Destabilizers of the thymidylate synthase homodimer accelerate its proteasomal degradation and inhibit cancer growth
Figures
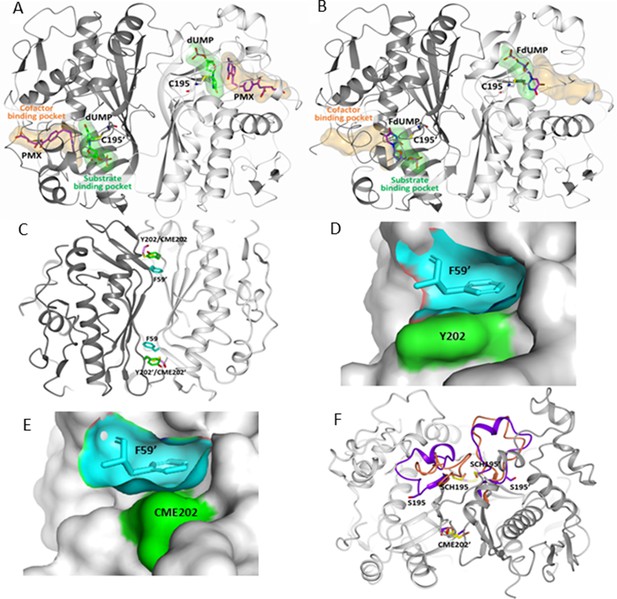
Drug target sites on hTS: the active-site (A,B) and the Y202 pocket (C–F).
Drugs that are directed to the active site of hTS can mimic either the cofactor MTHF or the substrate dUMP and bind in their binding pockets. (A) hTS (in cartoon with the A and B subunits colored white and grey, respectively) inhibited by the cofactor analogue pemetrexed (PMX, in sticks, purple carbons), which occupies the cofactor binding pocket by establishing a π-π interaction with the dUMP substrate (in sticks, green carbons) (PDB ID: 1JU6). The substrate and cofactor binding sites are represented as green and orange surfaces, respectively. The catalytic cysteine, C195, is highlighted in sticks. (B) The substrate analogue 5-fluoro-2’-deoxyuridine monophosphate (FdUMP) (in sticks, purple carbons) binds inside the catalytic cavity, mainly filling the substrate binding pocket (PDB ID: 3H9K, hTS variant R163K). (C) Location of F59, Y202 and CME202 residues at the hTS dimer interface by superimposition of native hTS and hTS-C195S-Y202C double mutant structures (CME: S,S(2-hydroxyethyl)cysteine; PDB-ID: 3N5G, 4O1X). (D) Surface of the Y202 pocket (cyan) filled by F59’ from the opposite monomer (cyan sticks). The surface of Y202 is highlighted in green (PDB-ID: 3N5G). (E) Surface of the C202 pocket (cyan) filled by F59’ from the opposite monomer (cyan sticks) in the C195S-Y202C double mutant (PDB-ID: 4O1X). The surface of C202 (oxidized as CME) is highlighted in green. (F) Superimposition of the hTS-C195S-Y202C and hTS-Y202C crystal structures with the catalytic loop in hTS-Y202C (inactive conformation, in orange) and in hTS-C195S-Y202C (active conformation, in violet); notice the different orientations of the catalytic residue couples S195/SCH195 and S195’/SCH195’ (PDB-ID: 4O1X, 4O1U) (SCH: S-methyl-thio-cysteine). The one-letter code is used for the standard amino-acids, while for the chemically modified amino-acid, a conventional three-letter code is used as described above.
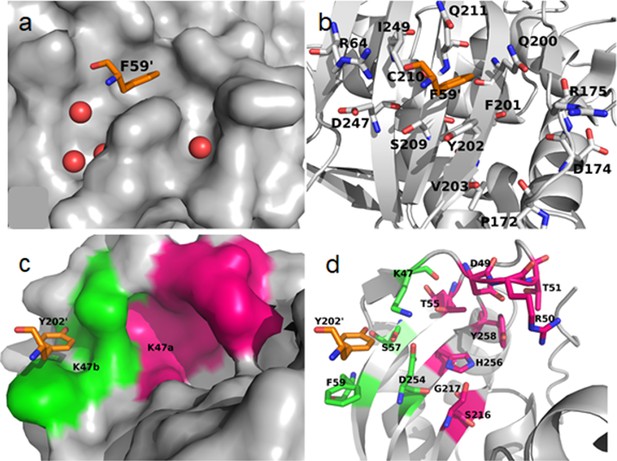
Y202 and K47 pockets at the hTS monomer-monomer interface (PDB ID: 1HVY, A chain) are represented.
(a) Surface presentation of the hTS monomer, with residue F59’ (in orange, shown as sticks) from the opposite monomer binding in the Y202 pocket (PDB ID: 1HVY, chain A). Crystallographic water molecules found in the Y202 pocket region are shown as red spheres. (b) Cartoon representation of the hTS monomer with F59’ (in orange, shown as sticks) from the opposite monomer binding in the Y202 pocket (PDB ID: 1HVY, chain A). Residues forming the Y202 pocket are shown in sticks. Atom colour code: carbon – grey/orange; nitrogen – blue; oxygen – red. (c) Surface representation of the K47 pocket region in the hTS monomer (PDB ID: 1HVY, chain A), with residue Y202’ (in orange, shown as sticks) from the opposite monomer, binding in the K47 subpocket K47b (green) adjacent to the K47a subpocket (pink). (d) Cartoon and sticks presentation of the K47 pocket region in the hTS monomer with residue Y202 (in orange, shown as sticks) from the opposite monomer, binding in the K47b subpocket (PDB ID: 1HVY, chain A). Residues K47, S57 and D254 form a dividing wall that is part of both sub-pockets K47a (pink) and K47b (green). Atom color code: carbon – grey/orange/green/pink; nitrogen – blue; oxygen – red.
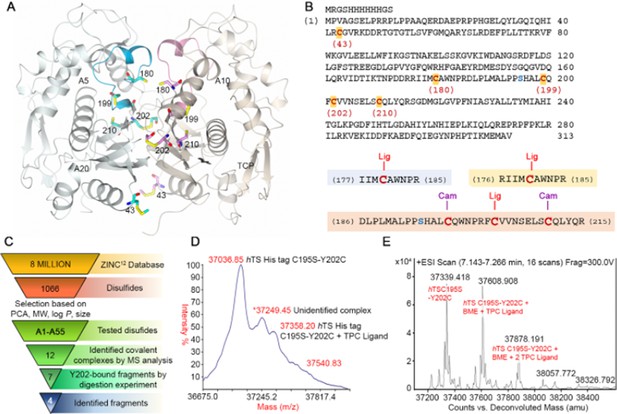
Disulfide library design and screening on the hTS C195S-Y202C variant.
(A) Crystal structure of the hTS C195S-Y202C dimer (in the cartoon, the A and B subunits are colored pale blue and grey, respectively; the catalytic loops, in active conformation, are highlighted in cyan and pink, respectively) showing the positions of the cysteine residues (in sticks, carbon atoms are colored cyan and pink in subunits A and B, respectively). Compounds A5, A10, A20 and the reference compound N-tosyl-D-prolinecysteamine (TPC) (Appendix 2—figure 1) were selected through the tethering experiments. They are indicated close to the peptide sequence found in the mass spectrometry identification process (Supplementary file 3B and C). Their chemical structures are reported in Supplementary file 3A. (B) Sequence of the hTS C195S-Y202C variant and cysteine-containing peptides generated by trypsin digestion. Cysteine residues are displayed in red (the residue number is reported below each cysteine) and the mutated S195 (the catalytic residue) in blue. Three cysteine-containing peptides were generated by proteolytic cleavage of the hTS C195S-Y202C variant: peptide I177-R185 and peptide R176-R185 containing C180 and peptide D186-R215, containing C199, C202, and C210. Ligand (Lig)-bound cysteines and those alkylated as carbamidomethyl (Cam)-cysteines are displayed. (C) Screening cascade from the 6 million compounds from the ZINC database (Irwin and Shoichet, 2005) to the final four fragments expected to bind in the Y202 pocket. (D, E) Representative MS spectra in the hTS-binding evaluation step. Maldi (D) and ESI (E) MS spectra for the reference compound TPC (Erlanson et al., 2000). The three labeled peaks are due to hTS C195S-Y202C and to the protein bound to BME and one and two molecules of TPC.
