Genetic variation in ALDH4A1 is associated with muscle health over the lifespan and across species
Figures
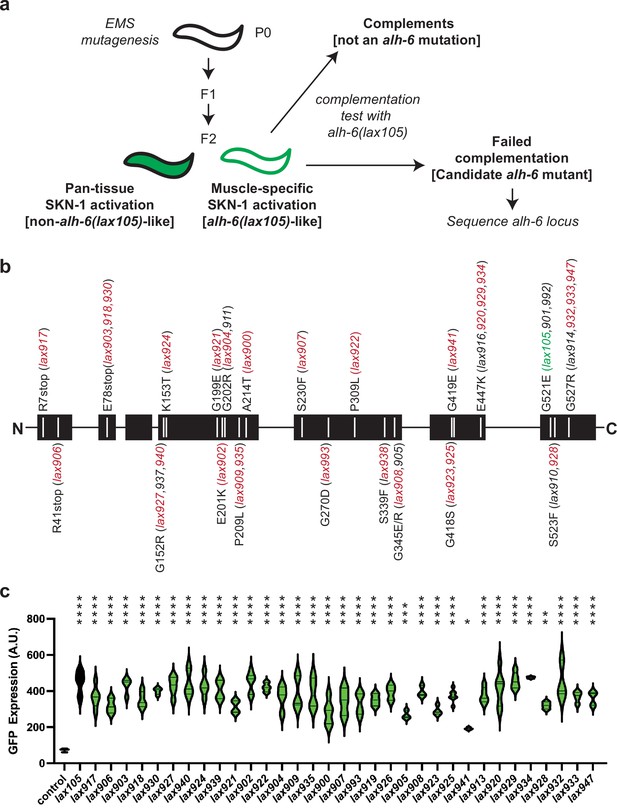
Mutation of alh-6 uniquely activates age-dependent and activation of the gst-4p::gfp oxidative stress reporter in muscle.
(a) Schematic representation of genetic screen for mutants that phenocopy alh-6(lax105). (b) Schematic representation of the ALH-6 protein with the molecular identity of mutants isolated and sequenced annotated. Alleles that were selected for additional functional tests of muscle function (Figure 4) are highlighted in red and the location of the canonical alh-6(lax105) allele is highlighted in green. These alleles represent all the sequenced mutations in alh-6 that were isolated from the ethyl methanesulfonate (EMS) screen. (c) Quantification of stress reporter activation in the muscle in the new alh-6 mutant alleles, as measured by the intensity of GFP fluorescence from the oxidative stress reporter gst-4p::gfp (see Figure 1—figure supplement 1 for representative images). t-Test relative to gst-4p::gfp reporter animals (control); *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
-
Figure 1—source data 1
Structure-function predictions of ALH-6 mutant proteins from computational modeling.
The impact of each missense mutation as predicted by Phyre and Missense3D (Kelley et al., 2015; Ittisoponpisan et al., 2019) and the corresponding phenotypic observations made in C. elegans.
- https://cdn.elifesciences.org/articles/74308/elife-74308-fig1-data1-v2.xlsx

Novel alleles of alh-6 induce muscle-specific activation of the gst-4p::gfp stress reporter reporter.
GFP fluorescence images of gst-4p::gfp animals harboring alh-6 mutations, as indicated. Scale bar = 100 μm. Quantification of fluorescence is shown in Figure 1c.
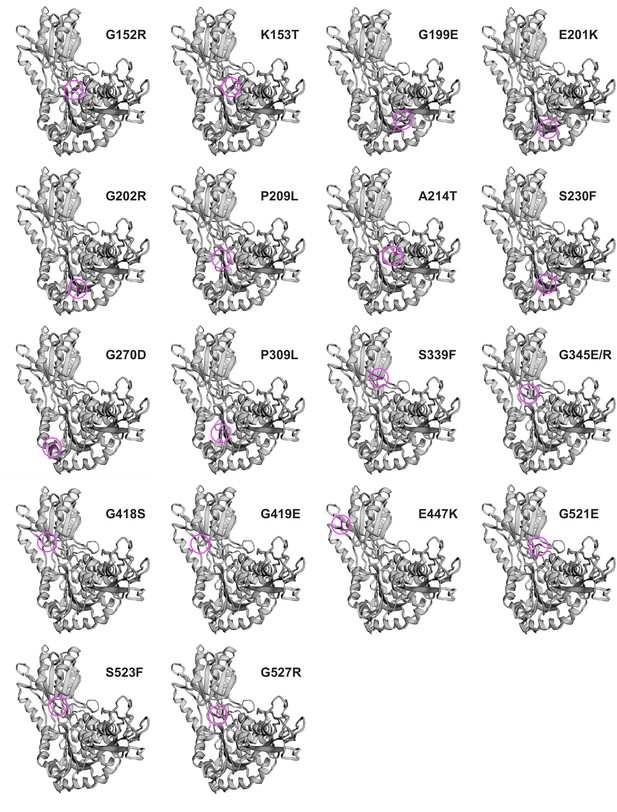
Location of amino acid substitutions in alh-6 mutants.
