Macrophage innate training induced by IL-4 and IL-13 activation enhances OXPHOS driven anti-mycobacterial responses
Figures
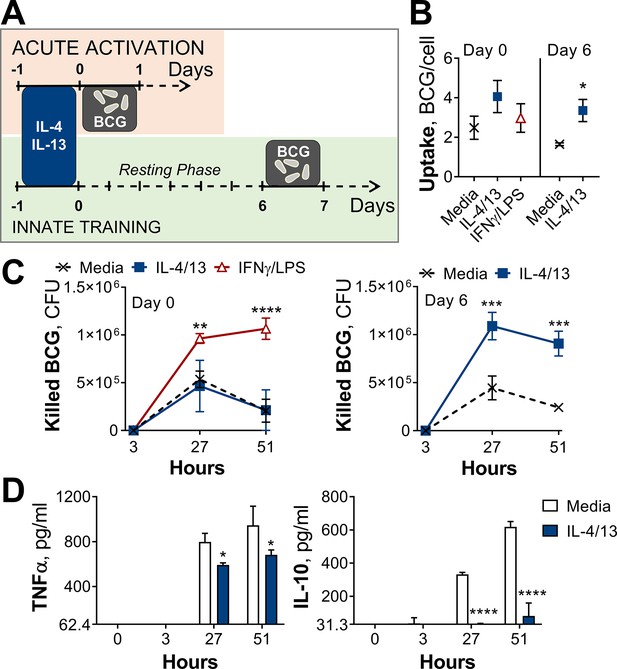
Training with IL-4 and IL-13 enhances BMDM mycobacterial killing capacity.
(A) Schematic of protocol for BCG infection following acute activation (Day 0) or training (Day 6) with IL-4 and IL-13. (B–C) BCG Denmark uptake (B) and killing after h as indicated (C) measured by difference in CFU after 3 hr per 0.5×106 BMDMs on Day 0 or Day 6. BMDMs were incubated with media, IL-4 with IL-13 or IFNγ with LPS for 24 hr on Day –1. (D) Secretion of indicated cytokines from BMDMs treated as in (C) standardized to 0.5×106 BMDMs. (B–D) Representative results (n=2 out of ≥3) showing mean ± SEM (B–C) or ± SD (D) and analyzed by student’s t-test (B) or multiple t-tests, with Holm-Sidak correction (C–D) compared with media control. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
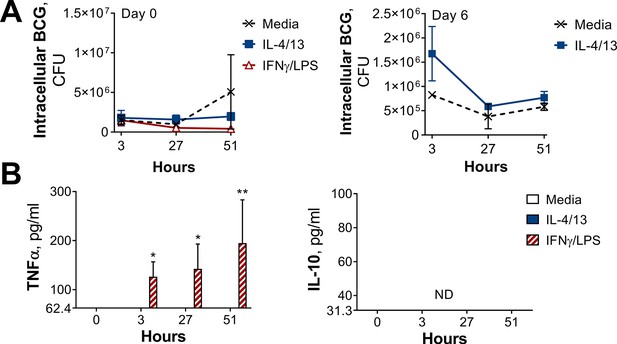
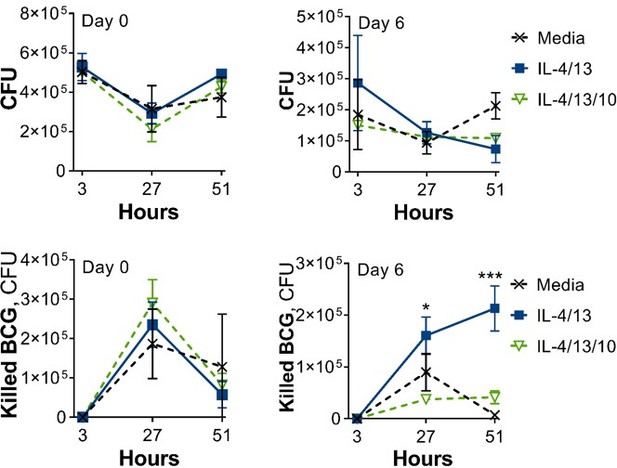
CFU counts and cytokine secretion from BCG killing experiments.
(A–B) BCG Denmark CFU counts (A) and cytokine secretion (B) on Day 0 or Day 6 after h following BMDM BCG infection. BMDMs were previously incubated with media control, IL-4 with IL-13 or IFNγ with LPS for 24 h on Day –1. Representative results (n=2 out of ≥3), standardized to 0.5×106 cells, shown as mean ± SEM (A–B) and analyzed by multiple t-test, with Holm-Sidak correction, compared with media control (B). * p≤0.05, ** p≤0.01.
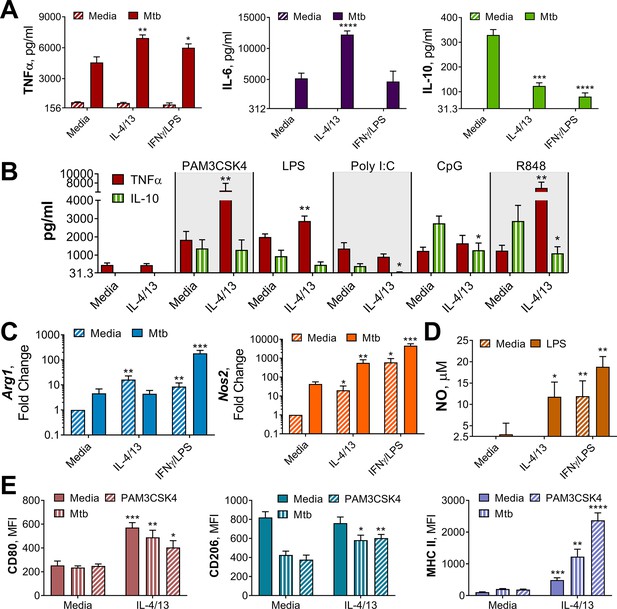
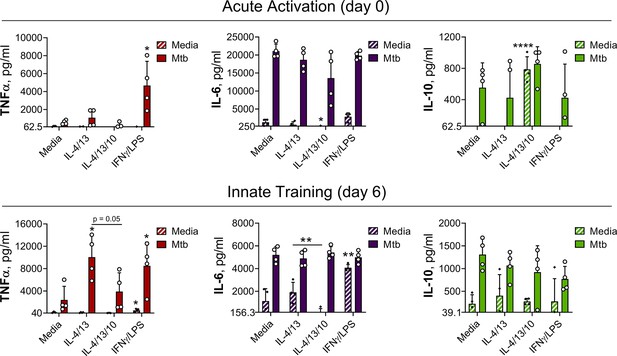
Innate training of BMDMs with IL-4 and IL-13 enhances pro-inflammatory responses.