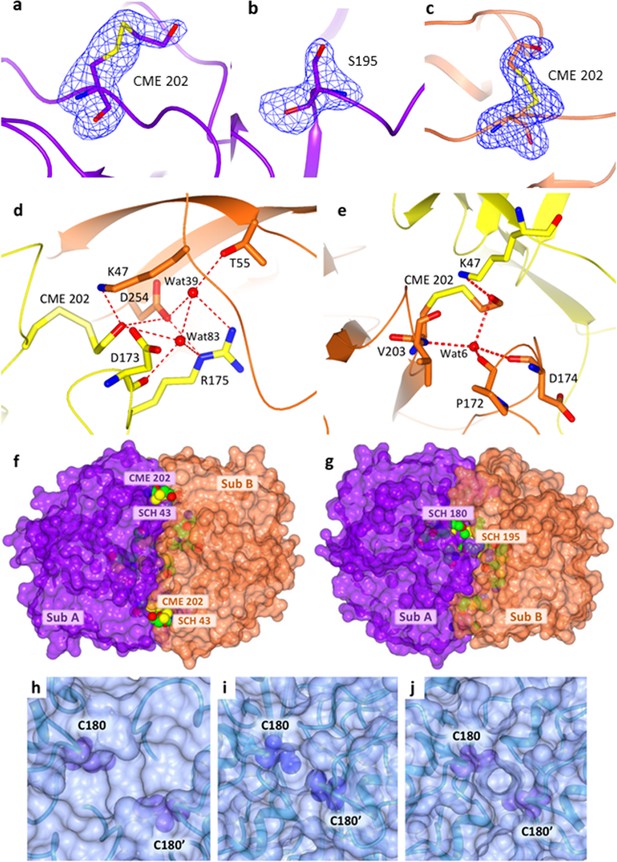
X-ray crystallographic structure of hTS-Y202C and hTS C195S-Y202C.
(a–c) Unbiased omit map (blue wired map contoured at 3.0 σ), showing the S,S(2-hydroxyethyl)cysteine (CME) chemical modification occurring at the mutated residue C202 (panel a), and the mutated residue S195 (panel b) in the hTS double mutant C195S-Y202C. Panel (c) reports the omit map (contoured at 3.0 σ) showing the S,S(2-hydroxyethyl)cysteine (CME) chemical modification occurring at the mutated residue C202 in the single hTS Y202C mutant. The mutated serine and the CME residues are represented as sticks. The hTS C195S-Y202C structure is drawn in violet whereas the single mutant structure is coloured in orange (cartoon representation). (d-e) H-bond interactions (represented as dashed lines) involving the CME202 residue in hTS Y202C subunits A (panel d) and B (panel e). Interacting residues are represented as sticks and water molecules as red spheres. Residues belonging to the A subunit are coloured in yellow while those of the B subunit are coloured in orange. (f–g) two views of the surface of the hTS Y202C dimer. The two symmetry-related subunits are shown in violet (subunit A) and orange (subunit B), while the CME and S-methyl-thio-cysteine (SCH) residues are represented as spheres. Only SCH43 (panel f), appears fully solvent accessible, with CME202, partially visible in its pocket, just below SCH43. Panel (g) shows that SCH180 is partially solvent accessible, while SCH195, CME199, and C210 are buried to different extents below the hTS surface. (h–j) surface representations showing the solvent accessibility of C180 residues in wt hTS and hTS double mutant (C195S-Y202C). The protein is shown in a transparent surface and the C180-C180’ residues from two facing subunits are represented by labeled blue-violet spheres. (h) wt hTS dimer in inactive conformation (PDB ID: 3N5G); (i) wt hTS in active conformation (PDB ID: 2RDA); (j) C195S-Y202C hTS mutant in active conformation (PDB ID: 4O1X). PISA server analysis indicates that C180-C180’ are buried in all three structures. However, as it can be appreciated by comparing the three figures, the active conformation increases the shielding of the facing C180-C180’ residues from solvent as their buried surface goes from about 30 Å2 (h) to 50–68 Å2 (i–j). Also, the crevice between subunits is much larger in inactive vs. active conformation (from about 15x6 Å in inactive to about 5x5 Å in active conformation).
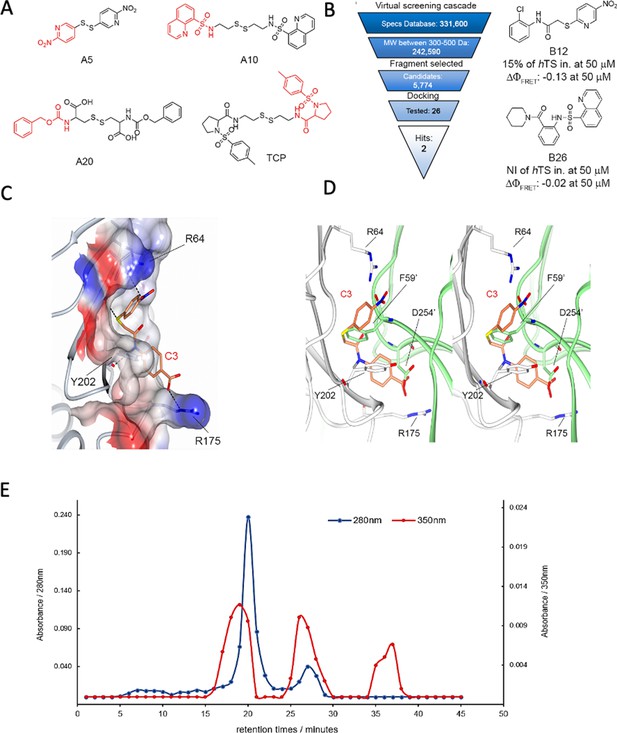
From hits to dimer-destabilizing compounds.
(A) Structures of the compounds identified in MS studies (in red the fragments used for the virtual screening approach to the design of the compound library without the disulfide bonds). (B) Virtual screening cascade and chemical structures of the identified compounds, B12 and B26. (C) Detail of the binding mode of compound C3 (orange sticks) with the hTS interface residues R64, Y202 and R175 (positive and negative potential regions are colored in blue and red, respectively) obtained with docking methods. (D) Stereo view of the superimposition of the calculated binding mode of C3 onto the first monomer (white cartoon) with the opposite monomer overlapping (lime cartoon). Atom color code: nitrogen – blue; oxygen – red; sulphur – yellow. (E) Results of anion exchange chromathography showing co-elution experiments of C3 with hTS dimer and monomer through anion exchange chromatography. Absorbances at 280 (blue) and 350 nm (red) of chromatographic fractions obtained by loading a 50 μM hTS and 4 μM C3 solution on an anion-exchange AEX HiPrep Q HP 16/10 mm column. (Figure 3—figure supplement 1, Figure 3—figure supplement 1—source data 1).
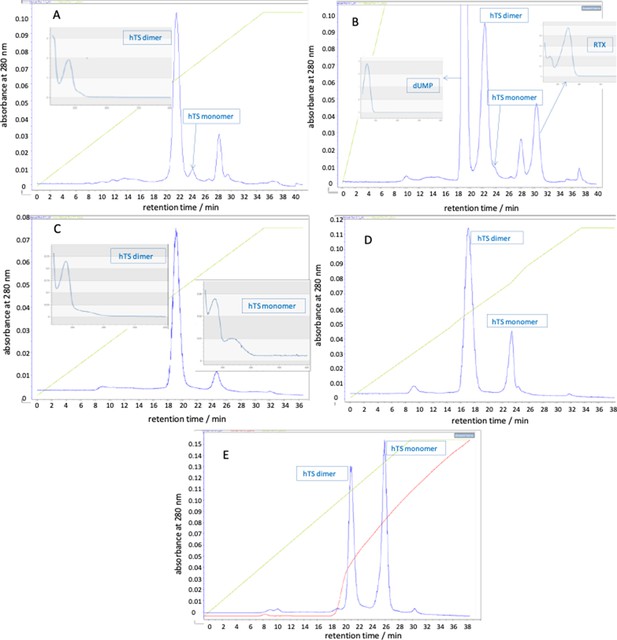
Anion-exchange chromatograms obtained with a 280 nm detection (blue traces) by loading on an AEX HiPrep Q HP 16/10 mm, Cytiva, column.
(A) hTS 300 μL, 50 μM. (B) hTS +250 μM dUMP and RTX. (C) hTS +4 μM C3. (D) hTS +40 μM C3. (E) hTS +80 μMC3. Green traces: NaCl gradient (% of buffer ‘B’), red traces: conductivity (mS/cm). The insets are the UV-Vis spectra of the indicated species.
-
Figure 3—figure supplement 1—source data 1
Anion-exchange chromatograms of hTS obtained with a 280 nm and 350 nm detection.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig3-figsupp1-data1-v2.xlsx
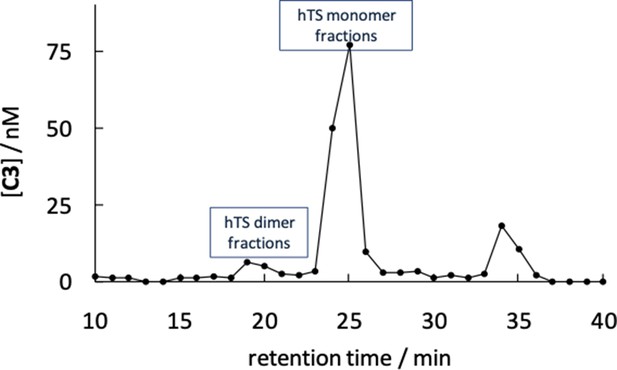
Profile of the variation in concentration of compound C3 in the AE chromatographic fractions determined by high-resolution MS (UltiMate 3000 UHPLC tandem Orbitrap Q-Ex).
