Diverse ancestry whole-genome sequencing association study identifies TBX5 and PTK7 as susceptibility genes for posterior urethral valves
Figures
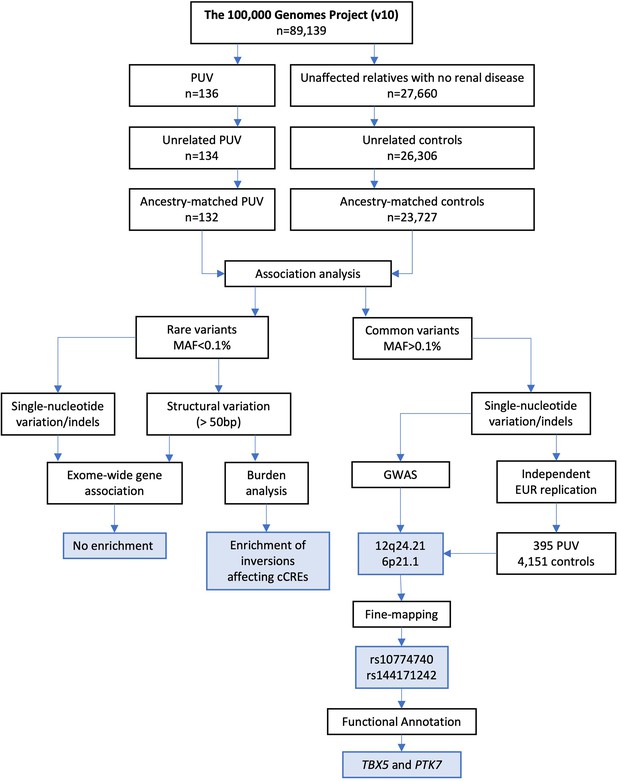
Study workflow.
The flowchart shows the number of samples included at each stage of filtering, the analytical strategies employed, and the main findings (blue boxes). PUV, posterior urethral valves; MAF, minor allele frequency; GWAS, genome-wide association study; EUR, European; cCRE, candidate cis-regulatory element.
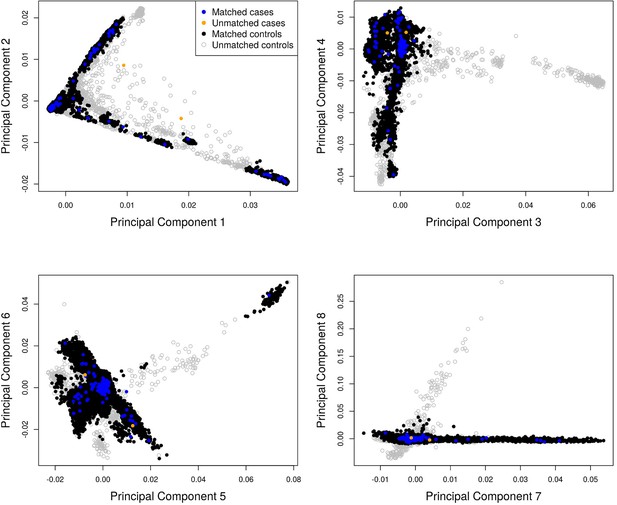
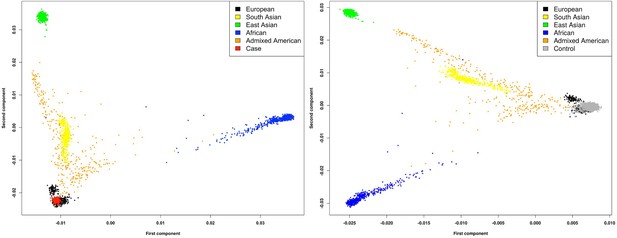
Principal component analysis (PCA) showing the first eight principal components for matched cases (blue) and controls (black) and unmatched cases (orange) and controls (grey).
Two cases and 2579 controls were excluded from downstream analyses.