Location of individual missense mutations of ALH-6 mutants on the predicted structure of the wild-type ALH-6 protein by Phyre2 (Kelley et al., 2015). Mutated residues are colored (purple) and circled.
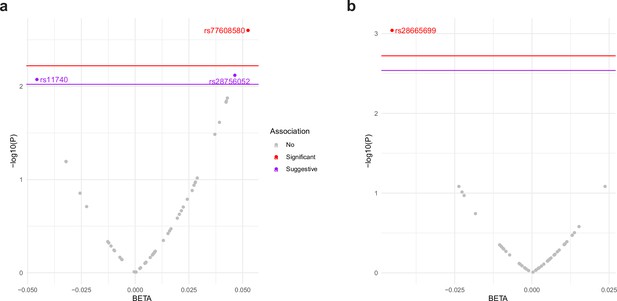
ALDH4A1 variants associate with human age-related phenotypes for change in muscle function.
Plot of association between variants in the ALDH4A1 gene and normative aging-related muscle decline in (a) gait speed and (b) grip strength in the US Health and Retirement Study (HRS). The x-axis shows the beta estimate for the effect of each SNP, represented by a dot, on the phenotype. The y-axis shows the log of the p value for the association between the SNP and the phenotype. SNPs that surpassed the empirical p value threshold, shown as a red line, for decline in gait speed (empirical p value = 0.006) and grip strength (empirical p value = 0.0019) are depicted as red dots. SNPs that surpassed a suggestive threshold (p value = 0.009 for gait speed) are depicted as purple dots.
-
Figure 2—source data 1
Details for phenotypes calculated from the US Health and Retirement Study.
- https://cdn.elifesciences.org/articles/74308/elife-74308-fig2-data1-v2.docx
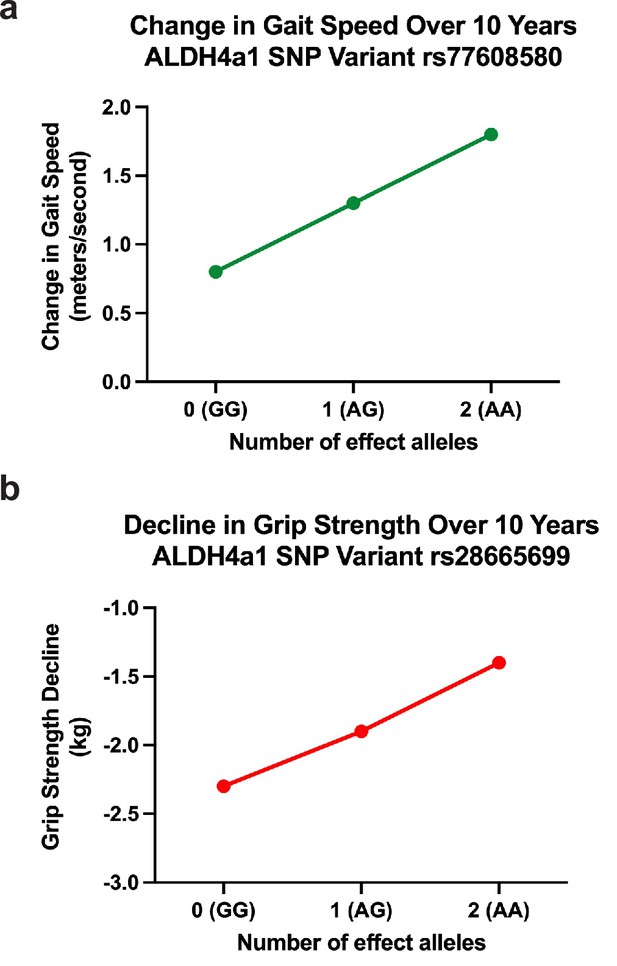
Effects of ALDH4A1 variation on phenotypes representing association with change in aging-related function in a normative, population-based sample of older adults.