Comparison with Figure 3E shows that most ligand is co-eluted with the hTS monomer, around a 25 min retention time. The much larger amount of compound C3 in the protein monomer fractions (23–27 min) relative to the dimer containing fractions (18–22 min) possibly results from preferential co-precipitation of the ligand with the protein dimers relative to the monomers prior to injection into the mass spectrometer.
-
Figure 3—figure supplement 2—source data 1
Profile of the variation in concentration of compound C3 in the AE chromatographic fractions determined by high-resolution MS (UltiMate 3000 UHPLC tandem Orbitrap Q-Ex).
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig3-figsupp2-data1-v2.xlsx
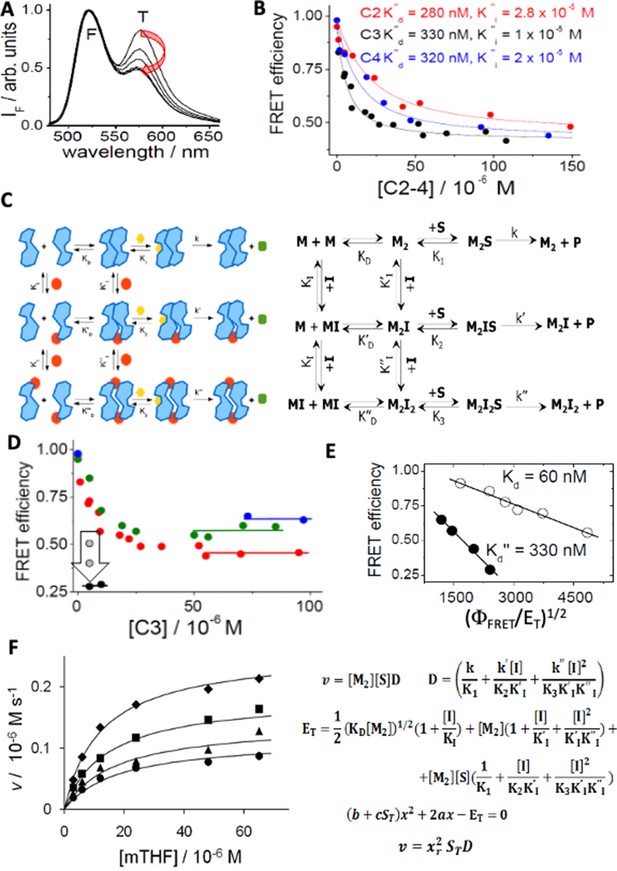
Spectroscopic and mechanistic analysis of hTS dissociative inhibition for compounds C2, C3 and C4.
(A) Emission spectra of fluorescein(F)- and tetramethylrhodamine (T)-labelled hTS (λexc = 450 nm): the decreasing emission contribution of T relative to F with increasing concentrations of C3 (0, 4.5, 9.5, 15, 25, 100 μM) parallels the decrease in the hTS dimer mole fraction. (B) Dependence of the observed FRET efficiency on the concentrations of three inhibitors (C3, black; C4, blue; C2, red); the hTS dimer concentration was 100 nM. The lines represent the best fittings of the experimental results to Equation S3 in the Appendix 4. The most significant fitting parameters are reported. (C) Cartoon, and standard chemical representations of the dissociative inhibition mechanism of hTS. Blue shapes represent enzyme monomers (M) in the active conformation bound to dUMP; red dots indicate a dissociative inhibitor (I), yellow hexagons, the folate substrate (S), green octagons, the product. (D) Dependence of FRET efficiency on the concentration of C3 with total protein concentrations (ET) of 490 nM (blue circles), 250 nM (green circles), 100 nM (red circles) and 50 nM (black circles). The horizontal segments represent the values of the efficiency in the limit of high inhibitor concentration. At ET = 50 nM and IT = 5 μM the vertical arrow indicates the evolution of the measured value of the FRET efficiency 5, 7, and 12 minu after inhibitor addition (grey and black circles). (E) Analysis of the hTS monomer-dimer equilibrium according to eq. ΦFRET = 1–0.5(ΦFRET/ET)1/2K1/2 (Genovese et al., 2010) and corresponding equilibrium constants without C3 (white circles, K=KD) and at saturating C3 concentrations (black circles, K=K’’D, ΦFRET = FRET efficiency). (F) Dependence of the initial reaction rate (v) on mTHF concentration (free substrate, [S], and total, ST) at four different C3 concentrations (from top to bottom, [I]=0, 12, 24, and 36 μM). The fitting curves represent equation v = xr2STD derived in the Appendix 4 and computed with ET (total enzyme dimer concentration)=300 nM, k=k’=0.9 s–1, k’’=0.1 s–1, K1=K2=10–5 M, K3=2 × 10–5 M, KI = 10–6 M, KI’ = KI’’=10–5 M,x = [M2]1/2 and using for xr the only acceptable root of the quadratic equation in panel 4 F (Figure 4—source data 2).
-
Figure 4—source data 1
Spectroscopic and mechanistic analysis of hTS dissociative inhibition for compounds C2, C3 and C4.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Inhibition of hTS by compound C3.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig4-data2-v2.xlsx
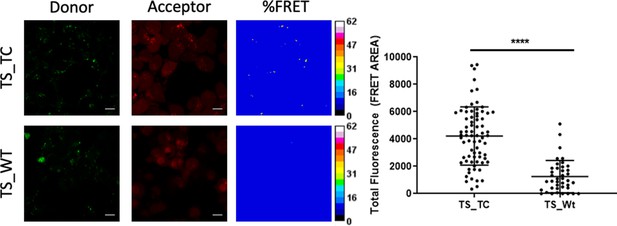
Fluorescence microscopy analysis of hTS dissociative inhibition.
Representative fluorescence microscopy images of TS-TC- and WT-TS-expressing HT-29 cells stained with red-emitting ReAsH and incubated with green-emitting E5-FITC. From left to right: green-emission and red-emission channels, and FRET efficiencies depicted according to the colour scale (%) in the right bars. Right: comparison of the average FRET areas for cells overexpressing TS with (TS_TC) or without (TS_Wt) ReAsH binding sites, obtained using PixFRET within ImageJ. Each dot represents the total FRET area of an individual field with at least 10 cells. The statistical analysis was performed using unpaired two tailed t-test (p-value >0.0001) using Prism 8 for windows (version 3.1.1).
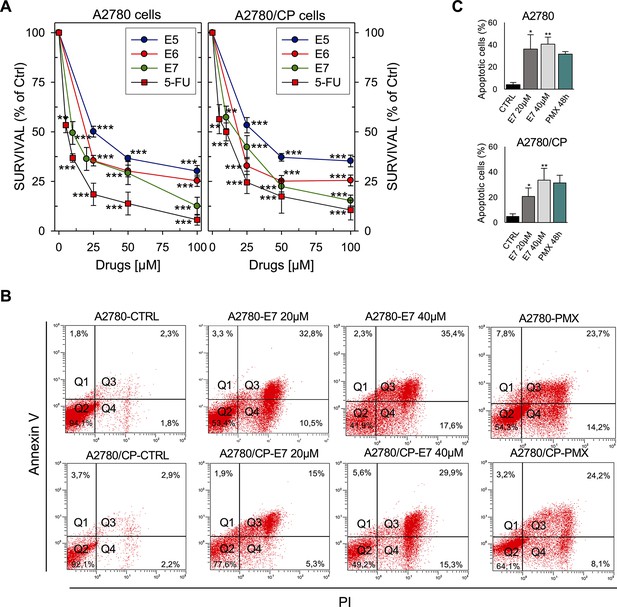
Effects of hTS inhibitors in human cancer cell lines.
(A) Dose-response curves for E5–E7 and 5-FU against A2780 and A2780/CP cell growth. (B) Flow cytometric analysis of apoptosis of A2780 and A2780/CP cells treated with E7 or PMX (Annexin V−/PI−: live cells, Annexin V+/PI−: early apoptotic cells, Annexin V+/PI+: late apoptotic cells, Annexin V−/PI+: necrotic cells). (Figure 6—source data 1). (C) Quantification of Annexin V-positive cells (Q2 + Q3) performed using the Annexin V/PI kit. Data indicate mean values and standard deviation of at least two biological repeats performed in duplicate. p Values were calculated with two-sided Student’s t-test and ANOVA followed by the Tukey’s multiple comparison. * p<0.05; ** p<0.01; *** p<0.001.
-
Figure 6—source data 1
Quantification of Annexin V-positive cells of apoptosis of A2780 and A2780/CP cells treated with E7 or PMX.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig6-data1-v2.docx
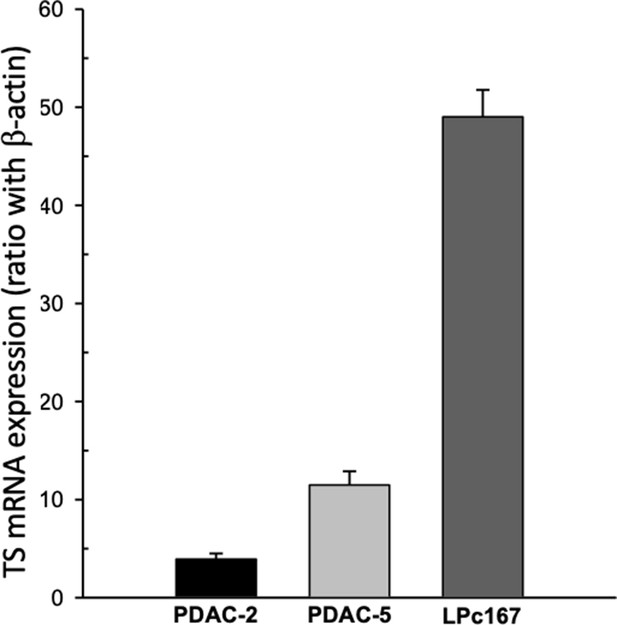
TS mRNA expression in the primary pancreatic cancer cells.