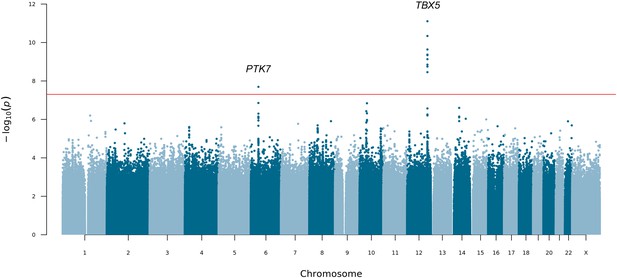
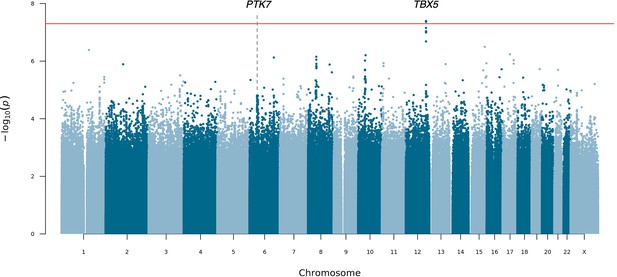
Manhattan plot for mixed-ancestry sequencing-based genome-wide association study (seqGWAS).
A genome-wide single-variant association study was carried out in 132 unrelated posterior urethral valves (PUV) cases and 23,727 controls for 19,651,224 variants with minor allele frequency (MAF) >0.1%. Chromosomal position (GRCh38) is denoted along the x axis and strength of association using a –log10(p) scale on the y axis. Each dot represents a variant. The red line indicates the Bonferroni adjusted threshold for genome-wide significance (p<5 × 10–8). The gene in closest proximity to the lead variant at significant loci is listed.
-
Figure 2—source data 1
Joint analysis of common and low-frequency variation (minor allele frequency [MAF] >0.1%) by gene across the exome.
Data generated using the genome-wide association study (GWAS) summary statistics as input to MAGMA. Gene names are given as ENSEMBL Gene IDs and SYMBOL. CHR, chromosome; NSNPS, number of variants annotated to that gene; NPARAM, number of parameters used in the model; N, sample size; ZSTAT, Z-value for the gene based on permutation p-value; P, p-value.
- https://cdn.elifesciences.org/articles/74777/elife-74777-fig2-data1-v2.txt
-
Figure 2—source data 2
Joint analysis of common and low-frequency variation (minor allele frequency [MAF] >0.1%) by gene set.
Data generated using the genome-wide association study (GWAS) summary statistics as input to MAGMA.
- https://cdn.elifesciences.org/articles/74777/elife-74777-fig2-data2-v2.txt
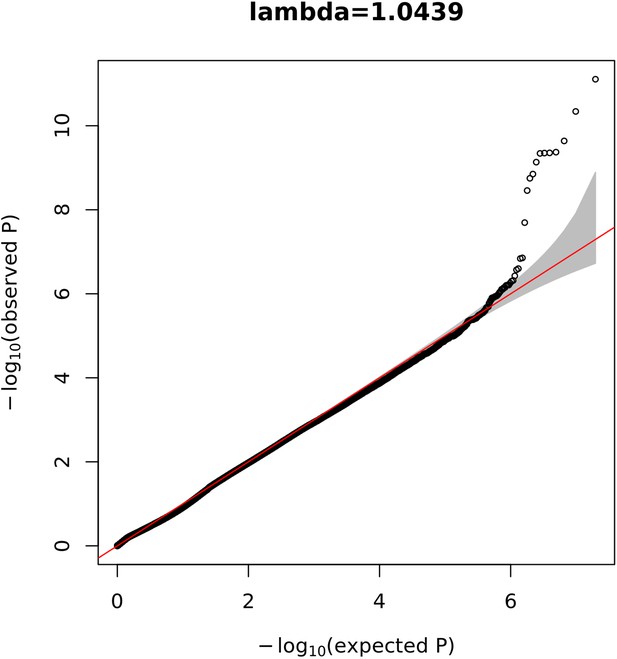
Quantile-quantile (Q–Q) plot for the mixed-ancestry genome-wide association study (GWAS) displaying the observed vs. the expected –log10(p) for each variant tested.
The grey shaded area represents the 95% confidence interval of the null distribution.