(a) Change in gait speed over 10 years. Effect of SNP rs77608580 on aging-related changes in gait speed (b = 0.052, p = 0.0025). Over the span of one decade, on average, those with one or two effect alleles will have faster gait speeds with a difference of 0.52 and 1.04 m/s, respectively, compared to those without an effect allele. (b) Decline in grip strength over 10 years. Variation in ALDH4A1 (SNP rs28665699) is inversely associated with decline in aging-related grip strength (b = −0.045, p = 0.0009). Individuals with one or two effect alleles have slower progression of weakened grip strength over 10 years by 0.5 and 1.0 kg, respectively, compared to the same aged individuals without the effect allele.
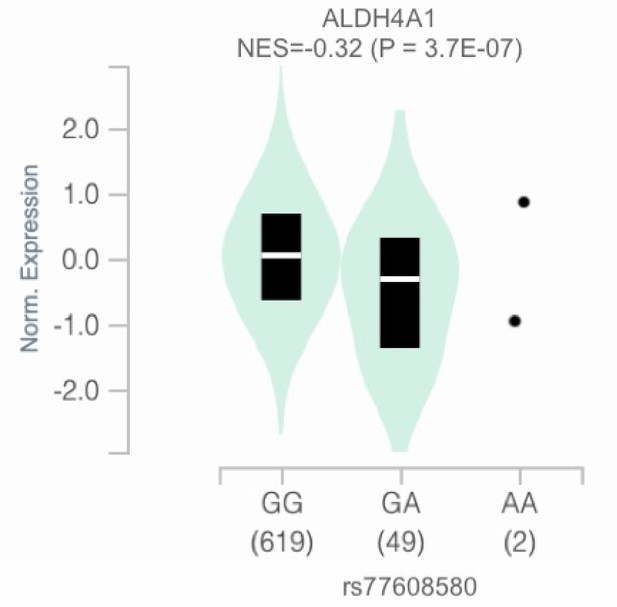
Association between rs77608580 and ALDH4A1 gene expression levels in whole blood.
Normalized gene expression levels for ALDH4A1 (y-axis) by SNP genotype (x-axis) are shown by violin plot. Also on the x-axis is the sample size by genotype in parenthesis. Violin plots show the density plot of the data (green cloud) with the median of the data shown by the white line of the black box plot within, the lower and upper border of the box plot corresponding to the first and third quartiles, respectively. The black dots represent sample points for the genotype in which there were too few samples to depict by box plot. A linear regression model was used to estimate the mean difference in expression levels, calculated as a normalized effect size (NES) to compare the alternative allele, G, to the minor allele, A, figure and data source: GTEx Analysis Release V8 (dbGaP Accession phs000424.v8.p2) (Battle et al., 2017).
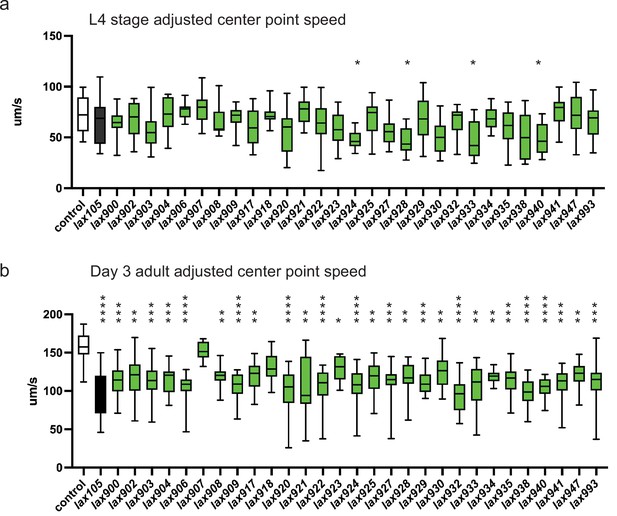
alh-6 mutations accelerate loss of muscle function.
WormLab software analysis of adjusted center point speed of individual animals of the given genotypes at the L4 stage (a) or day 3 of adulthood (b). Brown–Forsythe and Welch analysis of variance (ANOVA) test with Dunnett’s T3 multiple comparisons test, with individual variances computed for each comparison. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
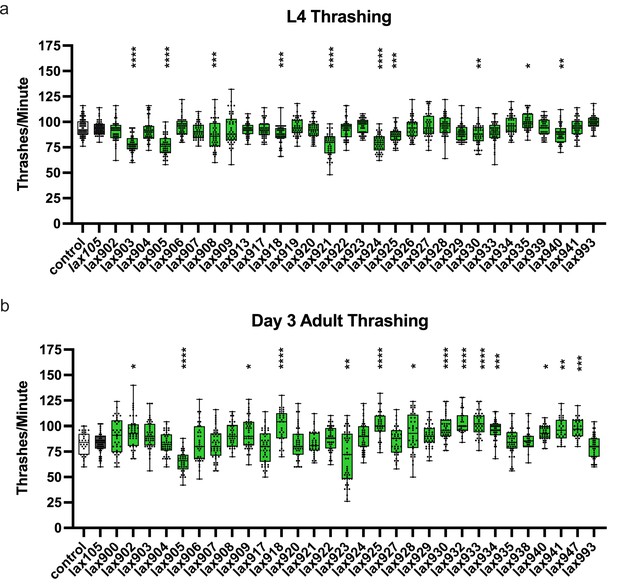
alh-6 mutations accelerate loss of muscle function.
