Pervasive translation in Mycobacterium tuberculosis
Figures
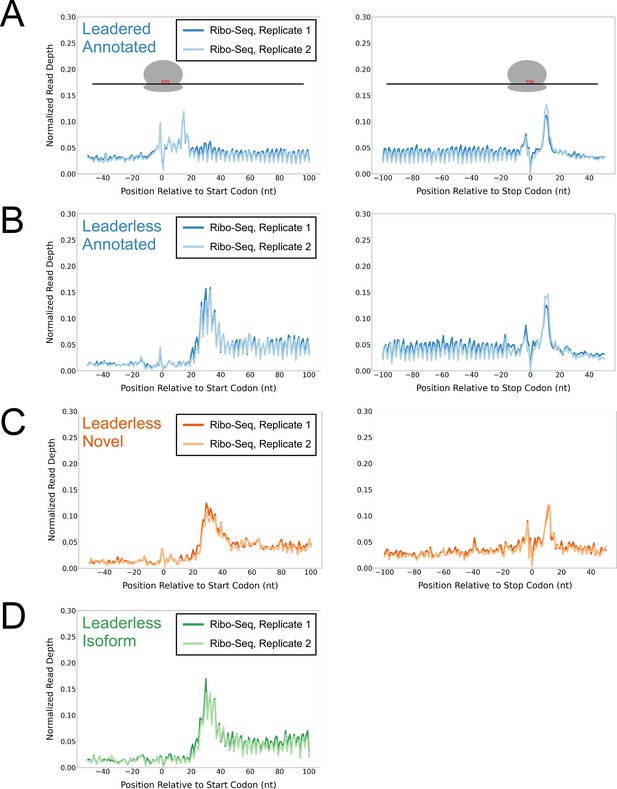
Ribo-seq data support the translation of hundreds of isoform and novel ORFs from leaderless mRNAs.
(A) Metagene plot showing normalized Ribo-seq sequence read coverage for untreated cells in the regions around start (left graph) and stop codons (right graph) of previously annotated, leadered ORFs. Note that sequence read coverage is plotted only for the 3’ ends of reads, since these are consistently positioned relative to the ribosome P-site (Woolstenhulme et al., 2015). Data are shown for two biological replicate experiments. The schematics show the position of initiating/terminating ribosomes, highlighting the expected site of ribosome occupancy enrichment at the downstream edge of the ribosome. (B) Equivalent data to (A) but for putative annotated, leaderless ORFs. (C) Equivalent data to (A) but for putative novel, leaderless ORFs. (D) Equivalent data to (A) but for putative isoform, leaderless ORFs. Only data for start codons are shown because the same stop codon is used by both an annotated and isoform ORF.
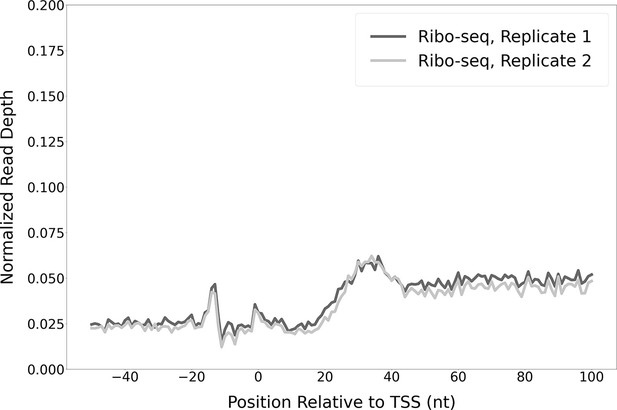
Modest enrichment of Ribo-seq signal downstream of the transcription start sites (TSSs) of non-leaderless RNAs.
Metagene plot showing normalized Ribo-seq sequence read coverage (data indicate the position of ribosome footprint 3’ ends) in the region from –50 to +100 nt relative to the TSSs of RNAs that are not leaderless mRNAs.
