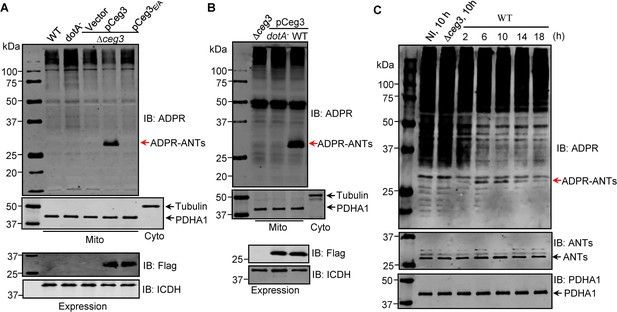
ADP-ribosylation of ADP/ATP translocases by Ceg3 occurs in cells infected with Legionella pneumophila.
(A) An intact mART motif in Ceg3 is required for its modification of ADP/ATP translocases during L. pneumophila infection. Bacteria of the indicated L. pneumophila strains were opsonized prior to infecting HEK293T cells transfected to express the FcγII receptor at an MOI of 100 for 2 hr. Samples of the mitochondrial fraction were probed for ADPR after SDS-PAGE (top). PDHA1 and tubulin were probed as controls to monitor the success of cell fractionation. One cytosolic fraction sample was included as an additional control (middle). The expression of Flag-Ceg3 in bacteria was analyzed with the Flag antibody, the metabolic enzyme isocitrate dehydrogenase (ICDH) was probed as a loading control (bottom). (B) A functional Dot/Icm system is required for Ceg3-induced ADP-ribosylation of ADP/ATP translocases in infected cells. HEK293T cells expressing the FcγII receptor were infected with opsonized bacteria expressing Flag-Ceg3 at an MOI of 100 for 2 hr. Isolated mitochondrial proteins resolved by SDS-PAGE were probed for ADPR modification (top). The quality of cell fractionation was determined by probing for PDHA1 and tubulin (middle), respectively. The expression of Ceg3 in bacteria was detected with the Flag antibody, and ICDH was probed as a loading control (bottom). (C) ADP-ribosylation of ADP/ATP translocases is detectably induced by wild-type L. pneumophila at 6 hr post-infection. HEK293T cells expressing the FcγII receptor were infected with opsonized bacteria for the indicated periods of time at an MOI of 30. Mitochondrial proteins were analyzed by anti-ADPR and anti-PDHA1 Western blot. MOI, multiplicity of infection; NI, no infection.
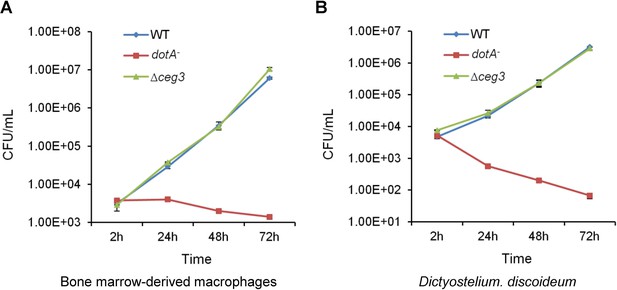
Deletion of ceg3 did not detectably affect intracellular growth of Legionella pneumophila.
BMDMs (A) or Dictyostelium discoideum (B) were infected with the indicated bacterial strains and intracellular bacteria were determined at the indicated time points. Each strain was done in triplicate and similar results were obtained in three independent experiments. Errors were derived from three technical replicates (mean±s.e. from three replicates). BMDM, bone marrow-derived macrophage.
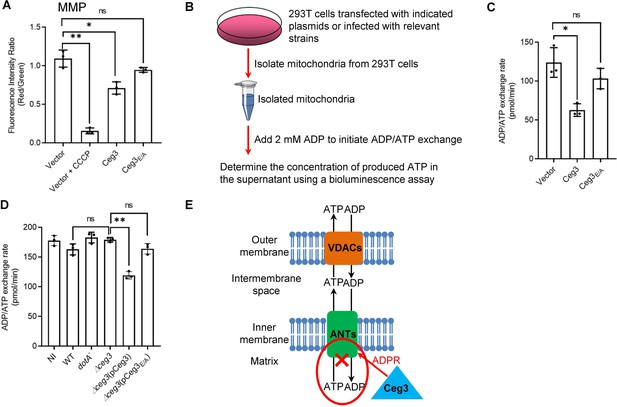
Ceg3 inhibits ADP/ATP exchange in mitochondria.