Rate of thrashing for individual animals of the given genotypes at the L4 stage (a) or day 3 of adulthood (b). Brown–Forsythe and Welch test with Dunnett’s T3 multiple comparisons test, with individual variances computed for each comparison. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Tables
Top SNPs associated with specific phenotypes.
| Phenotype | SNP name | Location | Ref. allele | Minor allele | Freq.* | Scan N† | Effect size‡ | p value |
|---|---|---|---|---|---|---|---|---|
| Grip strength decline | rs28665699 | 19200185 | A | A | 0.014 | 5228 | −0.045 | 9.1E−04 |
| Gait speed decline | rs77608580 | 19196968 | A | G | 0.017 | 3319 | 0.052 | 2.5E−03 |
-
*
Freq = frequency of minor allele as reported by 1000 genomes.
-
†
N = sample size of scan for the phenotype and SNP.
-
‡
Effect sizes provided are standardized regression coefficients.
SNPs remaining after filtering for minor allele frequency and pruning based on linkage disequilibrium.
| SNP | Location | Reference allele | Minor allele frequency |
|---|---|---|---|
| rs28652778 | 19194995 | A | 0.20 |
| rs28405179 | 19195143 | A | 0.03 |
| rs111289603 | 19195492 | G | 0.03 |
| kgp2515954 | 19195951 | A | 0.02 |
| rs77608580 | 19196968 | A | 0.04 |
| rs9699485 | 19197237 | G | 0.02 |
| rs3935824 | 19197849 | G | 0.18 |
| rs28665699 | 19200185 | A | 0.03 |
| rs28493067 | 19203333 | A | 0.35 |
| rs6426814 | 19204173 | A | 0.19 |
| rs35285457 | 19205258 | A | 0.14 |
| rs7365978 | 19206020 | A | 0.21 |
| rs28508407 | 19210018 | A | 0.27 |
| rs113232075 | 19211163 | G | 0.02 |
| rs9426718 | 19213022 | A | 0.02 |
| rs4911985 | 19215440 | G | 0.22 |
| rs28582076 | 19217295 | G | 0.02 |
| rs11484743 | 19219987 | C | 0.02 |
| rs17492518 | 19221621 | A | 0.04 |
| rs4912044 | 19230263 | A | 0.18 |
| rs79251057 | 19231130 | A | 0.04 |
Replication across ethnic subsamples in the HRS.
| Phenotype | SNP name | Location | Minor allele | European ancestry(N)* | African ancestry(N)* | Hispanicancestry(N)* | Fixed effect p value† | Random effectp value‡ | Fixed effect†:OR or beta | Random effect‡:OR or beta | Q§ | I¶ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grip strength decline | rs28665699 | 19200185 | A | 5228 | – | 409 | 0.00150 | 0.00150 | −0.0418 | −0.0418 | 0.3341 | 0.00 |
| Gait speed decline | rs77608580 | 19196968 | A | 3319 | 381 | 237 | 0.00775 | 0.72900 | 0.0424 | 0.0146 | 0.0577 | 64.95 |
-
*
N: sample size by group included in the meta-analysis.
-
†
Fixed effect: p value and effect size.
-
‡
Random effect: p value and effect size.
-
§
Cochrane’s Q statistic: indicator of variance across sample effect sizes.