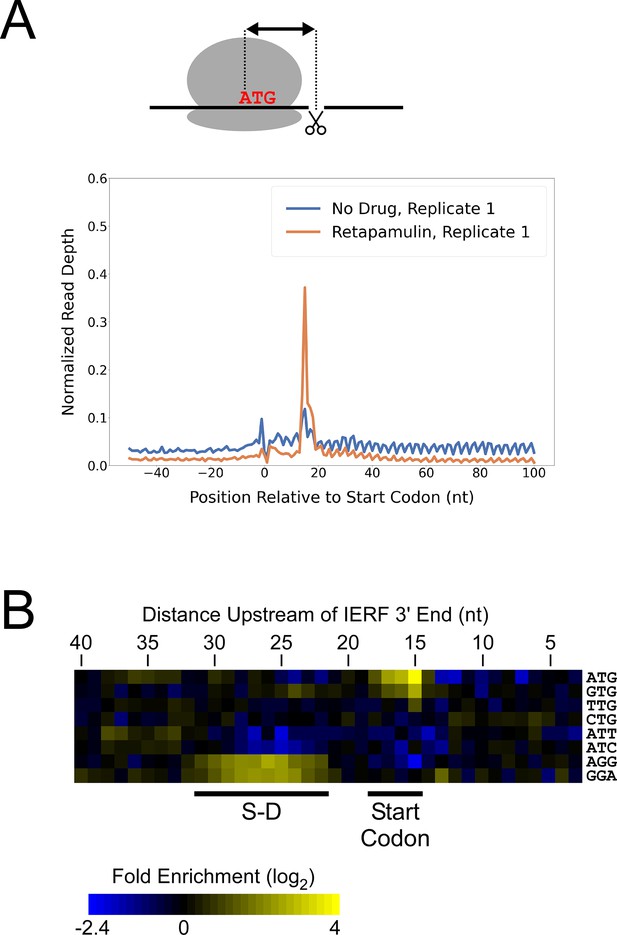
Ribo-RET of M. tuberculosis identifies sites of translation initiation.
(A) Metagene plot showing normalized Ribo-seq and Ribo-RET sequence read coverage (single replicate for each; data indicate the position of ribosome footprint 3’ ends) in the region from –50 to +100 nt relative to the start codons of annotated, leadered ORFs. (B) Heatmap showing the enrichment of eight selected trinucleotide sequences, for regions upstream of IERFs, relative to control regions. Expected positions of start codons and S-D sequences are indicated below the heatmap.
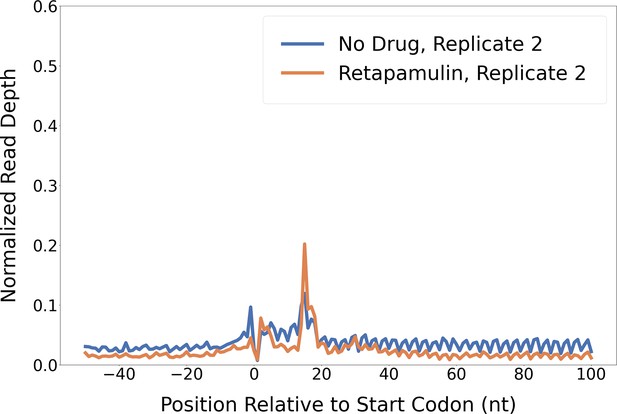
Retapamulin treatment traps initiating ribosomes.
Metagene plot showing normalized Ribo-seq and Ribo-RET sequence read coverage (single replicate for each; data indicate the position of ribosome footprint 3’ ends) in the region from –50 to +100 nt relative to the start codons of annotated, leadered ORFs. Figure 2A shows data for the other replicate datasets.
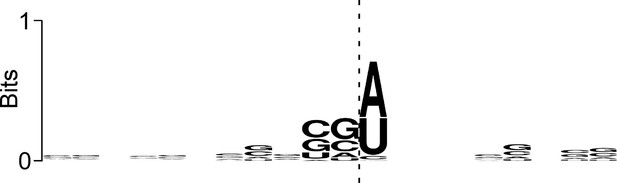
Sequence bias associated with the 3’ ends of ribosome-protected RNAs at IERFs.
Logo showing sequence bias around the 3’ ends of Ribo-RET RNA fragments associated with IERFs. The cleavage site at the 3’ end of the aligned RNA fragments is indicated by a vertical dashed line.