-
Figure 6—figure supplement 1—source data 1
TS mRNA expression in the primary pancreatic cancer cells.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig6-figsupp1-data1-v2.xls
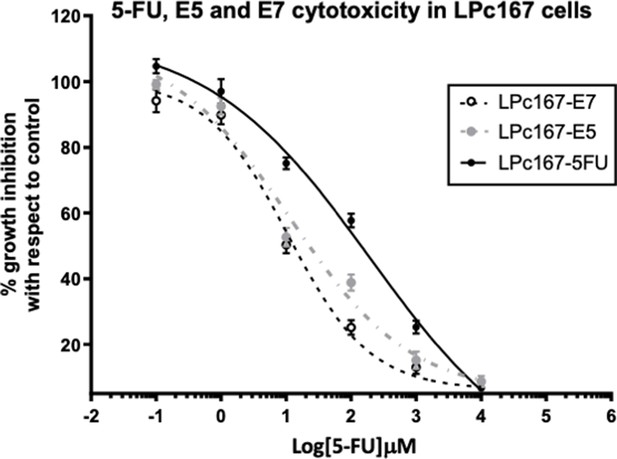
Dose-response curve of Pancreatic cancer cells LPc167 growth inhibition for compounds E7, E5, and 5FU.
-
Figure 6—figure supplement 2—source data 1
Pancreatic cancer cells LPc167 growth inhibition for compounds E7, E5, and 5FU.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig6-figsupp2-data1-v2.xlsx
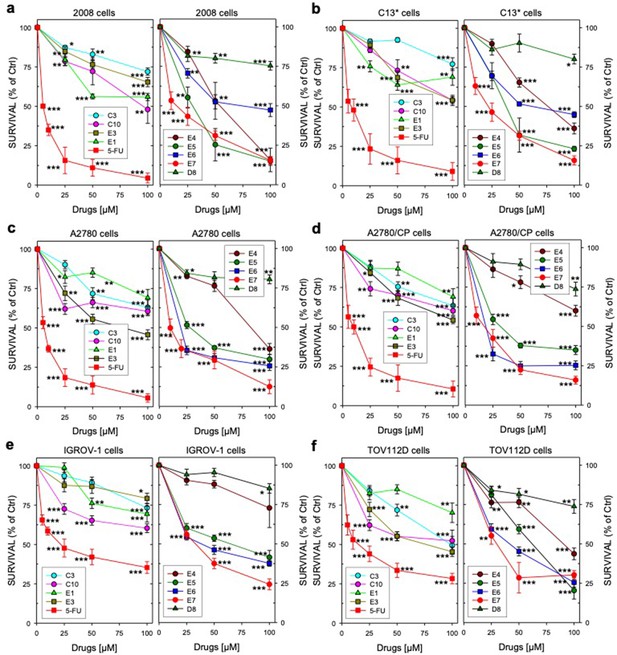
Ovarian cancer cell growth inhibition".
-
Figure 6—figure supplement 3—source data 1
Ovarian cancer cell growth inhibition all compounds.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig6-figsupp3-data1-v2.xlsx
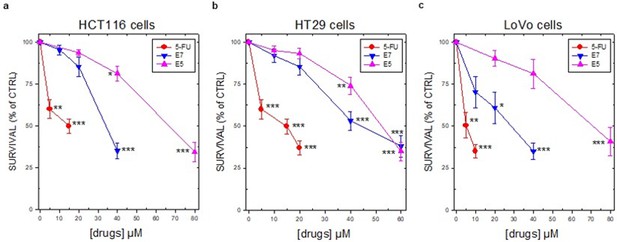
Colorectal cancer cell survival percentages as the mean ± S.E.M. of three separate experiments performed in duplicate.
(a) is referred to HCT116 cells. (B) is referred to HT29 cells and (c) is referred to LoVo cells. Data indicate mean values and standard deviation from three experiments performed in duplicate. p values were calculated with two-sided Student’s t-test and ANOVA followed by the Tukey’s multiple comparison. * p<0.05; ** p<0.01; *** p<0.001, n=3. E7 was the most interesting among these compounds, being active on all the cancer cells tested with IC50 values between 10 and 37 µM after 72 hr of exposure. It showed a lower or, in a few cases, comparable activity with respect to 5-FU.
-
Figure 6—figure supplement 4—source data 1
Colon cancer cell inhibition E7 and E5.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig6-figsupp4-data1-v2.xlsx
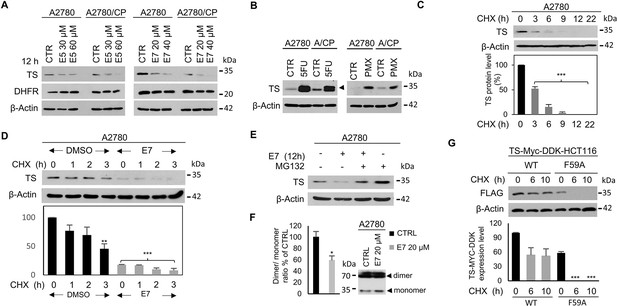
Dimer destabilizers effects on wild type and mutant hTS levels in human cancer cell lines.
(A) Effects of E5 and E7 on hTS and DHFR protein levels in A2780 and A2780/CP cells. (B) Increase of hTS protein level in A2780 and A2780/CP cells following treatment with 5-FU (5 µM, 72 hr) (ternary-complex, arrowhead) or PMX (5 µM, 48 hr). (C) hTS half-life determination in A2780 cells after treatment with CHX (90 µg/ml) for 0–22 hr. (D) Stability of hTS in A2780 cells treated with E7 for 12 hr, followed by treatment with CHX for the indicated times. (E) hTS levels inA2780 cells treated with E7 for 12 hr, then with 10 µM MG132 for 5 hr. (F) Effects of a 3 hr exposure to E7 on hTS dimer/monomer equilibrium in A2780 cells. (G) Half-life determination of exogenous hTS protein (anti-FLAG stain) in HCT116 cells transfected with TS-Myc-DDK (CHX, 0, 6, and 10 hr) or with the F59A mutant (CHX 0, 6, and 10 hr) tagged vector. Data indicate mean values and standard deviation of biological repeats performed in duplicate. p Values were calculated with two-sided Student’s t-test and ANOVA followed by the Tukey’s multiple comparison. * p<0.05; ** p<0.01; *** p<0.001.
-
Figure 7—source data 1
Effects of E5 on hTS and DHFR protein levels in A2780 and A2780/CP cells.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data1-v2.zip
-
Figure 7—source data 2
Effects of E5 on hTS and DHFR protein levels in A2780 and A2780/CP cells_TS and DHFR.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data2-v2.zip
-
Figure 7—source data 3
Effects of E7 on hTS and DHFR protein levels in A2780 and A2780/CP cells.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data3-v2.zip
-
Figure 7—source data 4
Effects of E7 on hTS and DHFR protein levels in A2780 and A2780/CP cells_TS and DHFR.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data4-v2.zip
-
Figure 7—source data 5
hTS protein level in A2780 and A2780/CP cells treated with 5-FU and PMX (PMX and 5FU).
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data5-v2.zip
-
Figure 7—source data 6
hTS protein level in A2780 and A2780/CP cells treated with PMX.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data6-v2.zip
-
Figure 7—source data 7
Stability of hTS in A2780 cells treated with E7 for 12 hr, followed by treatment with CHX for the indicated times.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data7-v2.zip
-
Figure 7—source data 8
Stability of hTS in A2780 cells treated with E7 for 12 hr, followed by treatment with CHX for the indicated times.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data8-v2.zip
-
Figure 7—source data 9
hTS levels in A2780 cells treated with E7 for 12 hr, with MG132 for 5 h.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data9-v2.zip
-
Figure 7—source data 10
hTS levels in A2780 cells treated with E7 for 12 hr, with MG132 for 5 hr.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data10-v2.zip
-
Figure 7—source data 11
Effects of a 3 hr exposure to E7 on hTS dimer/monomer equilibrium in A2780 cells.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data11-v2.zip
-
Figure 7—source data 12
Effect of Dimer destabilizers and hTS levels and half-life in human cancer cell lines (Figure C, D, G).
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data12-v2.docx
-
Figure 7—source data 13
Half-life determination of exogenous hTS protein (anti-FLAG stain) in HCT116 cells transfected with TS-Myc-DDK (CHX) or with the F59A mutant (CHX).