(A–B) Cytokine secretion following BMDM 24 h incubation with irradiated M. tuberculosis (Mtb) (A) TLR agonists (B) or media on Day 6 as indicated. BMDMs were previously incubated with media, IL-4 with IL-13 or IFNγ with LPS on Day –1 for 24 hr (n=3). (C) qPCR of indicated mRNA in BMDMs treated as in (A) standardized to BMDMs incubated with media Day –1 and Day 6 (n=4). (D) Nitric oxide (NO) secretion from BMDMs treated as in (B) (n=3). (E) Expression of CD80, CD206 and MHC II (gating strategy Figure S2A) on BMDMs treated as in (A–B) (n=3). Mean ± SD are shown and analyzed by student’s t-test, compared with media control. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
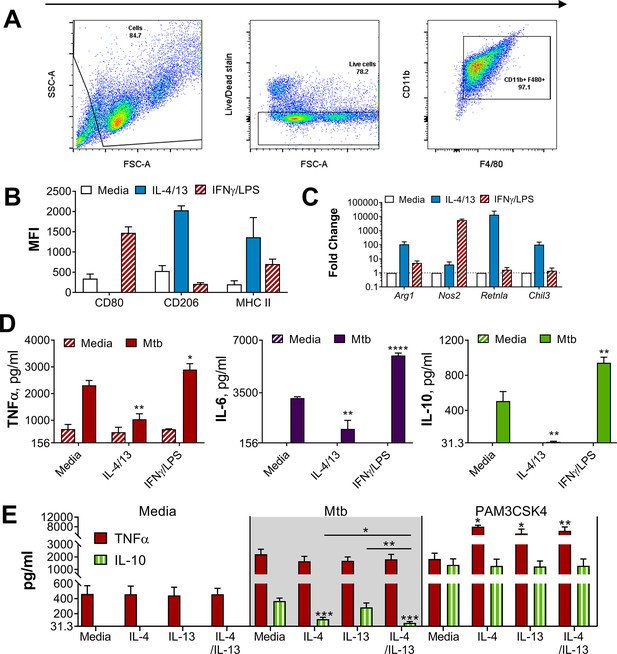
BMDMs either classically activated with IFNγ and LPS or alternatively activated with IL-4 and IL-13.
(A) Gating strategy for flow cytometry analysis, gating on cells, live cells and CD11b+F480+BMDMs. (B) Surface marker expression of CD80, CD206 and MHC II on BMDMs incubated with media as a control, IL-4 with IL-13 or IFNγ with LPS for 24 hr (n=3). (C) qPCR of indicated mRNA in BMDMs treated as in (B), standardized to media control (n=4). (D–E) Cytokine secretion from BMDMs incubated with media, IL-4, IL-13, IL-4 with IL-13 or IFNγ with LPS as indicated for 24 hr on Day –1 and stimulated with media, irradiated M. tuberculosis H37Rv (Mtb) or PAM3CSK4 for 24 hr on Day 0 (D) or 6 (E) (n=3). Mean ± SD are shown and analyzed by student’s t-test, compared to media control or as indicated. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
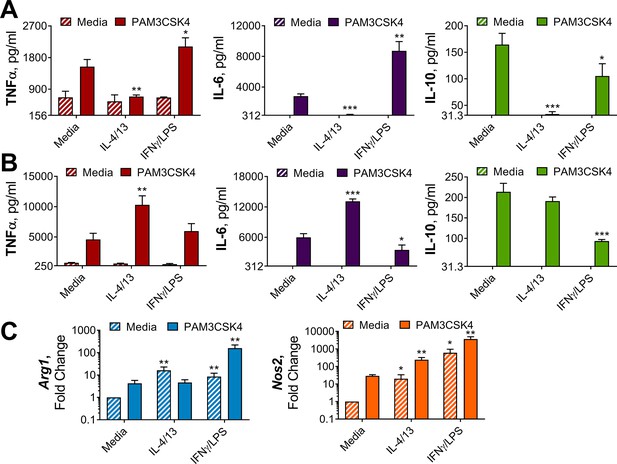
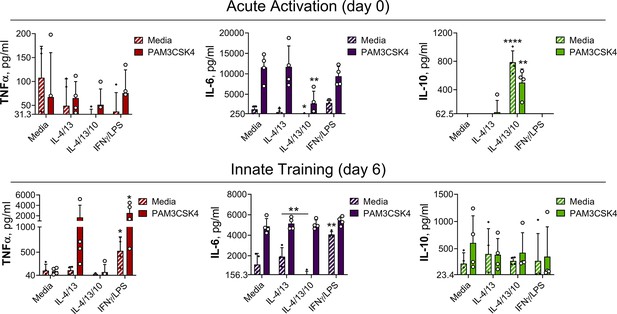
IL-4 and IL-13 innate training enhances inflammatory and bactericidal responses following TLR 1/2 stimulation.
(A–B) Cytokine secretion from BMDMs following incubation with media control, IL-4 with IL-13 or IFNγ with LPS for 24 hr on Day –1 and incubated with media or PAM3CSK4 on Day 0 (A) or Day 6 (B) for 24 hr (n=3). (C) qPCR of indicated mRNA in BMDMs treated as in (B), standardized to BMDMs incubated with media Day –1 and Day 6 (n=4). Mean ± SD are shown and analyzed by student’s t-test, compared with media control. * p≤0.05, ** p≤0.01, *** p≤0.001.
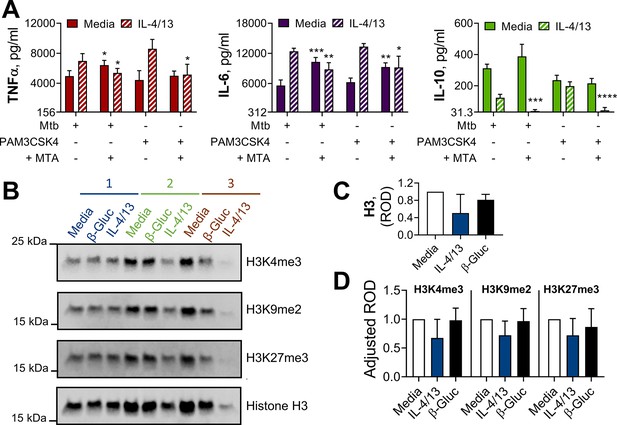
Methylation contributes to IL-4 and IL-13 innate training.