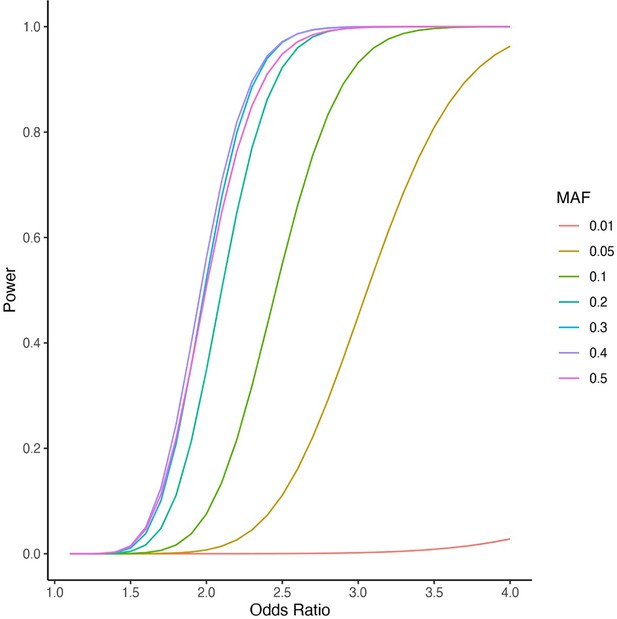
Power calculations for the mixed-ancestry genome-wide association study (GWAS) were performed at various minor allele frequencies (MAF) using 132 cases and 23,727 controls under an additive genetic model to achieve genome-wide significance of p<5 × 10–8.
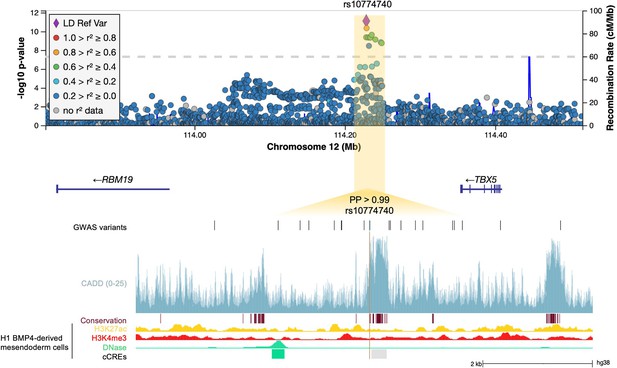
12q24.21.
Regional association plot with chromosomal position (GRCh38) denoted along the x axis and strength of association using a –log10(p) scale on the y axis. The lead variant (rs10774740) is represented by a purple diamond. Variants are coloured based on their linkage disequilibrium (LD) with the lead variant using 1000 Genomes data from all population groups. Functional annotation of the lead prioritized variant rs10774740 is shown, intersecting with CADD score (version 1.6), PhastCons conserved elements from 100 vertebrates, and ENCODE H3K27ac ChIP-seq, H3K4me3 ChIP-seq, and DNase-seq from mesendoderm cells. ENCODE cCREs active in mesendoderm are represented by shaded boxes; low DNase (grey), DNase-only (green). Genome-wide association study (GWAS) variants with p<0.05 are shown. Note that rs10774740 has a relatively high CADD score for a non-coding variant and intersects with a highly conserved region. PP, posterior probability derived using PAINTOR; cCRE, candidate cis-regulatory element.
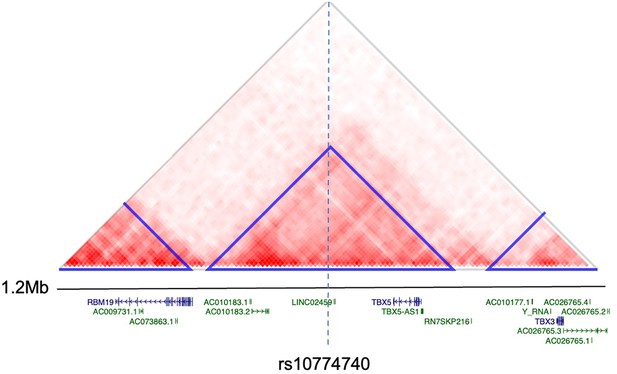
Heatmap of Hi-C interactions from H1 BMP4-derived mesendoderm cells demonstrating that rs10774740 is located within the same topologically associating domain (TAD) as TBX5.