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data13-v2.zip
-
Figure 7—source data 14
Half-life determination of exogenous hTS protein (anti-FLAG stain) in HCT116 cells transfected with TS-Myc-DDK (CHX) or with the F59A mutant (CHX).
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data14-v2.zip
-
Figure 7—source data 15
Effects of E5 on hTS and DHFR protein levels in A2780 and A2780/CP cells.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data15-v2.zip
-
Figure 7—source data 16
Effects of E5 on hTS and DHFR protein levels in A2780 and A2780/CP cells_TS and DHFR.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data16-v2.zip
-
Figure 7—source data 17
Effects of E7 on hTS and DHFR protein levels in A2780 and A2780/CP cells.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data17-v2.zip
-
Figure 7—source data 18
Effects of E7 on hTS and DHFR protein levels in A2780 and A2780/CP cells_TS and DHFR.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data18-v2.zip
-
Figure 7—source data 19
hTS protein level in A2780 and A2780/CP cells treated with 5-FU and PMX (PMX and 5FU).
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data19-v2.zip
-
Figure 7—source data 20
hTS protein level in A2780 and A2780/CP cells treated with PMX.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data20-v2.zip
-
Figure 7—source data 21
Stability of hTS in A2780 cells treated with E7 for 12 hr, followed by treatment with CHX for the indicated times.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data21-v2.zip
-
Figure 7—source data 22
Stability of hTS in A2780 cells treated with E7 for 12 hr, followed by treatment with CHX for the indicated times.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data22-v2.zip
-
Figure 7—source data 23
hTS levels in A2780 cells treated with E7 for 12 hr, with MG132 for 5 h.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data23-v2.zip
-
Figure 7—source data 24
hTS levels in A2780 cells treated with E7 for 12 hr, with MG132 for 5 hr.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data24-v2.zip
-
Figure 7—source data 25
Effects of a 3 hr exposure to E7 on hTS dimer/monomer equilibrium in A2780 cells.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data25-v2.zip
-
Figure 7—source data 26
Half-life determination of exogenous hTS protein (anti-FLAG stain) in HCT116 cells transfected with TS-Myc-DDK (CHX) or with the F59A mutant (CHX)_actin.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data26-v2.zip
-
Figure 7—source data 27
Half-life determination of exogenous hTS protein (anti-FLAG stain) in HCT116 cells transfected with TS-Myc-DDK (CHX) or with the F59A mutant (CHX).
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-data27-v2.zip
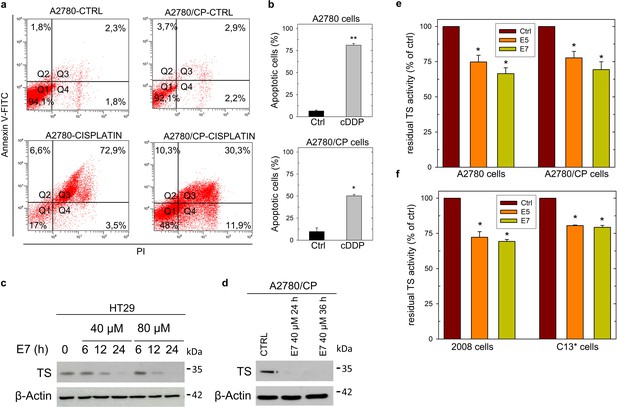
Effect of cisplatin on cell death, effect of dimer destabilizers on hTS activity and hTS protein levels in cancer cells.
(a) Flow cytometric analysis of apoptosis of A2780 and A2780/CP cells treated with cisplatin (13 µM, 72 hr) (Annexin V-/PI-: live cells, Annexin V+/PI-: early apoptotic cells, Annexin V+/PI+: late apoptotic cells, Annexin V-/PI+: necrotic cells). (b) Quantification of Annexin V-positive cells (Q2 +Q3) was performed using Annexin V/PI kit. (c) Effects of E7(different times) on hTS protein level in HT29 cells, and on A2780/CP cells. (e), cellular extracts from A2780 and A2780/CP cell lines processed for TS activity after incubation with E5 and E7 at concentrations corresponding to IC50 values (28.8 and 34.3 µM for E5; 10.1 and 21.5 µM for E7). (f) Cellular extracts from 2008 and C13* cell lines processed for TS activity after incubation with E5 and E7 at concentrations corresponding to IC50 values (29.8 and 38.3 µM for E5; 14.6 and 21.1 µM for E7). All the results are the mean of three experiments performed in duplicate. Data indicate mean values and standard deviation. p Values were calculated with two-sided Student’s t-test and ANOVA followed by the Tukey’s multiple comparison. * p<0.05; ** p<0.01; *** p<0.001.
-
Figure 7—figure supplement 1—source data 1
Quantification of Annexin V-positive cells by flow cytometric analysis of apoptosis of A2780 and A2780/CP cells treated with cisplatin, E7 and PMX.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-figsupp1-data1-v2.docx
-
Figure 7—figure supplement 1—source data 2
Effect of E7 on hTS protein levels in HT29 cells _actin.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-figsupp1-data2-v2.zip
-
Figure 7—figure supplement 1—source data 3
Effect of E7 on hTS protein levels in HT29 cells.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-figsupp1-data3-v2.zip
-
Figure 7—figure supplement 1—source data 4
Effect of E7 on hTS protein levels in HT29 cells _actin.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-figsupp1-data4-v2.zip
-
Figure 7—figure supplement 1—source data 5
Effect of E7 on hTS protein levels in HT29 cells.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-figsupp1-data5-v2.zip
-
Figure 7—figure supplement 1—source data 6
Effect of dimer destabilizers on hTS protein levels in A2780/CP cells, E7 and actin.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-figsupp1-data6-v2.zip
-
Figure 7—figure supplement 1—source data 7
Effect of dimer destabilizers on hTS protein levels in A2780/CP cells, E7 and actin.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-figsupp1-data7-v2.zip
-
Figure 7—figure supplement 1—source data 8
TS activity assay for A2780 and A2780/CP cell lines after treatment with E5 and E7.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig7-figsupp1-data8-v2.xlsx
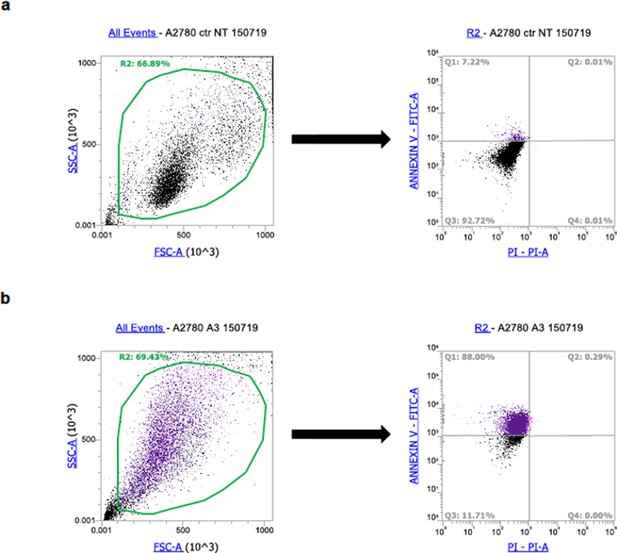
An example of gating strategy used for cell death analysis.
Gating strategy (representative flow cytometry profiles) used to assess the percentage of apoptotic cells (Annexin V/PI) shown in Figure 6B in the main text. In panel a, the untreated (ctr) A2780 cells are reported. The left panel represents the Forward scatter (FSC-A) vs Side scatter (SSC-A). The right panel is the representative flow cytometry analysis plots of Annexin V/PI staining (Parameters: PI-PI-BL2-A vs ANNEXIN V-FITC-BL1-A; Gate: all events). In panel b, the cisplatin treated cells (13 μM, 72 hr) are shown. In the left panel, FSC-A vs SSC-A are reported. In the right panel, a representative flow cytometry analysis plots of Annexin V/PI staining is reported (Parameters: PI-PI-BL2-A vs ANNEXIN V-FITC-BL1-A; Gate: all events).
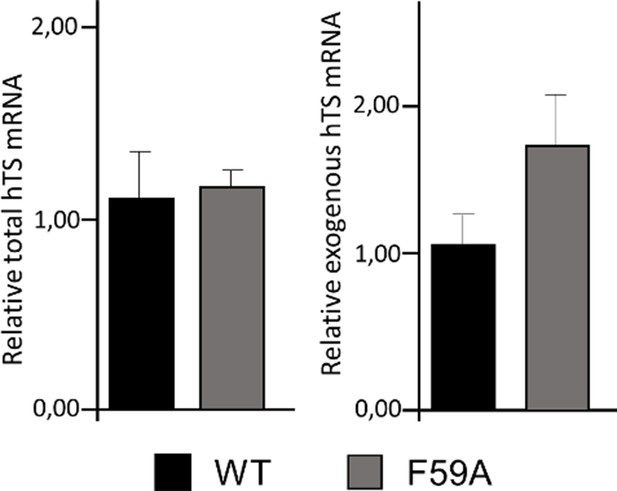
Expression levels of total (exogenous plus endogenous) and exogenous hTS mRNAs 24 hr after transfection of HCT116 cells with wild-type hTS-Myc- DDK tagged or mutant hTS-Myc-DDK-F59A vectors.