(A) Cytokine secretion following BMDM incubation with media control or IL-4 with IL-13 for 24 hr on Day –1, with or without 1 hr pre-incubation with methylation inhibitor MTA, and incubation with irradiated M. tuberculosis H37Rv (Mtb) or PAM3CSK4 for 24 hr on Day 6 (n=4). (B–D) Protein extract (B) and densitometry values (C–D) from western blot analysis of histone methylation in BMDMs incubated with media control, yeast β-glucan (β-Gluc) or IL-4 with IL-13 on Day –1 and lysed on Day 6 (n=3). (B) Each replicate is labelled 1–3. (A, C–D) Mean ± SD are shown and analyzed by student’s t-test, comparing with or without MTA (A) or with media control (C–D). * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
-
Figure 2—figure supplement 3—source code 1
Unedited and labelled western blots.
Unedited western blots for histone H3, H3K4me3, H3K27me3 and H3K9me2, accompanying ponceau stains and figure where size and wells are labelled.
- https://cdn.elifesciences.org/articles/74690/elife-74690-fig2-figsupp3-code1-v2.zip
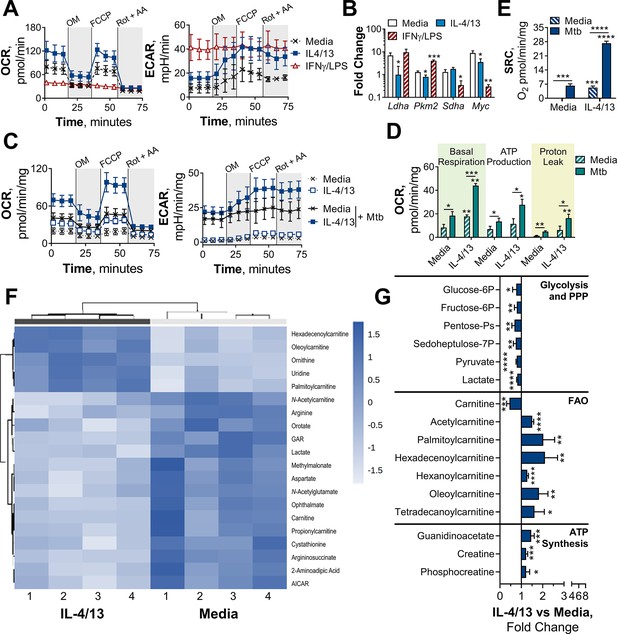
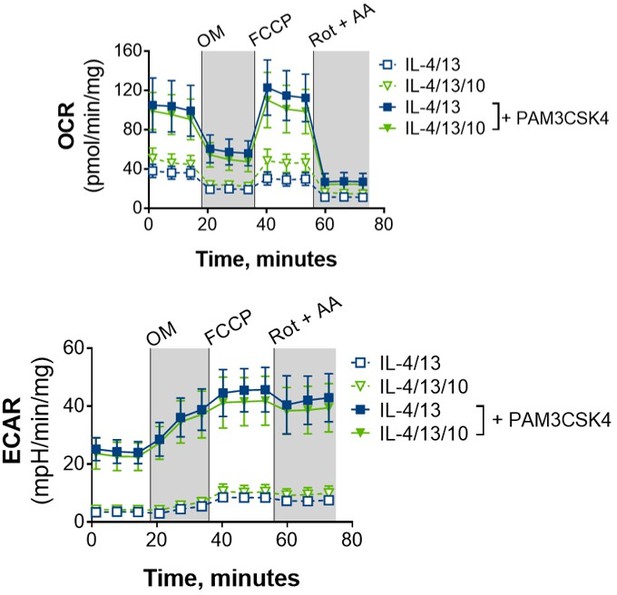
BMDMs trained with IL-4 and IL-13 retain M2-typical metabolism upon mycobacterial challenge.
(A) Extracellular flux analysis of BMDMs, following 24 hr incubation with IL-4 and IL-13, IFNγ and LPS or media (n=3). Mitochondrial stress test inhibitors: oligomycin (OM; blocks ATP synthase), FCCP (uncouples the electron transport chain [ETC] from ATP synthesis), rotenone (Rot; inhibits complex I of the ETC) and antimycin-A (AA; inhibits complex III of the ETC). (B) qPCR of indicated mRNA in BMDMs following incubation with media, IL-4 with IL-13 or IFNγ with LPS for 24 hr on Day –1 and stimulated with irradiated M. tuberculosis (Mtb) for 6 hr (Pkm2) or 24 hr (Ldha, Sdha and Myc), standardized to BMDMs given media on Day –1 and Day 6 (n=3). (C–E) Extracellular flux analysis (C) basal respiration, ATP production, proton leak (D) and spare respiratory capacity (SRC) (E) of BMDMs treated as in (B) with incubation with media or IL-4 with IL-13 on Day –1 and stimulation with media or Mtb for 24 hr on Day 6 (n=3). (F–G) Metabolites from BMDMs treated as in (B) incubated with media or IL-4 with IL-13 on Day –1 and stimulation with Mtb for 24 hr on Day 6 (n=4). MetaboAnalyst generated heatmap representing hierarchical clustering of the top 20 most up/down regulated metabolites (F). Fold change compared with media control (=1) (G). Mean ± SD are shown and analyzed by student’s t-test. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001. Abbreviations: AICAR, aminoimidazole carboxamide ribonucleotide; ECAR, extracellular acidification rate; FAO, fatty acid oxidation; GAR, glycinamide ribonucleotide; OCR, oxygen consumption rate; P, phosphate; PPP, pentose phosphate pathway.
-
Figure 3—source data 1
MetaboAnalyst R-history.
Statistical analysis carried out by MetaboAnalyst. Raw data can be found in Source data 1.
- https://cdn.elifesciences.org/articles/74690/elife-74690-fig3-data1-v2.txt
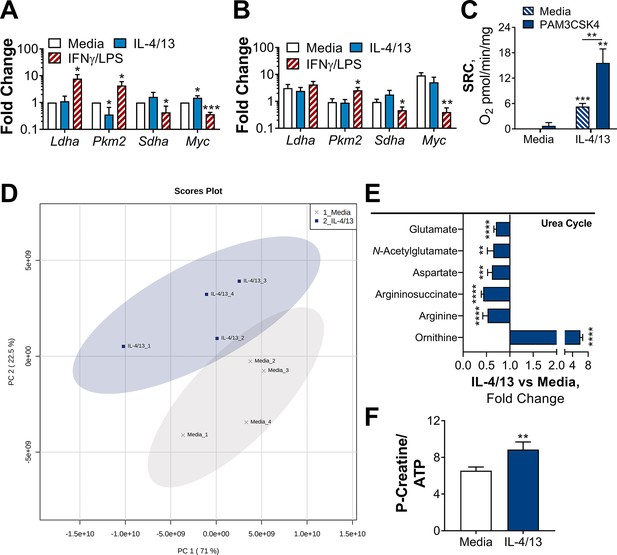
Challenged BMDMs previously trained with IL-4 and IL-13 retain M2-typical metabolism.