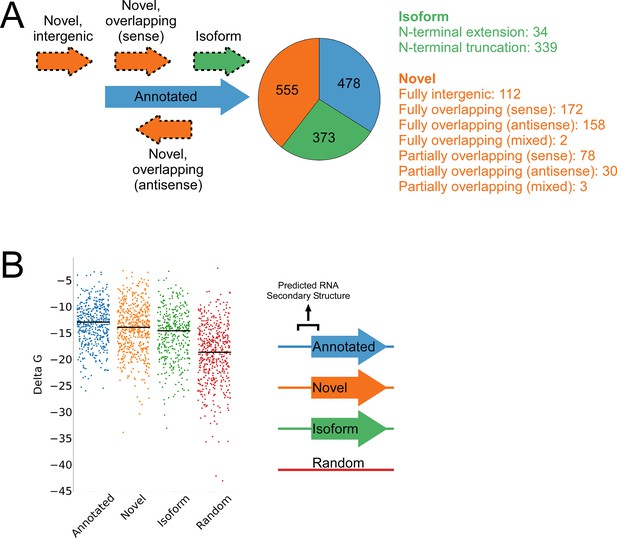
Features of higher-confidence ORFs identified by Ribo-RET.
(A) Distribution of different classes of ORFs identified by Ribo-RET. The pie-chart shows the proportion of identified ORFs in each class. Isoform ORFs are further classified based on whether they are longer (‘N-terminal extension’) or shorter (‘N-terminal truncation’) than the corresponding annotated ORF. Novel ORFs are further classified based on their overlap with annotated genes. ‘Sense’, ‘antisense’, and ‘mixed’ refer to whether the overlapping gene(s) is/are in the sense, antisense, or both (multiple overlapping genes) orientations with respect to the novel ORF. ‘Fully’ and ‘Partially’ indicate whether all or only some of the novel ORF overlaps annotated genes. (B) Strip plot showing the ΔG for the predicted minimum free energy structures for the regions from –40 to +20 nt relative to putative start codons for the different classes of ORF, and for a set of 500 random sequences. Median values are indicated by horizontal lines.
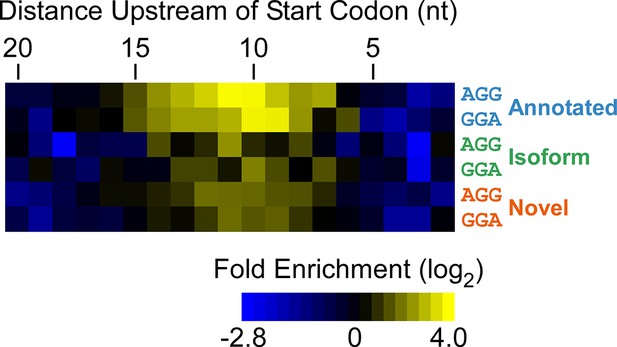
Enrichment of SD-like sequences upstream of higher-confidence ORFs identified by Ribo-RET.
Heatmap showing the enrichment of AGG and GGA trinucleotide sequences relative to control regions, for positions upstream of the start codons of annotated, isoform, and novel ORFs identified by Ribo-RET (higher-confidence set of ORFs).
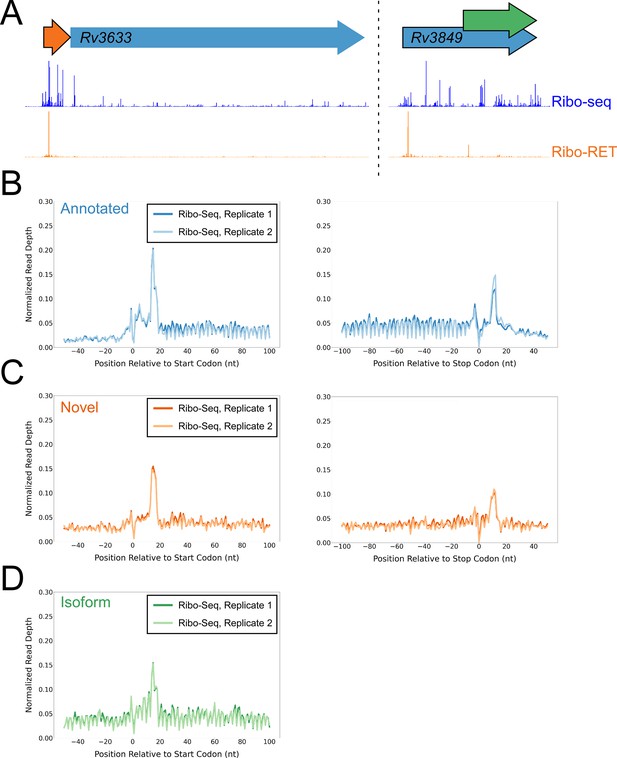
Ribo-seq data support the translation of hundreds of isoform and novel ORFs identified by Ribo-RET.