Quantitative Real Time RT-PCR results showing the expression of total and exogenous hTS mRNAs in HCT116 cells transfected with wild- type hTS-Myc-DDK tagged (WT) or mutant hTS-Myc-DDK-F59A vectors. The mRNA expression was calculated with the ΔΔCt method (calibrator: average of HCT116 cells transfected with WT hTS- Myc-DDK-tagged vector) with GAPDH as endogenous control. Columns and bars indicate mean values + S.E.M. (n=6 for two independent experiments).
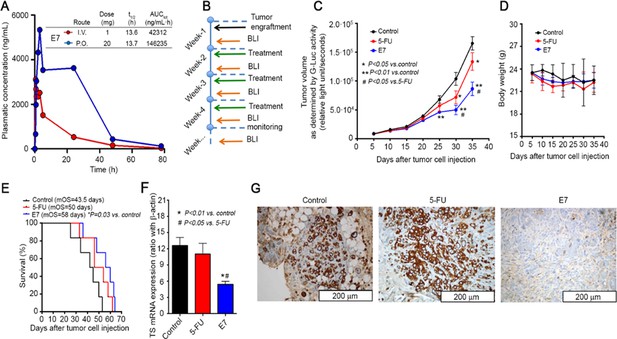
Effects of E7 in vivo pancreatic cancer model.
(A) PK data on healthy mice for E7. (B) Set-up of in vivo experiments, using mice injected orthotopically with PDAC-2-primary cells, and monitored every 5 days by bioluminescence imaging (BLI). (C) Effects of E7 and 5-FU on tumor growth, as detected in blood samples with the G-luc (proportional to the number of cancer cells, carrying G-luc). Points indicate mean values obtained from the analysis of the six mice in each group. Bars indicate standard deviation. p Values were calculated with two-sided Student’s t-test and ANOVA followed by the Tukey’s multiple comparison. (D) Effects of E7 and 5-FU on the weight of the mice, demonstrating that tumor shrinkage induced by these treatments was not accompanied by severe toxicity. Points indicate mean values obtained from the analysis of the six mice in each group. (E) Survival curves in the groups of mice treated with E7 and 5-FU. Median overall survival (mos) is reported. Statistically significant differences were determined by two-sided log-rank test. (F) Quantitative PCR results (calculated with the standard curve method, using the ratio with the housekeeping gene β-actin) showing the reduced expression of hTS mRNA in lysates from frozen tissues from PDAC-2 mice treated with E7 (24 hr before their sacrifice) compared with untreated control mice. Columns and bars indicate mean values and standard deviation. p Values were calculated with two-sided Student t test and ANOVA followed by the Tukey’s multiple comparison. (G) Representative immunohistochemical images showing a weak staining for hTS in the tissues from mice treated with E7 compared to the strong staining in 5-FU-treated and control mice. (Figure 8—source data 1).
-
Figure 8—source data 1
Orthotopic pancreatic cancer in vivo.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig8-data1-v2.xlsx
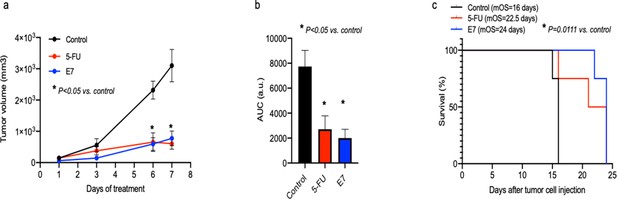
In vivo effects of E7 on Athymic Nude-Foxn1nu female mice.
(a) Tumor growth kinetics at the indicated time points. Athymic Nude-Foxn1nu female mice were subcutaneously inoculated with 2×106 A2780 cells. Implanted xenotransplanted mice, chosen randomly, were subjected to E7, 5FU and vehicle treatment, as indicated in the materials and methods section; n=4 mice per group. * E7-treatment is statistically different to control (vehicle) for the tumor volume at 6th and 7th day (t-test p<0.05). Error bars indicate SEM. (b) Quantitative analysis of the AUC of tumor growth kinetics in control, E7 and 5FU-treated mice. Bars are representative of the mean ± SEM obtained from all mice treated (t-test, * p<0.05). (c) Survival curves (Kaplan Meier) showing the percentage of survival of xenotransplanted mice, placebo control versus E7 and 5FU-treated mice (Log-rank (Mantel-Cox) test p-value = 0.0024; n=4).
-
Figure 8—figure supplement 1—source data 1
In vivo effects of E7 on Athymic Nude-Foxn1nu female mice.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig8-figsupp1-data1-v2.xlsx
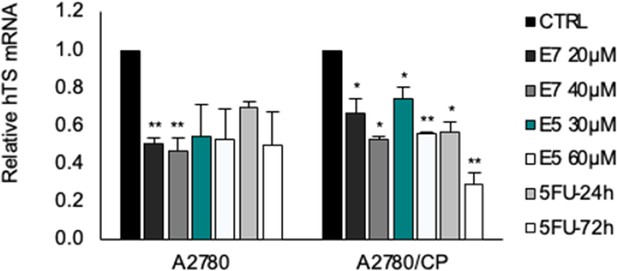
Real time PCR (qPCR) experiments.
Quantitative PCR (qPCR) measurements were performed for hTS transcripts in A2780 and A2780/CP cells treated with E7 and E5 for 12 hr and 5FU (5 μM) for 24 and 72 hr. Data indicate mean values and standard deviation of biological repeats performed in duplicate. p Values were calculated with two-sided Student’s t-test and ANOVA followed by the Tukey’s multiple comparison. * p<0.05; ** p<0.01.
-
Figure 8—figure supplement 2—source data 1
Real-time PCR (qPCR).
Quantitative PCR (qPCR) measurements of hTS transcripts in A2780 and A2780/CP cells treated with E7, E5, and 5FU.
- https://cdn.elifesciences.org/articles/73862/elife-73862-fig8-figsupp2-data1-v2.pdf
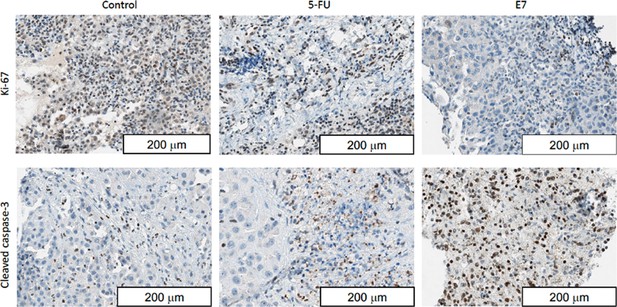
Representative immunohistochemical images showing extensive caspase- 3 activation after treatment with E7, which was also able to significantly reduced cell proliferation (Ki67 staining).
Conversely, only a limited caspase-3 staining was found in tumors treated with 5-FU, while most of the untreated tumor tissues showed no caspase-3 activation.
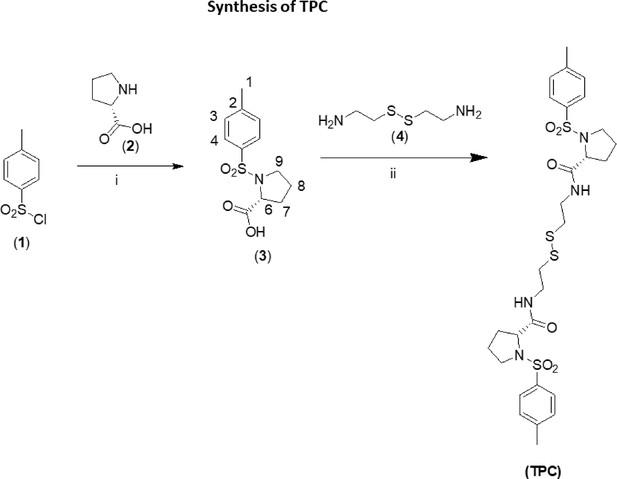
Synthesis of TPC.
(i) Diethyl ether, NaOH 2 M, RT, 4 h; HCl conc.; (ii) HOBT, EDAC, TEA, DCC.