(A–B) qPCR of indicated mRNA in BMDMs following incubation with media control, IL-4 with IL-13 or IFNγ with LPS on for 24 h Day –1 and stimulated with media (A) or PAM3CSK4 (B) on Day 6 for 6 hr (Pkm2) and 24 h (Ldha, Sdha and Myc). Fold change was standardized to BMDMs incubated with media Day –1 and Day 6 (n=3). (C) Spare respiratory capacity (SRC) of BMDMs treated as in (A–B), with incubation of media or IL-4 with IL-13 on Day –1 (n=3). (D–F) Metabolites isolated from BMDMs following incubation with either media control (untrained) or IL-4 with IL-13 (trained) for 24 hr on Day –1 and stimulation with Mtb for 24 hr on Day 6 (n=4). Metaboanalyst principal component analysis (PCA) (D) fold change of metabolites from trained BMDMs compared with media control (=1) (E) and calculated ratio of phosphocreatine (P-creatine) to ATP (F) are shown. Mean ± SD are shown and analyzed by student t-test, compared with media control. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
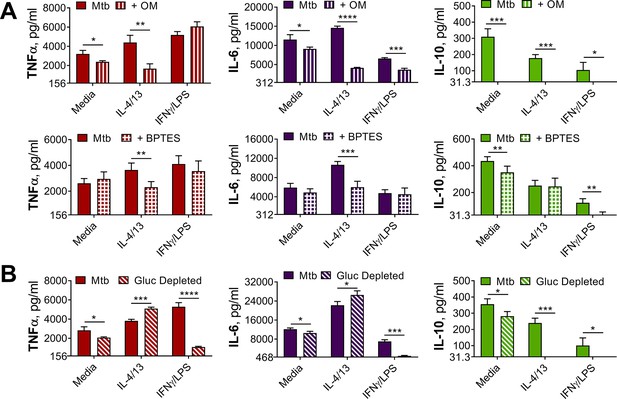
OXPHOS drives inflammatory cytokine responses in IL-4/13 trained BMDMs.
(A) Cytokine secretion from BMDMs following incubation with media, IL-4 with IL-13 or IFNγ with LPS for 24 hr on Day –1 and incubation with irradiated M. tuberculosis (Mtb) on Day 6 for 24 hr, with or without pre-incubation (Day 6) of oligomycin (OM) or BPTES (n=4). (B) Cytokine secretion from BMDMs treated as in (A) with or without glucose depleted conditions between Day –1–7 (n=3). Mean ± SD are shown and analyzed by student’s t-test as indicated. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
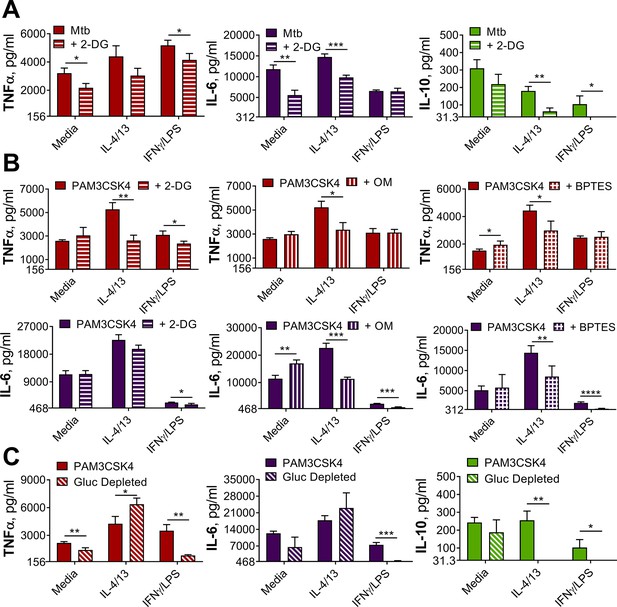
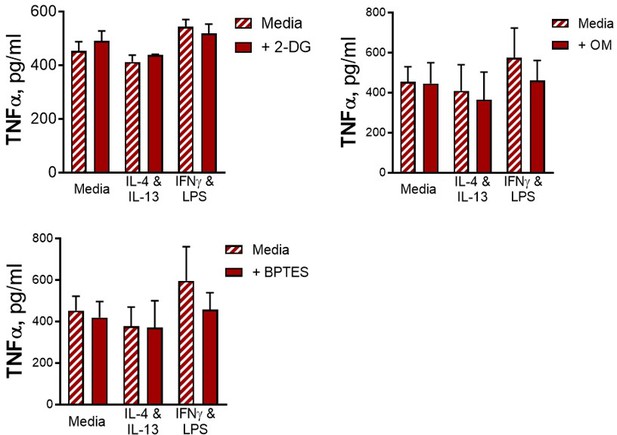
Innate training with IL-4 and IL-13 enhancement of pro-inflammatory responses is not dependent upon glucose metabolism.
(A–B) Cytokine secretion from BMDMs following incubation with media control, IL-4 with IL-13 or IFNγ with LPS for 24 hr on Day –1 and incubation with irradiated M. tuberculosis H37Rv (Mtb) (A) or PAM3CSK4 (B) for 24 hr on Day 6, with or without pre-incubation of metabolic inhibitors oligomycin (OM) (n=3), 2-deoxy glucose (2-DG) (n=3) or BPTES (n=4). (C) Cytokine secretion from BMDMs following incubation with media control, IL-4 with IL-13 or IFNγ with LPS for 24 hr on Day –1 and incubation with media or PAM3CSK4 for 24 hr on Day 6, with or without glucose depleted conditions between Day –1–7 (n=3). Mean ± SD are shown and analyzed by student’s t-test as indicated. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
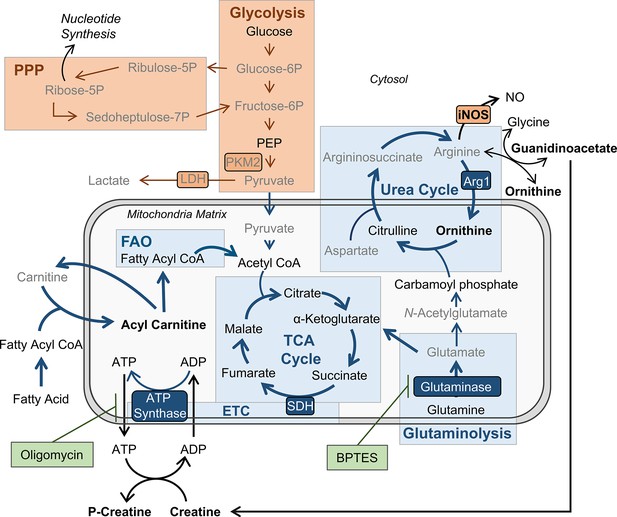
BMDMs trained with IL-4 and IL-13 retain M2-typical metabolism: schematic summary of results.