(A) Ribo-seq and Ribo-RET sequence read coverage (read 3’ ends) across two genomic regions, showing examples of putative ORFs in the annotated (blue arrow), novel (orange arrow), and isoform (green arrow) categories. ORFs identified by Ribo-RET shown with a black outline. (B) Metagene plot showing normalized Ribo-seq sequence read coverage (data indicate the position of ribosome footprint 3’ ends) for untreated cells in the regions around start (left graph) and stop codons (right graph) of ORFs predicted from Ribo-RET profiles, that correspond to previously annotated genes. (C) Equivalent data to (B) but for putative novel ORFs identified from Ribo-RET data. (D) Equivalent data to (B) but for putative isoform ORFs identified from Ribo-RET data. Only data for start codons are shown because the same stop codon is used by both an annotated and isoform ORF.
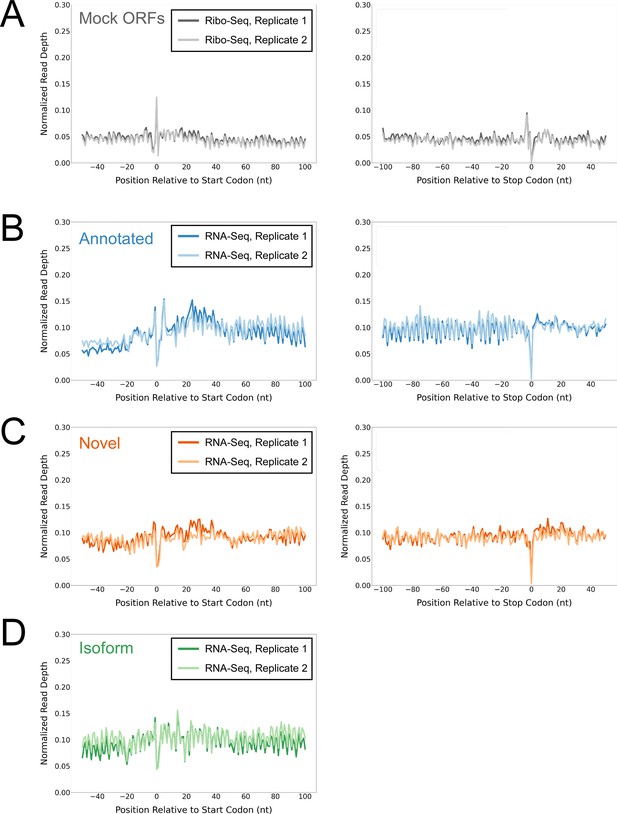
Control analyses using mock ORFs or RNA-seq data.
(A) Metagene plot showing normalized Ribo-seq sequence read coverage (data indicate the position of RNA fragment 3’ ends) for untreated cells in the regions around start (left graph) and stop codons (right graph) of mock ORFs. (B) Metagene plot showing normalized RNA-seq sequence read coverage (read 3’ ends) for untreated cells in the regions around start (left graph) and stop codons (right graph) of annotated ORFs identified from Ribo-RET data. (C) Equivalent data to (B) but for putative novel ORFs identified from Ribo-RET data. (D) Equivalent data to (B) but for putative isoform ORFs identified from Ribo-RET data. Only data for start codons are shown because the same stop codon is used by both an annotated and isoform ORF.
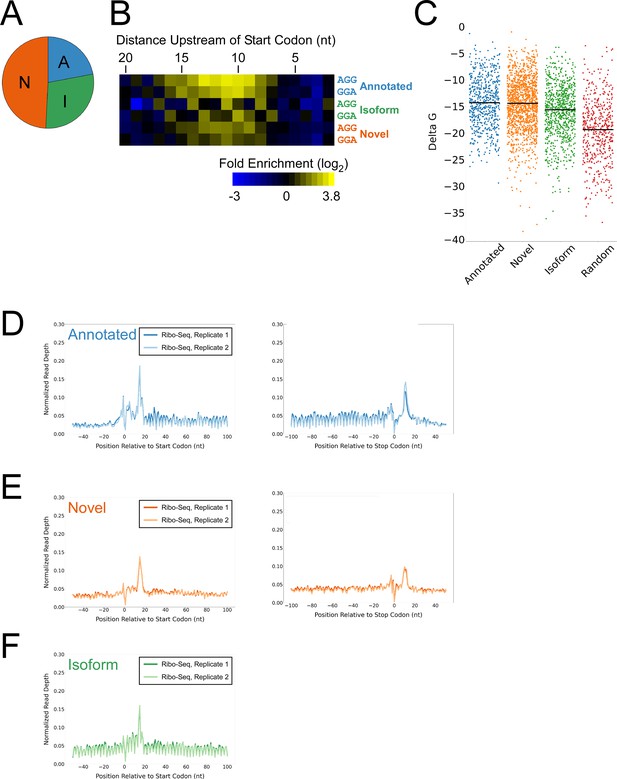
Features of lower-confidence ORFs identified by Ribo-RET.