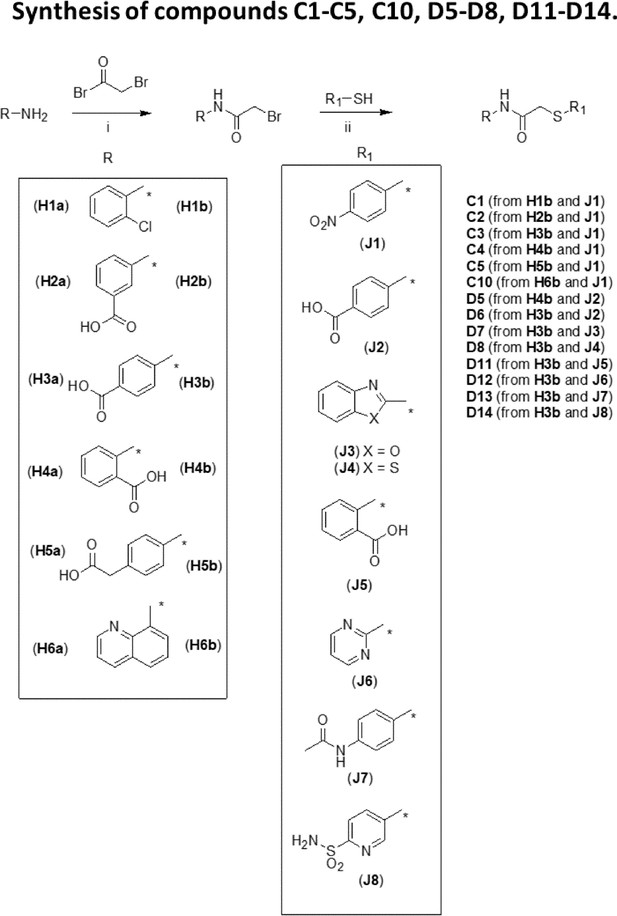
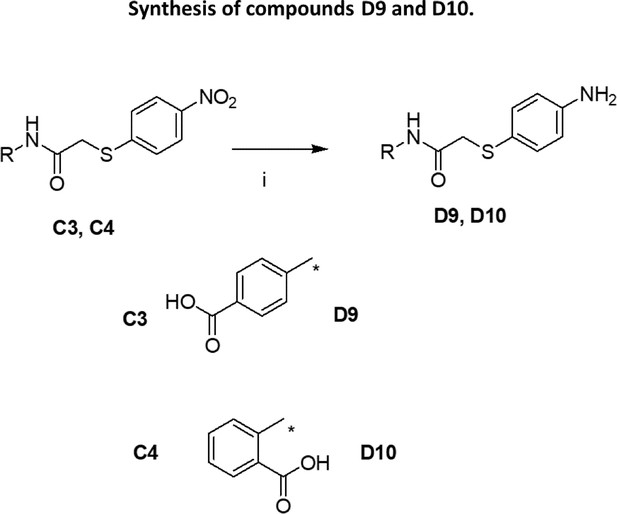
Synthesis of compounds C1-C5, C10, D5-D8, D11-D14.
(i): TEA/CH2Cl2 (for H1b, H2b) or Na2CO3/H2O (for H2b, H3b, H5b, H6b); (ii): K2CO3/acetone (for C2-C5, D5-D8, D11-D14) or TEA/THF (for C1, C10).
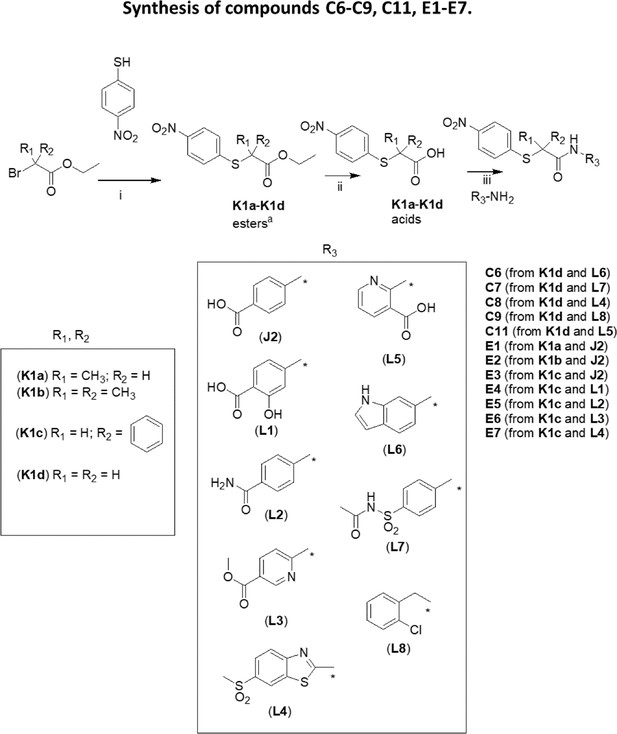
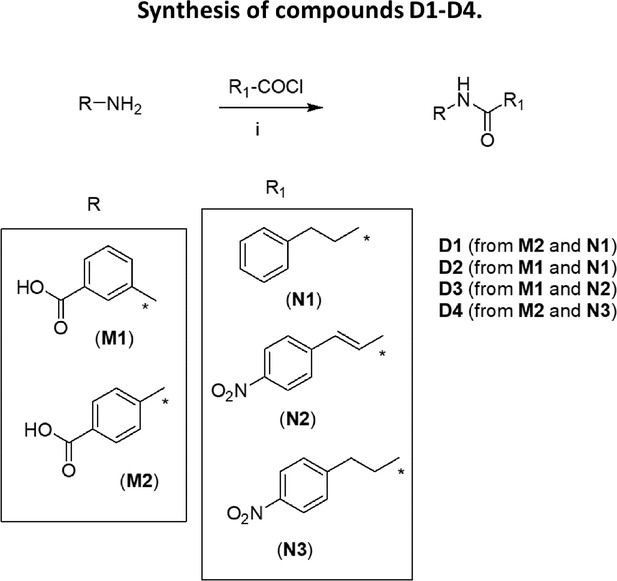
Synthesis of compounds C6-C9 and C11, E1-E7.
(i): K2CO3/acetone. In the case of K1d, methylbromoacetate was used instead of ethylbromoacetate; (ii): NaOH/EtOH; (iii): (COCl)2/THF then TEA (for E1-E6, C6, C7, C9, C11) or WSC/DMF (for C8, E7).
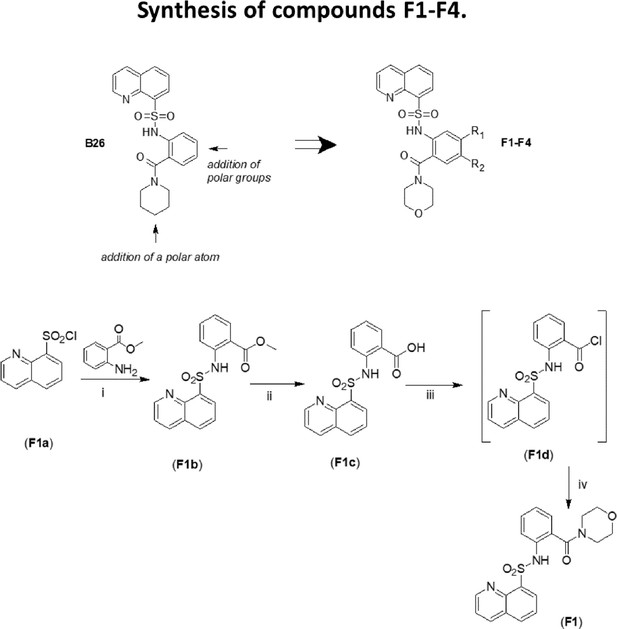
Design of compounds F (upper panel).
Synthesis of compound F1 (bottom panel). (i): CH2Cl2/pyridine/DMAP, RT; (ii): THF/LiOH 2 M; (iii): THF/DMF, (COCl)2; (iv): morpholine, CH2Cl2/TEA 0 °C.
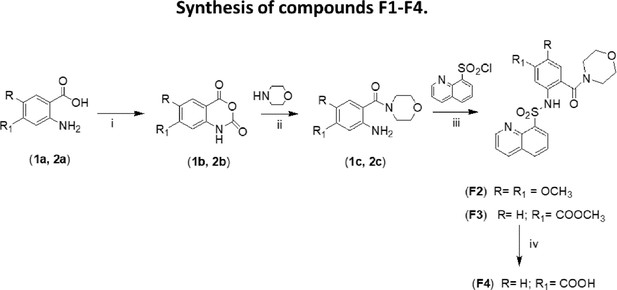
Synthesis of compounds F2-F4.
(i): THF/triphosgene, r.t.; (ii): DMF/DMAP; (iii): CH2Cl2/pyridine/DMAP; (iv): THF/LiOH 2 M.
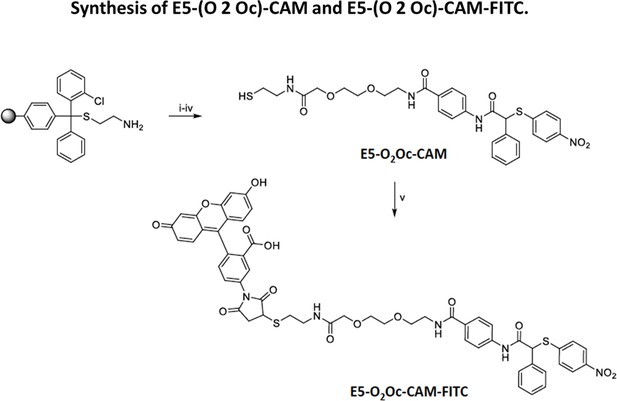
Synthesis of compounds E5-O2Oc-CAM and E5-O2Oc-CAM-FITC.