Pathways have been simplified. Key: metabolic pathways strongly upregulated by M1/M2 macrophage activation are highlighted by red/blue colored boxes, respectively. Inhibitors are indicated by green boxes. Arrow width represents which pathways are implicated (thicker) or not (narrower) in trained M(4/13) following stimulation with irradiated M. tuberculosis (Mtb). Metabolites (measured by LC-MS) or enzymes (measured by qPCR) written in bold or in grey text are enhanced or reduced respectively compared with untrained macrophages. Trained M(4/13) do not employ classical activated macrophage metabolism – aerobic glycolysis and pentose phosphate pathway (PPP) – and instead employ alternative activated macrophage metabolism, characterized by production of ATP through the tricarboxylic acid (TCA) cycle, coupled with the electron transport chain (ETC) via oxidative phosphorylation (OXPHOS), as well as enhanced use of the urea cycle. Glutaminolysis, FAO and ATP synthesis regulation are implicated. This is demonstrated by inhibitor experiments and by changed expression of metabolites. Abbreviations: LDH, lactate dehydrogenase; NO, nitric oxide; P, phosphate; PEP, Phosphoenolpyruvic acid; PKM2, pyruvate kinase M2; SDH, succinate dehydrogenase.
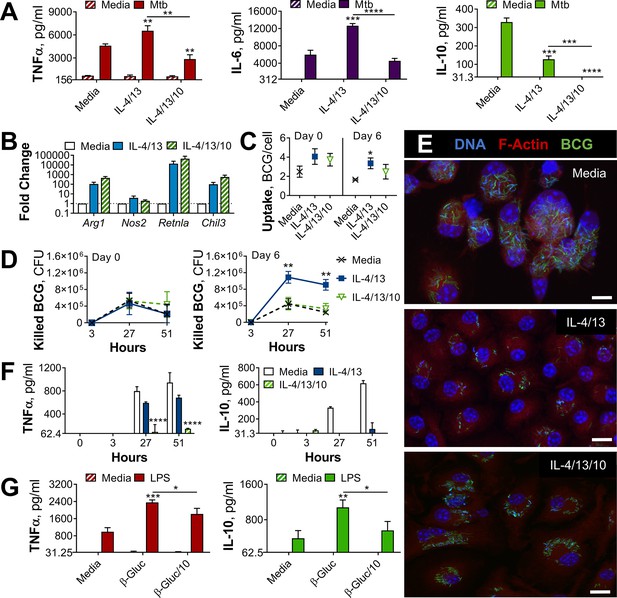
IL-10 inhibits bactericidal and pro-inflammatory training induced by IL-4 and IL-13 and alters yeast β-glucan training.
(A) Cytokine secretion following BMDM incubation with media or IL-4 and IL-13, with or without IL-10, for 24 hr on Day –1 and incubated for 24 hr with media or irradiated M. tuberculosis (Mtb) on Day 6. Mean ± SD (n=3) are shown and analyzed by student’s t-test, compared to media control or as indicated. (B) qPCR of indicated mRNA in BMDMs following 24 hr incubation with media or IL-4 and IL-13, with or without IL-10, standardized to media control. Mean ± SD (n=4) are shown. (C–F) BCG Denmark uptake (C) killing after h as indicated (D) as measured by difference in CFU after 3 hr per 0.5×106 BMDMs on Day 0 or Day 6. Representative images of Hoechst- (blue), modified auramine-O- (green) and phalloidin (red)-stained BMDMs were taken on Day 6, 27 hr after BCG incubation, where scale bars are, from top to bottom, 8- 10- and 10 μm (E). Cytokine secretion on Day 6, at h indicated (F). BMDMs were previously incubated with media or IL-4 and IL-13, with or without IL-10, for 24 hr on Day –1. (C–D, F) representative results (n=2 of ≥3) are shown as mean ± SD (C, F) or SEM (D) and analyzed by student’s t-test compared with media (C) or multiple t-tests, with Holm-Sidak correction, comparing with or without IL-10 (D, F). (G) Cytokine secretion following BMDM incubation with media or β-glucan (β-Gluc), with or without IL-10, for 24 hr on Day –1 and incubated for 6 hr (TNFα) or 24 hr (IL-10) with media or LPS on Day 6. (A, G) Mean ± SD (n=3) are shown and analyzed by student’s t-test. * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
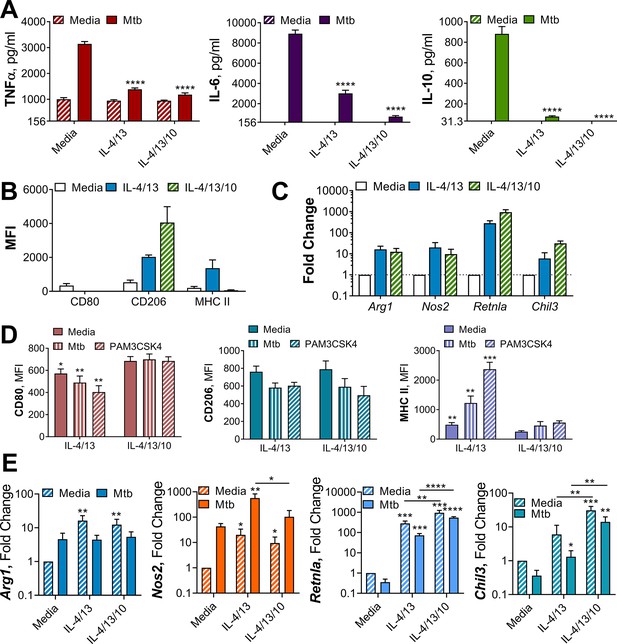
Alternative macrophage activation with or without IL-10.