(A) Distribution of different classes of lower-confidence ORFs identified by Ribo-RET. (B) Heatmap showing the enrichment of AGG and GGA trinucleotide sequences relative to control regions, for positions upstream of the start codons of lower-confidence annotated, isoform, and novel ORFs identified by Ribo-RET. (C) Strip plot showing the ΔG for the predicted minimum free energy structures for the regions from –40 to +20 nt relative to start codons for the different classes of lower-confidence ORF, and for a set of 500 random sequences. Median values are indicated by horizontal lines. (D) Metagene plot showing normalized Ribo-seq sequence read coverage (data indicate the position of ribosome footprint 3’ ends) for untreated cells in the regions around start (left graph) and stop codons (right graph) of lower-confidence annotated ORFs identified from Ribo-RET data. (E) Equivalent data to (D) but for lower-confidence novel ORFs identified from Ribo-RET data. (F) Equivalent data to (D) but for lower-confidence isoform ORFs identified from Ribo-RET data. Only data for start codons are shown because the same stop codon is used by both an annotated and isoform ORF.
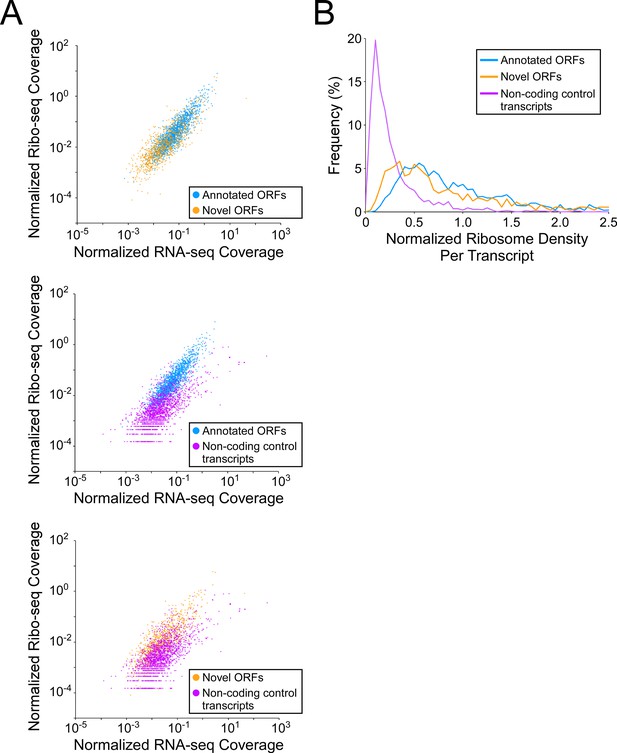
Novel ORFs are efficiently translated.
(A) Pairwise comparison of normalized RNA-seq and Ribo-seq coverage for annotated, novel and non-coding control transcripts. Reads are plotted as RPM per nucleotide using a single replicate of each dataset for reads aligned to the reference genome at their 3’ ends. The categories compared are: (i) annotated ORFs (higher-confidence and lower-confidence ORFs detected by Ribo-RET, and leaderless ORFs; blue datapoints), (ii) novel ORFs (higher-confidence and lower-confidence ORFs detected by Ribo-RET and leaderless ORFs, for regions at least 30 nt from an annotated gene; orange datapoints), and (iii) a set of 1854 control transcript regions that are expected to be non-coding (see Materials and methods; purple datapoints). ORF/transcript sets are plotted in pairs to aid visualization. (B) Normalized ribosome density per transcript (ratio of Ribo-seq coverage to RNA-seq coverage) for the same sets of ORFs/transcripts. The graph shows the frequency (%) of ORFs/transcripts within each group for bins of 0.05 density units.