(A) Ceg3 interferes with the mitochondrial membrane potential (MMP). HEK293T cells transfected to express Ceg3 or its inactive mutant Ceg3E/A were used to determine MMP by the JC-10 dye. Samples treated with 20 μM CCCP for 1 hr were included as a positive control for loss of MMP integrity. Quantitation shown was from three independent experiments done in triplicate. Error bars: standard error of the mean (SEM). Statistical analysis was determined by two-tailed t-test. ns, not significant; *, p<0.05; **, p<0.01. (B) The workflow for measuring ADP/ATP exchange rates. (C) Ceg3 interferes with the ADP/ATP exchange by mitochondria. Mitochondria isolated from HEK293T cells expressing the indicated proteins were suspended in a reaction buffer containing 10 mM HEPES (pH 7.4), 250 mM sucrose, and 10 mM KCl. 2 mM ADP was added to initiate the ADP/ATP exchange process. After 5 min incubation, the concentrations of ATP transported from mitochondria were determined to calculate the ADP/ATP exchange rates. Quantitation shown was from three independent experiments. Error bars: standard error of the mean (SEM). Statistical analysis was determined by two-tailed t-test. ns, not significant; *, p<0.05. (D) Ceg3 perturbs ADP/ATP exchange in cells infected with Legionella pneumophila. Opsonized bacteria of the indicated L. pneumophila strains were used to infect HEK293T cells expressing the FcγII receptor at an MOI of 100 for 2 hr. Mitochondria isolated from the infected cells were used to determine ADP/ATP exchange rates. Results shown were from three independent experiments. Error bars: standard error of the mean (SEM). Statistical analysis was determined by two-tailed t-test. ns, not significant; *, p<0.05; **, p<0.01. (E) A diagram depicting the inhibition of mitochondrial ADP/ATP exchange by Ceg3. Ceg3-induced modification of ANTs by ADPR in the inner membrane blocks the ADP/ATP transport activity of the translocases. MOI, multiplicity of infection; NI, no infection.
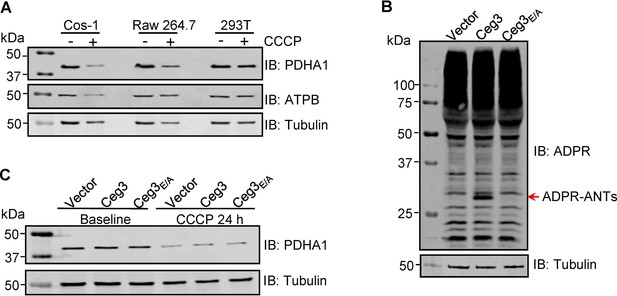
ADP-ribosylation of ANTs by Ceg3 does not affect mitophagy induction.
(A) CCCP induces mitophagy in COS-1 cells. Lysates of the indicated cell lines that have been treated with 20 μM CCCP for 24 hr were resolved by SDS-PAGE and then probed for PDHA1 (mitochondrial matrix protein) and ATPB (mitochondrial inner membrane protein). Tubulin was probed as a loading control. (B) Ceg3 expressed in COS-1 cells by lentiviral transduction induced ADP-ribosylation of ADP/ATP translocase. Lysates of COS-1 cells transduced with a lentivirus harboring the indicated plasmid at an MOI of 10 for 2 days were probed for ADPR modification, tubulin was probed as a loading control. (C) Ceg3 expressed in COS-1 cells does not affect the protein level of PDHA1 regardless of CCCP treatment. COS-1 cells expressing the indicated proteins for 1 day were treated with 20 μM CCCP for another 24 hr, cells treated with DMSO were used as controls. PDHA1 was probed to evaluate mitophagy, tubulin was detected as a loading control. MOI, multiplicity of infection.
Additional files
-
Supplementary file 1
Identification of ADP/ATP translocases as the targets of Ceg3 in samples obtained by Af1521-pulldown.
The protein band specifically present in Af1521 pulldown samples from expressing Ceg3 was analyzed by mass spectrometry; the 10 proteins with the most hits were listed.
- https://cdn.elifesciences.org/articles/73611/elife-73611-supp1-v2.docx
-
Supplementary file 2
Bacterial strains, antibodies, plasmids and primers used in this study.
- https://cdn.elifesciences.org/articles/73611/elife-73611-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/73611/elife-73611-transrepform1-v2.docx
-
Source data 1
Figures with the uncropped gels or blots with the relevant bands clearly labeled.
- https://cdn.elifesciences.org/articles/73611/elife-73611-supp3-v2.pdf
-
Source data 2
The original files of the raw unedited gels or blots.
- https://cdn.elifesciences.org/articles/73611/elife-73611-supp4-v2.zip