(A) Cytokine secretion from BMDMs incubated with media as a control or IL-4 and IL-13, with or without IL-10 for 24 hr on Day-1, then incubated with media or Mtb for 24 hr on Day 0 (n=4). (B) Surface marker expression of CD80, CD206 and MHC II on BMDMs incubated with media as a control or IL-4 and IL-13, with or without IL-10 for 24 hr (n=3) (gating strategy shown Figure 2—figure supplement 1A). (C) qPCR of indicated mRNA in BMDMs treated as in (B) standardized to media control (n=4). (D) Surface marker expression of CD80, CD206, and MHC II on BMDMs following incubation with IL-4 and IL-13, with or without IL-10, for 24 hr on Day –1 and incubated with media, irradiated M. tuberculosis H37Rv (Mtb) or PAM3CSK4 for 24 hr on Day 0 (n=3). (E) qPCR of indicated mRNA in BMDMs treated as in (A) standardized to BMDM incubated with media on Day –1 and Day 0 (n=4). Mean ± SD are shown (A–E) and analyzed by student’s t-test, compared with media control (A, E) comparing with or without IL-10 (D, E). * p≤0.05, ** p≤0.01, *** p≤0.001, **** p≤0.0001.
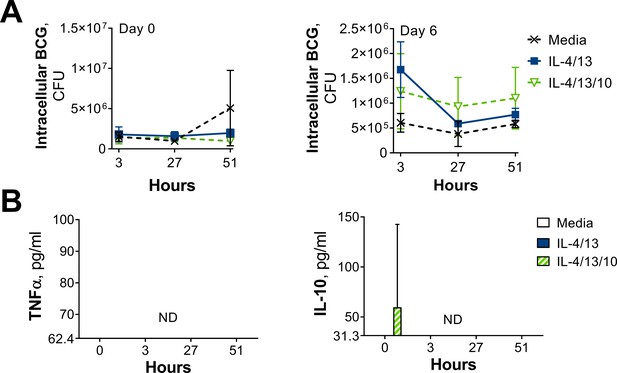
CFU counts and cytokine secretion from BCG killing experiments.
(A–B) BCG Denmark CFU counts (A) and cytokine secretion (B) on Day 0 or Day 6 after h following BMDM BCG infection. BMDMs were previously incubated with media control or IL-4 and IL-13, with or without IL-10, for 24 hr on Day –1. Representative results (n=2 out of ≥3), standardized to 0.5×106 cells, shown as mean ± SEM (C–D) and analyzed by multiple t-test, with Holm-Sidak correction, compared with media control (D) * p≤0.05, ** p≤0.01.
IL-4 and IL-13 innate training enhances MoDM secreted TNFα following Mtb stimulation.
Cytokine secretion following MoDM 24 hr incubation with irradiated M. tuberculosis (Mtb) or media on Day 0 or on Day 6 as indicated. MoDM were previously incubated with media, IL-4 with IL-13 – with or without the addition of IL-10 – or IFNγ with LPS on Day -1 for 24 hr (n = 4, each rep shown by a symbol). Mean ± SD are shown and analyzed by student’s t-test, compared with media control or comparing IL-4/IL-13 training with or without IL-10, as indicated. * p ≤ 0.05, ** p ≤ 0.01, **** p ≤ 0.0001.
IL-4 and IL-13 innate training alters MoDM responses following PAM3CSK4 stimulation.
Cytokine secretion following MoDM 24 hr incubation with PAM3CSK4 or media on Day 0 or on Day 6 as indicated. MoDM were previously incubated with media, IL-4 with IL-13 – with or without the addition of IL-10 – or IFNγ with LPS on Day –1 for 24 hr (n=4, each rep shown by a symbol). Mean ± SD are shown and analyzed by student’s t-test, compared with media control or comparing IL-4/IL-13 training with or without IL-10, as indicated. * p≤0.05, ** p≤0.01, **** p≤0.0001.
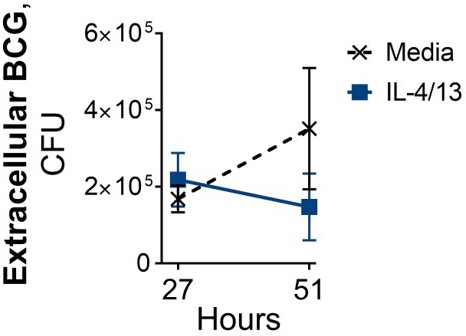
Following BCG infection on Day 6, where BMDMs were incubated with media control or IL-4/13 on Day -1 for 24 hours.
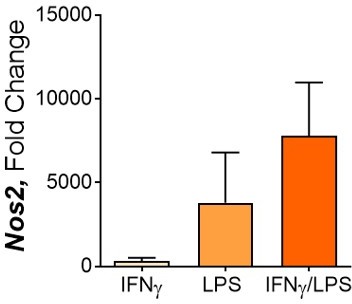
100 ng/ml IFNγ, 50 ng/ml LPS, 50 ng/ml each.