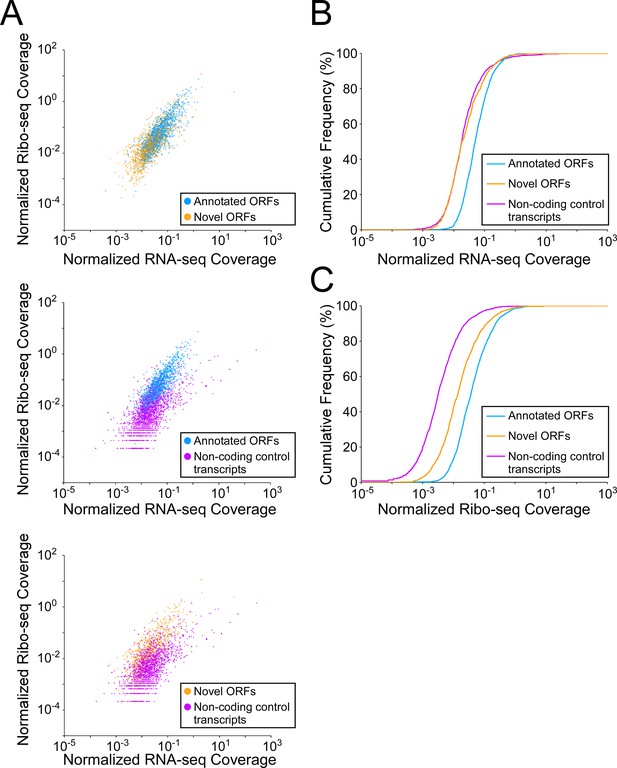
Novel and isoform ORFs are expressed at lower levels than annotated ORFs.
(A) Pairwise comparison of normalized RNA-seq and Ribo-seq coverage for annotated, novel, and non-coding control transcripts. Each data-point represents one transcript. Values are plotted as RPM per nucleotide using a single replicate of each dataset for reads aligned to the reference genome at their 3’ ends (c.f. Figure 5, which shows data for the other replicate for each dataset). The categories compared are: (i) annotated ORFs (higher-confidence and lower-confidence ORFs detected by Ribo-RET, and leaderless ORFs; blue datapoints), (ii) novel ORFs (higher-confidence and lower-confidence ORFs detected by Ribo-RET and leaderless ORFs, for regions at least 30 nt from an annotated gene; orange datapoints), and (iii) a set of 1854 control transcript regions that are expected to be non-coding (see Materials and methods; purple datapoints). ORF/transcript sets are plotted in pairs to aid visualization. (B) Cumulative frequency distributions of normalized RNA-seq coverage for annotated, novel, and non-coding control transcripts. Coverage values (x-axis) are the average of two replicate datasets. Values on the y-axis indicate the percentage of transcripts with coverage less than or equal to a given value on the x-axis. (C) Cumulative frequency distributions of normalized Ribo-seq coverage for annotated, novel, and non-coding control transcripts.
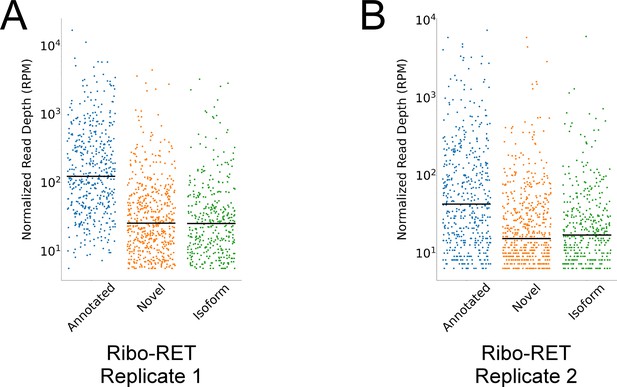
Novel and isoform ORF start codons have lower ribosome occupancy than annotated ORF start codons in Ribo-RET data.
(A) Strip plot showing the normalized sequencing read depth for a single Ribo-RET replicate dataset, at start codons of higher-confidence annotated, isoform, and novel ORFs identified by Ribo-RET. Median values are indicated by horizontal lines. (B) Equivalent to (A) but for a second replicate Ribo-RET dataset.