(i) Cysteamine 2-chlorotrityl resin (1 eq.), Fmoc-O2Oc-OH (4 eq.), DIC (4 eq.), HOBt (4 eq.), DMF, r.t., 30 min; (ii) 40% piperidine in DMF, r.t., 5 min; (iii) E3 (2 eq.), HATU (3 eq.) DIPEA (2 eq.), DMF, r.t., 3 h; (iv) trifluoroacetic acid (TFA)/H2O/phenol/triisopropylsilane 88:5:5:2 v/v (10 mL / 0.2 g of resin); r.t., 30 min; (v) E5-(O2Oc)-CAM (1 eq.), Fluorescein-5-Maleimide (0.06 eq.), CH3CN/H2O, r.t., 5 min.
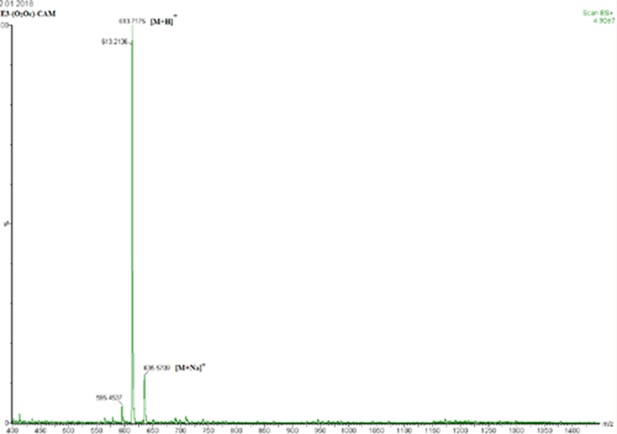
Mass spectrum of E5-(O2Oc)-CAM ESI-HRMS calcd for C29H33N4O7S2+ (M+H+): 613.1785.
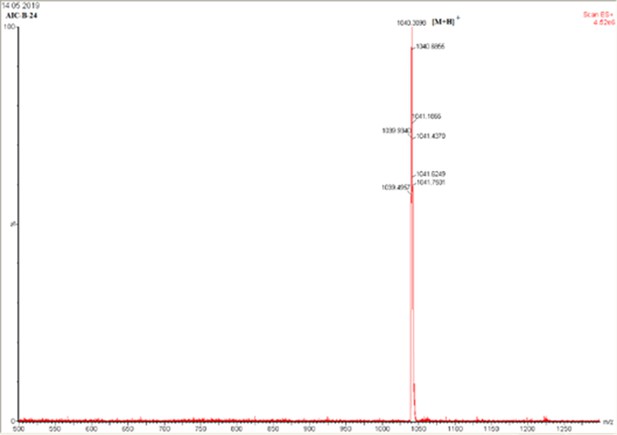
Mass spectrum of E5-(O2Oc)-CAM-FITC.
ESI-HRMS calcd for C53H46N5O14S2+ (M+H+):1040.2477.
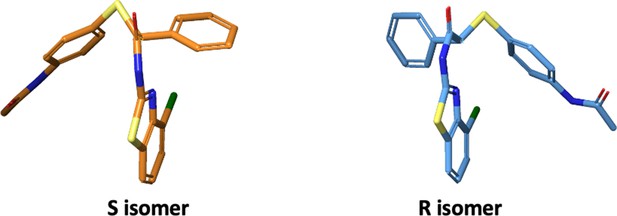
Structure 2D of the E7 enantiomers.
(A) Representation of S and R isomers of compound E7 obtained with (B) Structural modification of compound B12. The changes in chemical structure and the associated IC50s for recombinant-enzyme inhibition, DFFRET, abilities to dissociate the hTS dimers in cells, cellular growth inhibition and in vivo data of the key compounds C3, E5 and E7 of the sub-series are reported.
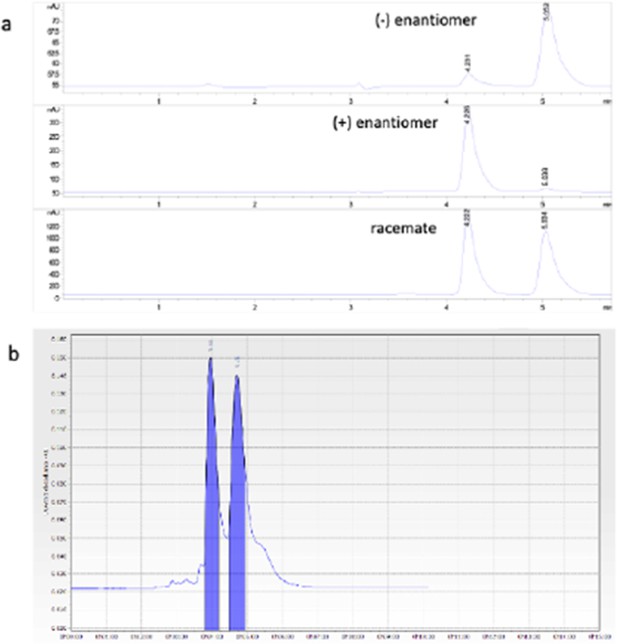
Chromatograms of single (+/-)-E7 enantiomers.
Column: Chiracel OD(250x4.6 mm I.D., 5 mm), mobile phase: water:acetonitrile 2:98 (v/v), flow rate:1 mL/min; l=254 nm. (b) Semipreparative enentiseparation of (+/-)-E7. Column: Chirasphere OD, mobilephase water:acetonitrile 2:98 (v/v), flow rate:5 mL/min, l:254 nm, room temperature.
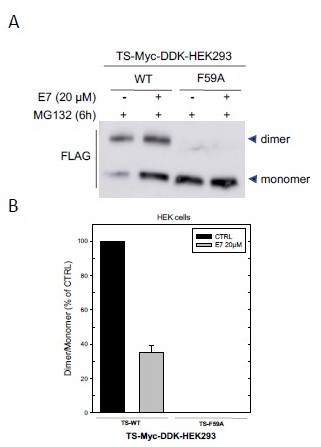
(A) Effects on exogenous hTS dimer/monomer equilibrium in HEK293 cells after a 3 hour exposure to E7 (20 μM), in the presence of MG132 (6 hour).
Next the cells have been lysed and crosslinked (BS3), and western blot performed by using the anti-Flag antibody. (B) Quantification of the immunoblot shown in (A) of exogenous hTS dimer/monomer equilibrium (anti-FLAG stain) in HEK293 cells transfected with TS-Myc-DDK (24 hour) or with the F59A mutant (24 hour) tagged vector.
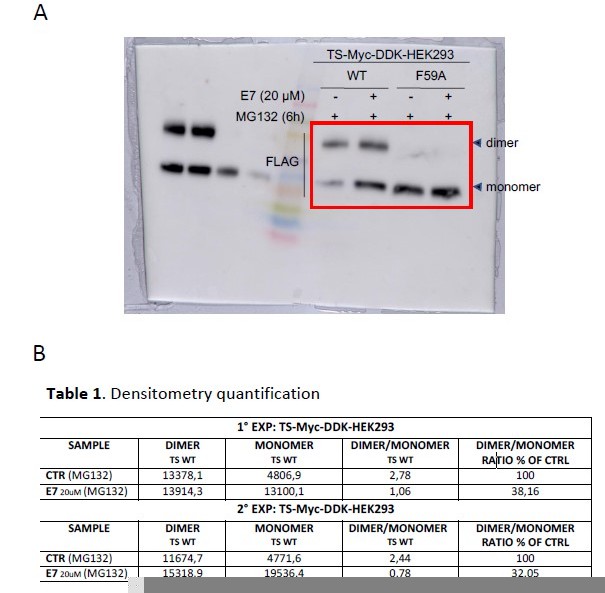
Original blots (A) and densitometry quantification (B).
(A) Effects on exogenous hTS dimer/monomer equilibrium in HEK293 cells after a 3 hours exposure to E7 (20 µM), in the presence of MG132 (6 hour). Next the cells have been lysed and crosslinked (BS3), and western blot performed by using the anti-Flag antibody. (B) Quantification of the immunoblot shown in (A) of exogenous hTS dimer/monomer equilibrium (anti-FLAG stain) in HEK293 cells transfected with TSMyc-DDK (24 hour) or with the F59A mutant (24 hour) tagged vector.
Additional files
-
Supplementary file 1
Computational protein-pocket identification for tethering.
- https://cdn.elifesciences.org/articles/73862/elife-73862-supp1-v2.docx
-
Supplementary file 2
X-ray crystallography.
- https://cdn.elifesciences.org/articles/73862/elife-73862-supp2-v2.docx
-
Supplementary file 3
Mass spectrometry data.
- https://cdn.elifesciences.org/articles/73862/elife-73862-supp3-v2.docx
-
Supplementary file 4
Compounds FRET and inhibition data.
- https://cdn.elifesciences.org/articles/73862/elife-73862-supp4-v2.docx
-
Supplementary file 5
Mass Spectrometry analysis of the fractions eluted from AEX.
- https://cdn.elifesciences.org/articles/73862/elife-73862-supp5-v2.docx
-
Supplementary file 6
Cancer cell growth inhibition and pharmacokinetic.
- https://cdn.elifesciences.org/articles/73862/elife-73862-supp6-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/73862/elife-73862-transrepform1-v2.pdf