-
¶
I: heterogeneity index to quantify dispersion across samples.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (C. elegans) | N2 | Caenorhabditis Genetics Center (CGG) | Laboratory reference strain (wild type) | |
| Strain, strain background (C. elegans) | SPC321 | PMID:24440036 | Genotype: alh-6(lax105) | |
| Strain, strain background (C. elegans) | CL2166 | Caenorhabditis Genetics Center (CGG) | Genotype: gst4-p::gfp | |
| Strain, strain background (C. elegans) | SPC223 | PMID:24440036 | Genotype: alh-6(lax105);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC542 | This paper | Genotype: alh-6(lax917);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC531 | This paper | Genotype: alh-6(lax906);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC528 | This paper | Genotype: alh-6(lax903);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC552 | This paper | Genotype: alh-6(lax927);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC561 | This paper | Genotype: alh-6(lax937);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC564 | This paper | Genotype: alh-6(lax940);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC549 | This paper | Genotype: alh-6(lax924);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC566 | This paper | Genotype: alh-6(lax945);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC540 | This paper | Genotype: alh-6(lax915);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC562 | This paper | Genotype: alh-6(lax938);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC563 | This paper | Genotype: alh-6(lax939);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC546 | This paper | Genotype: alh-6(lax921);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC527 | This paper | Genotype: alh-6(lax902);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC529 | This paper | Genotype: alh-6(lax904);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC536 | This paper | Genotype: alh-6(lax911);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC534 | This paper | Genotype: alh-6(lax909);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC559 | This paper | Genotype: alh-6(lax935);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC532 | This paper | Genotype: alh-6(lax907);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC569 | This paper | Genotype: alh-6(lax993);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC544 | This paper | Genotype: alh-6(lax919);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC562 | This paper | Genotype: alh-6(lax938);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC551 | This paper | Genotype: alh-6(lax926);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC530 | This paper | Genotype: alh-6(lax905);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC533 | This paper | Genotype: alh-6(lax908);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC548 | This paper | Genotype: alh-6(lax923);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC550 | This paper | Genotype: alh-6(lax925);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC565 | This paper | Genotype: alh-6(lax941);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC538 | This paper | Genotype: alh-6(lax913);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC543 | This paper | Genotype: alh-6(lax918);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC541 | This paper | Genotype: alh-6(lax916);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC545 | This paper | Genotype: alh-6(lax920);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC554 | This paper | Genotype: alh-6(lax929);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC558 | This paper | Genotype: alh-6(lax934);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC526 | This paper | Genotype: alh-6(lax901);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC568 | This paper | Genotype: alh-6(lax992);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC535 | This paper | Genotype: alh-6(lax910);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC553 | This paper | Genotype: alh-6(lax928);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC539 | This paper | Genotype: alh-6(lax914);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC556 | This paper | Genotype: alh-6(lax932);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC557 | This paper | Genotype: alh-6(lax933);gst-4p::gfp | |
| Strain, strain background (C. elegans) | SPC567 | This paper | Genotype: alh-6(lax947);gst-4p::gfp | |
| Sequence-based reagent | pMiniT 2.0 vector and cloning kit | New England Biolabs | #E1202S | |
| Software, algorithm | MBF Bioscience Wormlab | https://www.mbfbioscience.com/wormlab | ||
| Other | US Health and Retirement Study (HRS) | https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000428.v2.p2 | National Center for Biotechnology Information Genotypes and Phenotypes Database dbGaP Study Accession: phs000428.v2.p2 | |
| Strain, strain background (Escherichia coli) | OP50-1 | Caenorhabditis Genetics Center (CGG) | RRID:WB-STRAIN:WBStrain00041971 | Standard E. coli B diet Streptomycin resistant |
| Software, algorithm | GraphPad Prism | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 | Version 6 |
| Software, algorithm | ImageJ | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Software, algorithm | Phyre2 | http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index | ||
| Software, algorithm | MisSense3D | http://missense3d.bc.ic.ac.uk/missense3d/ |
| phenotype | SNPname | location | minor allele | European AncestryN | African AncestryN | HispanicN | Fixed EffectP-value | Random EffectP-value | Fixed Effect:OR or β | Random Effect:OR or β | Q | I |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Walking across a room | Rs 111289603 | 19195492 | G | 9907 | -- | 1067 | 0.00050 | 0.00050 | 1.4664 | 1.4664 | 0.5825 | 0.00 |
| Grip strength decline | Rs 28665699 | 19200185 | A | 5228 | -- | 409 | 0.00150 | 0.00150 | -0.0418 | -0.0418 | 0.3341 | 0.00 |
| IADL decline (3 tasks) | rs111289603 | 19195492 | G | 9041 | -- | 935 | 0.00316 | 0.07460 | 0.0644 | 0.0642 | 0.0984 | 63.39 |
| Gait speed decline | rs77608580 | 19196968 | A | 3319 | 381 | 237 | 0.00775 | 0.72900 | 0.0424 | 0.0146 | 0.0577 | 64.95 |
| SNP name | 0 | 1 | 2 |
|---|---|---|---|
| Rs77608580 | GG | AG | AA |
| Rs28665699 | GG | AG | AA |
| Rs111289603 | AA | AG | GG |