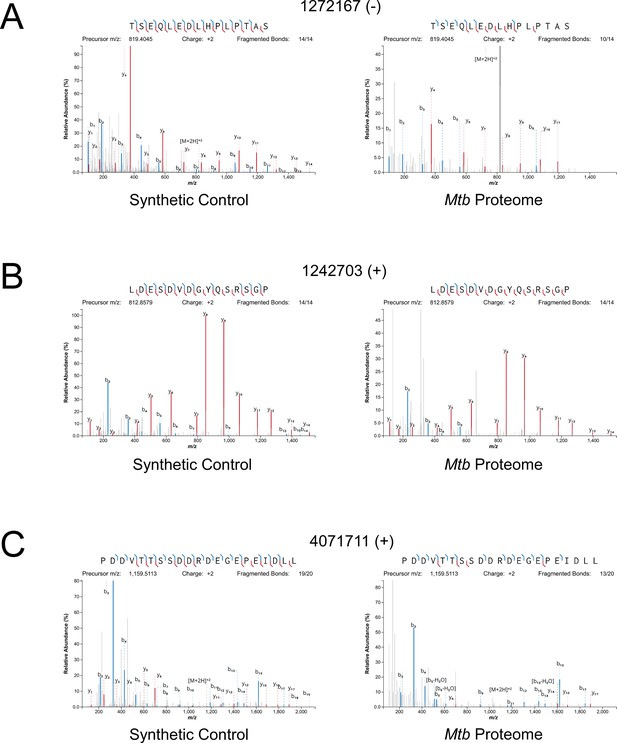
Mass spectrometry validation of selected ORFs.
MS/MS spectra from novel ORFs measured with a synthetic peptide compared to spectra measured from the Mtb proteome. The genome coordinate and strand of each selected novel ORF start codon is indicated. (A) Leaderless ORF 1272167 (-) was identified from amino-acids 2–24. The y4 and parent m/z ions are off-scale. (B) Leaderless ORF 1242703 (+) was observed from amino acids 46–61. (C) Leadered ORF 4071711 (+) was observed from amino acids 4–26. The b3 ion is off-scale. Measured b-ions are in blue, and y-ions are in red. The nearly complete spectrum obtained for each peptide and the fragment-mass balance clearly indicate that these sORFs are identical to their synthetic cognates.
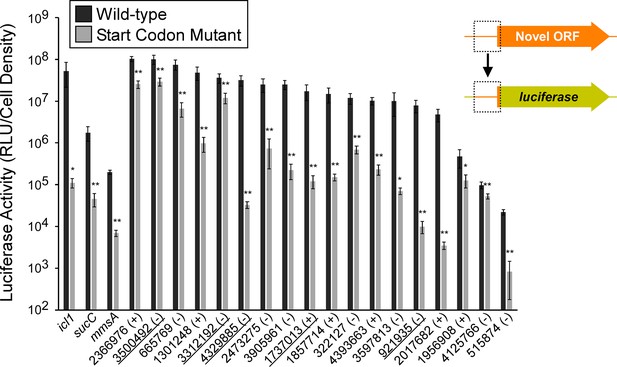
Validation of selected novel and isoform ORFs using luciferase reporter fusions.
Luciferase reporter assays for constructs consisting of the region from position –25 up to the Ribo-RET-predicted start codon fused translationally to a luciferase reporter gene, as illustrated in the schematic. Fusions were tested for 18 putative novel ORFs identified from Ribo-RET data, and three previously annotated ORFs that serve as positive controls. Wild-type and mutant start codon reporter construct pairs were separately integrated into the M. smegmatis chromosome to quantify the net contribution of translation from the predicted start codon. The genome coordinate and strand of each selected novel ORF start codon is indicated. Underlined coordinates indicate novel ORFs identified from a single Ribo-RET replicate dataset.