Nos2 expression after 24 hours.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli, serotype R515) | Lipopolysaccharide, LPS | Enzo | Cat# ALX-581–007 L002 | |
| Strain, strain background (H. sapiens) | Primary cell isolation | Buffy packs from Irish Blood Transfusion Service | N/A | |
| Strain, strain background (M. musculus, C57BL/6JOlaHsd) | Primary cell isolation | In-house colonies | C57BL/6JOlaHsd | Both sexes employed |
| Strain, strain background (Mycobacterium bovis) | Bacille Calmette-Guérin (BCG) Denmark 1331 | Gift to Prof. Gordon from Prof. Behr, McGill University, Canada | N/A | |
| Strain, strain background (Mycobacterium tuberculosis, strain H37Rv) | Irradiated whole cells of M. tuberculosis | BEI resources | NR-49098 | Non-viable bacteria |
| Cell line (M. musculus) | L929 | gift of Prof. Muñoz-Wolf, Trinity College, Dublin gift of Prof. Sheedy, Trinity College, Dublin | N/A | Cell lines maintained in E. Lavelle and F. Sheedy labs. |
| Antibody | Anti-CD11b-APC-eFluor 780 (Rat monoclonal) | Thermo Fisher Scientific | Cat# 47-0112-82, Clone M1/70 | FACS (0.1 μl per test) |
| Antibody | Anti-CD206-PE (Rat monoclonal) | BioLegend | Cat# 141706, Clone C068C2 | FACS (0.4 μl per test) |
| Antibody | Anti-CD80-FITC (Hamster monoclonal) | BD Biosciences | Cat# 561954, Clone 16–10 A1 | FACS (0.15 μl per test) |
| Antibody | Anti-F4/80-PerCP-Cy5.5 (Rat monoclonal) | Thermo Fisher Scientific | Cat# 45-4801-82, Clone BM8 | FACS (0.25 μl per test) |
| Antibody | Anti-Histone H3 (Mouse monoclonal) | Active Motif | Cat# 39763 | WB (1/3,000) |
| Antibody | Anti-H3K4me3 (Rabbit polyclonal) | Abcam | Cat# ab8580 | WB (1/1000) |
| Antibody | Anti- H3K9me2 (Mouse monoclonal) | Abcam | Cat# ab1220 | WB (1/1000) |
| Antibody | Anti-H3K27me3 (Mouse monoclonal) | Active Motif | Cat# 61017 | WB (1/1000) |
| Antibody | Anti-MHC class II-eFlour 450 (Rat monoclonal) | Thermo Fisher Scientific | Cat# 48-5321-82, Clone M5/114.15.2 | FACS (0.2 μl per test) |
| Antibody | Anti-mouse IgG (Goat monoclonal) | LI-COR | Cat# 925–32210 | WB (1/5000 dilution) |
| Antibody | Anti-Rabbit IgG (Goat monoclonal) | LI-COR | Cat# 926–32211 | WB (1/2500) |
| Antibody | Fc block: Anti-CD16/CD32 (Rat monoclonal) | BD Biosciences | Cat# 553142, Clone 2.4G2 | FACS (0.5 μl per test) |
| Sequence-based reagent | Actb _F | Primer BLAST | Invitrogen Custom DNA Oligos | GCTTCTTTGCAGCTCCTTCGT |
| Sequence-based reagent | Actb _R | Primer BLAST | Invitrogen Custom DNA Oligos | CGTCATCCATGGCGAACTG |
| Sequence-based reagent | Arg1 _F | Primer BLAST | Invitrogen Custom DNA Oligos | TACAAGACAGGGCTCCTTTCAG |
| Sequence-based reagent | Arg1 _R | Primer BLAST | Invitrogen Custom DNA Oligos | TGAGTTCCGAAGCAAGCCAA |
| Sequence-based reagent | Chitl3 _F | Primer BLAST | Invitrogen Custom DNA Oligos | AAGCTCTCCAGAAGCAATCC |
| Sequence-based reagent | Chitl3 _R | Primer BLAST | Invitrogen Custom DNA Oligos | AGAAGAATTGCCAGACCTGTGA |
| Sequence-based reagent | Ldha _F | Primer BLAST | Eurofins genomics (MWG) | GAGACTTGGCTGAGAGCATAA |
| Sequence-based reagent | Ldha _R | Primer BLAST | Eurofins genomics (MWG) | GATACATGGGACACTGAGGAA |
| Sequence-based reagent | Myc _F | Primer BLAST | Invitrogen Custom DNA Oligos | CAGCGACTCTGAAGAAGAGCA |
| Sequence-based reagent | Myc _R | Primer BLAST | Invitrogen Custom DNA Oligos | GACCTCTTGGCAGGGGTTTG |
| Sequence-based reagent | Nos2 _F | Primer BLAST | Invitrogen Custom DNA Oligos | TCCTGGACATTACGACCCCT |
| Sequence-based reagent | Nos2 _R | Primer BLAST | Invitrogen Custom DNA Oligos | CTCTGAGGGCTGACACAAGG |
| Sequence-based reagent | Pkm2 _F | Primer BLAST | Eurofins genomics (MWG) | TGTCTGGAGAAACAGCCAAG |
| Sequence-based reagent | Pkm2_R | Primer BLAST | Eurofins genomics (MWG) | CGAATAGCTGCAAGTGGTAGA |
| Sequence-based reagent | Retnla _F | Primer BLAST | Invitrogen Custom DNA Oligos | CAGCTGATGGTCCCAGTGAAT |
| Sequence-based reagent | Retnla _R | Primer BLAST | Invitrogen Custom DNA Oligos | AGTGGAGGGATAGTTAGCTGG |
| Sequence-based reagent | Sdha _F | Primer BLAST | Eurofins genomics (MWG) | GGAACACTCCAAAAACAGACC |
| Sequence-based reagent | Sdha _R | Primer BLAST | Eurofins genomics (MWG) | CCACCACTGGGTATTGAGTAGAA |
| Sequence-based reagent | Tbp _F | Primer BLAST | Invitrogen Custom DNA Oligos | CAGGAGCCAAGAGTGAAGAACA |
| Sequence-based reagent | Tbp _R | Primer BLAST | Invitrogen Custom DNA Oligos | AAGAACTTAGCTGGGAAGCCC |
| Peptide, recombinant protein | Heat-Shocked Bovine Serum Albumin (BSA) | Thermo Fisher Scientific | Cat# 12881630 | |
| Peptide, recombinant protein | M-MLV reverse transcriptase | Promega | Cat# M3683 | |
| Peptide, recombinant protein | Recombinant human IFNγ | Peprotech | Cat# 300–02 | |
| Peptide, recombinant protein | Recombinant human IL-10 | Peprotech | Cat# 200–10 | |
| Peptide, recombinant protein | Recombinant human IL-13 | Peprotech | Cat# 200–13 | |
| Peptide, recombinant protein | Recombinant human IL-4 | Peprotech | Cat# 200–04 | |
| Peptide, recombinant protein | Recombinant human M-CSF | Prospec Protein Specialists | Cat# CYT-308 | |
| Peptide, recombinant protein | Recombinant murine IFNγ | Peprotech | Cat# 315–05 | |
| Peptide, recombinant protein | Recombinant murine IL-10 | Peprotech | Cat# 210–10 | |
| Peptide, recombinant protein | Recombinant murine IL-13 | Peprotech | Cat# 210–13 | |
| Peptide, recombinant protein | Recombinant murine IL-4 | Peprotech | Cat# 214–14 | |
| Commercial assay or kit | BCA Protein Assay Kit (Pierce) | Thermo Fisher Scientific | Cat# 23225 | |
| Commercial assay or kit | Griess Reagent System kit | Promega | Cat# G2930 | |
| Commercial assay or kit | High Pure RNA Isolation Kit | Roche | Cat# 11828665001 | |
| Commercial assay or kit | Human IL-10 ELISA Kit uncoated | Invitrogen | Cat# 88710688 | |
| Commercial assay or kit | Human IL-6 ELISA Kit uncoated | Invitrogen | Cat# 88706688 | |
| Commercial assay or kit | Human TNFα ELISA Kit uncoated | Invitrogen | Cat# 88734688 | |
| Commercial assay or kit | Mouse IL-10 ELISA MAX | BioLegend | Cat# 431411 | |
| Commercial assay or kit | Mouse IL-6 ELISA MAX | BioLegend | Cat# 431301 | |
| Commercial assay or kit | Mouse TNFα DuoSet ELISA | R&D Systems | Cat# DY410 | |
| Chemical compound, drug | 2-deoxyglucose, 2-DG | Sigma Aldrich | Cat# D8375 | |
| Chemical compound, drug | Antimycin-A | Sigma Aldrich | Cat# A8674 | |
| Chemical compound, drug | BPTES | Sigma Aldrich | Cat# SML0601-5mg | |
| Chemical compound, drug | CpG | InvivoGen | Cat# ODN M362 | |
| Chemical compound, drug | FCCP | Sigma Aldrich | Cat# C2920 | |
| Chemical compound, drug | Glucose | Sigma Aldrich | Cat# G8270 | |
| Chemical compound, drug | L-Glutamine | Gibco | Cat# 25030–024 | |
| Chemical compound, drug | Glycerol | Sigma Aldrich | Cat# G2025 | |
| Chemical compound, drug | MTA | Sigma Aldrich | Cat# D5011-25MG | |
| Chemical compound, drug | Oligomycin | Sigma Aldrich | Cat# 75351 | |
| Chemical compound, drug | PAM3CSK4 | InvivoGen | Cat# tlrl-pms | |
| Chemical compound, drug | Poly I:C | InvivoGen | Cat# tlrl-pic | |
| Chemical compound, drug | Pyruvate | Sigma Aldrich | Cat# P5280 | |
| Chemical compound, drug | Resiquimod/R848 | InvivoGen | Cat# tlrl-r848 | |
| Chemical compound, drug | Rotenone | Sigma Aldrich | Cat# R8875 | |
| Chemical compound, drug | Sodium Chloride | Sigma Aldrich | Cat# S9888 | |
| Chemical compound, drug | Valine-d8 | CK isotopes | Cat# DLM-488 | |
| Chemical compound, drug | WGP Dispersible | InvivoGen | Cat# tlrl-wgp | |
| Software, algorithm | FlowJo 7 | FlowJo LLC, Franklin Lakes, New Jersey | https://www.flowjo.com/solutions/flowjo | |
| Software, algorithm | ImageJ | National Institutes of Health and the Laboratory for Optical and Computational Instrumentation | https://imagej.nih.gov/ij/ | |
| Software, algorithm | MetaboAnalyst 5.0 | Xia Lab @ McGill | https://www.metaboanalyst.ca/ | |
| Software, algorithm | Microsoft Office Excel | Microsoft, Redmond, Washington | https://products.office.com/en-au/excel | |
| Software, algorithm | Prism 8.2 | GraphPad Software, San Diego, California | https://www.graphpad.com/scientific-software/prism/ | |
| Software, algorithm | Tracefinder 5.0 | Thermo Fisher Scientific, Waltham, Massachusetts | https://www.thermofisher.com/ie/en/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/lc-ms-data-acquisition-software/tracefinder-software.html | |
| Other | 4% PFA in PBS | Santa Cruz Biotechnology | Cat# NC0238527 | Fixing buffer for flow cytometry and confocal microscopy |
| Other | Acetonitrile | Thermo Fisher Scientific | Cat# 10001334 | Extraction buffer for Metabolomics |
| Other | DMEM (high glucose) | Sigma Aldrich | Cat# D5671 | Cell culture media (BMDMs) |
| Other | DMEM (no glucose) | Gibco | Cat# 11966025 | Cell culture media (BMDMs) |
| Other | dNTP Mix | Meridian Bioscience | Cat# BIO-39028 | Nucleotides for cDNA synthesis |
| Other | FBS | Biosera | Batch# 015BS551 | Serum for cell culture media |
| Other | Fixable Viability Stain 510 | Invitrogen | Cat# 564406 | Viability stain used for FACS |
| Other | Hoechst 33342 | Thermo Fisher Scientific | Cat# 10150888 | DNA dye used for confocal microscopy |
| Other | KAPA SYBR FAST Rox low qPCR Kit Master Mix | Sigma Aldrich | Cat# KK4622 | Nucleic acid stain for qPCR |
| Other | Lymphoprep | Stemcell Technologies | Cat# 07851 | Density gradient medium for isolating PBMCs |
| Other | Methanol | Thermo Fisher Scientific | Cat# 10284580 | Extraction buffer for Metabolomics |
| Other | Middlebrook 7H11 powder | Sigma Aldrich | Cat# M0428 | For making Mycobacterial culture media (agar plates) |
| Other | Middlebrook 7H9 powder | Sigma Aldrich | Cat# M0178 | For making Mycobacterial culture media |
| Other | Modified Auramine-O stain and quencher | Scientific Device Laboratory | Cat# 345–04 L | Mycobacterial stain for confocal micrsocopy |
| Other | PBS, sterile | Gibco | Cat# 14190094 | Cell wash buffer |
| Other | Penicillin-Streptomycin | Gibco | Cat# 15-070-063 | Antibiotics for cell culture |
| Other | Phalloidin-Alexa Fluor 647 | Invitrogen | Cat# A22287 | Stain actin for confocal microscopy |
| Other | Radio-Immunoprecipitation Assay (RIPA) buffer | Sigma Aldrich | Cat# R0278-50ML | Cell lysis buffer |
| Other | Random Hexamer Primer Mix | Meridian Bioscience | Cat# BIO-38028 | Primers for cDNA synthesis |
| Other | Reverse Transcriptase Buffer | Promega | Cat# A3561 | Buffer for cDNA synthesis |
| Other | RNAseOUT | Invitrogen | Cat# 10777019 | RNAse inhibitor for cDNA synthesis |
| Other | RPMI 1640 Glutamax | Gibco | Cat# 21875034 | Cell culture media (MoDMs) |
| Other | Seahorse Calibration Fluid pH 7.4 | Agilent | Part# 100840–000 | Extracellular flux calibration fluid |
| Other | Seahorse XF DMEM Medium | Agilent | Cat# 103575–100 | Extracellular flux culture media |
| Other | Tween20 | Sigma Aldrich | Cat# P1379-1L | Detergent for cell lysis and ELISA wash buffer |
| Other | Vectashield mounting media | VWR | Cat# 101098–042 | Mounting media for confocal microscopy |
| Other | Water, sterile | Baxter | Cat# UKF7114 | Solvent |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/74690/elife-74690-transrepform1-v2.docx
-
Source data 1
Macrophage Innate Training Induced by IL-4 and IL-13 Activation Enhances OXPHOS Driven Anti-Mycobacterial Responses Dataset.
Raw data, calculations and results of statistical analyses for all included figures.
- https://cdn.elifesciences.org/articles/74690/elife-74690-data1-v2.xlsx