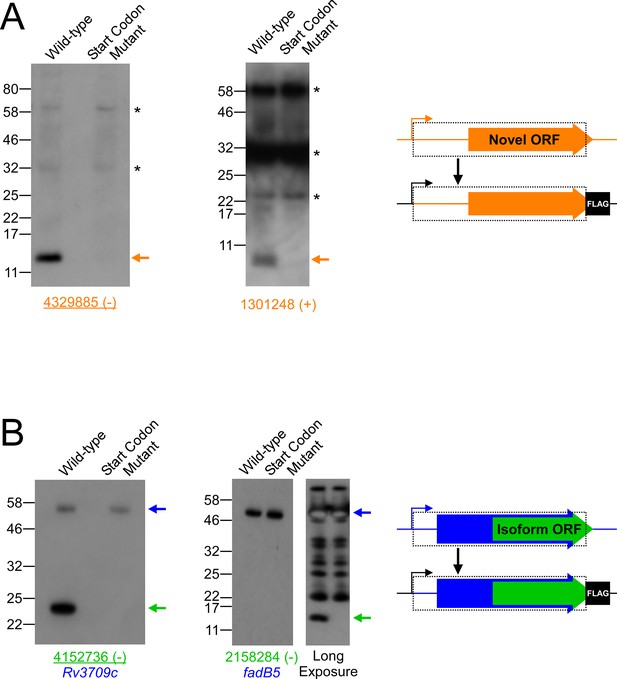
Validation of selected novel and isoform ORFs by western blot.
(A) Western blot with anti-FLAG antibody to detect FLAG-tagged novel ORFs integrated into the M. smegmatis chromosome with either an intact (wild-type) or mutated start codon. The integrated constructs included the entire 5’ UTR and open-reading frame (indicated by a dashed box), but not the native promoter. Bands corresponding to the tagged novel ORF are indicated with an orange arrow. Asterisks indicate the positions of common cross-reacting proteins. Novel ORF 4329885 (-) was identified from a single Ribo-RET replicate dataset. The positions of molecular weight marker bands are indicated. (B) Western blots with anti-FLAG antibody to detect FLAG-tagged isoform ORFs integrated into the M. smegmatis genome with either an intact (wild-type) or mutated start codon. The integrated constructs included the overlapping full-length, annotated ORF and its entire 5’ UTR. Bands corresponding to the tagged full-length and isoform ORFs are indicated with blue and green arrows, respectively. The western blot for the isoform ORF overlapping fadB5 was developed with a short (left panel) and a long (right panel) exposure due to the large difference in steady-state levels of the full-length and isoform proteins. Isoform ORF 4152736 (-) was identified from a single Ribo-RET replicate dataset.
-
Figure 6—figure supplement 2—source data 1
Images of full western blots are provided.
The zipped folder includes (i) individual files for each blot, and (ii) a summary file showing all blots, with boxes to show the regions used in Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/73980/elife-73980-fig6-figsupp2-data1-v3.zip
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mycobacterium tuberculosis) | mc27000 | DOI: 10.1016/j.vaccine.2006.05.097 | ΔpanCD ΔRD1 | |
| Strain, strain background (Mycobacterium smegmatis) | mc2155 | DOI: 10.1111/j.1365–2958.1990.tb02040.x | ||
| Antibody | Monoclonal anti-FLAG M2 antibody (Mouse monoclonal) | SIGMA | Catalog # F1804 | Used at (1:1,000) dilution for western blot |
| Recombinant DNA reagent | pRV1133C (plasmid) | This study | pRV1133C | Integrates at attP site; includes the metE promoter region |
| Recombinant DNA reagent | pGE450 (plasmid) | This study | pGE450 | Derivative of pRV1133C containing 3 x FLAG |
| Recombinant DNA reagent | pGE190 (plasmid) | This study | pGE190 | Derivative of pRV1133C containing the nLuc gene from pNL1.1 (Promega, cat no 1001) |
| Other | Micrococcal nuclease (S7) | SIGMA | Catalog # 10107921001 | |
| Other | Nano-Glo Luciferase Assay Reagent | Promega | Catalog # N1110 | |
| Chemical compound, drug | Retapamulin | SIGMA | Catalog # CDS023386 | |
| Software, algorithm | CLC Genomics Workbench | Qiagen | v8.5.1 | Alignment of sequence reads from.fastq files |
| Software, algorithm | RNAfold | DOI:10.1186/1748-7188-6-26 | v2.4.14 | ViennaRNA Package https://www.tbi.univie.ac.at/RNA/ |
Additional files
-
Supplementary file 1
Supplementary tables.
(A) List of putative leaderless ORFs. (B) List of IERFs. (C) List of ORFs identified by Ribo-RET. (D) Analysis of G/C skew for cys-rich regulatory ORFs. (E) Analysis of isoform ORFs and their position relative to overlapping annotated ORFs. (F) List of oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/73980/elife-73980-supp1-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/73980/elife-73980-transrepform1-v3.docx