TADs are represented by blue triangles. Protein-coding genes are denoted in blue, non-coding genes in green.
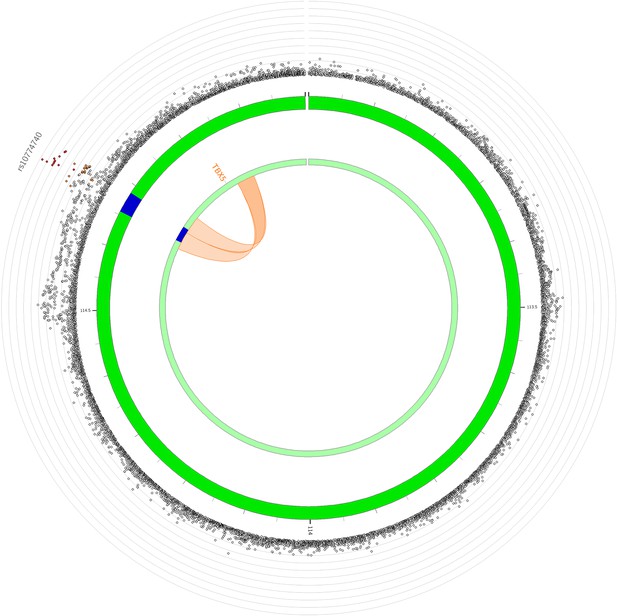
Circos plot illustrating significant chromatin interactions between 12q24.21 and the promoter of TBX5.
The outer layer represents a Manhattan plot with variants plotted against strength of association. Only variants with p<0.05 are displayed. Genomic risk loci are highlighted in blue in the second layer. Significant chromatin loops detected in H1 BMP4-derived mesendoderm cultured cells are represented in orange.
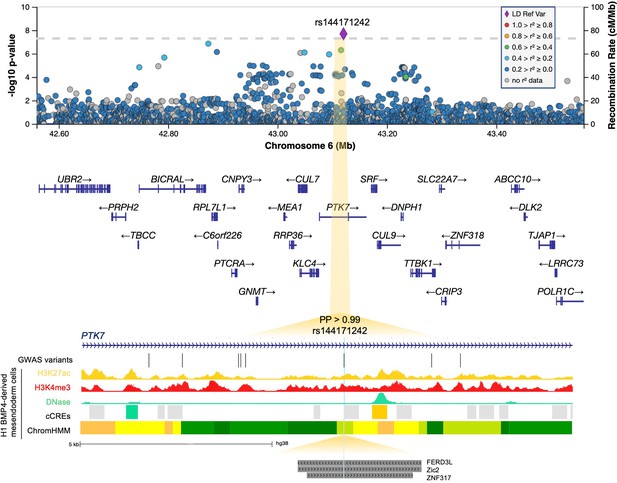
6p21.1.
Regional association plot with chromosomal position (GRCh38) along the x axis and strength of association using a –log10(p) scale on the y axis. The lead variant (rs144171242) is represented by a purple diamond. Variants are coloured based on their linkage disequilibrium (LD) with the lead variant using 1000 Genomes data from all population groups. Functional annotation of the lead prioritized variant rs144171242 is shown intersecting with ENCODE H3K27ac ChIP-seq, H3K4me3 ChIP-seq, and DNase-seq from mesendoderm cells. ENCODE cCREs active in mesendoderm are represented by shaded boxes; low DNase (grey), DNase-only (green), and distal enhancer-like (orange). ChromHMM illustrates predicted chromatin states using Roadmap Epigenomics imputed 25-state model for mesendoderm cells; active enhancer (orange), weak enhancer (yellow), strong transcription (green), transcribed and weak enhancer (lime green). Predicted transcription factor-binding sites (TFBS) from the JASPAR 2020 CORE collection (Fornes et al., 2020) are indicated by dark grey shaded boxes. Genome-wide association study (GWAS) variants with p<0.05 are shown. Note that rs144171242 intersects with both a predicted regulatory region and TFBS. PP, posterior probability derived using PAINTOR; cCREs, candidate cis-regulatory elements.
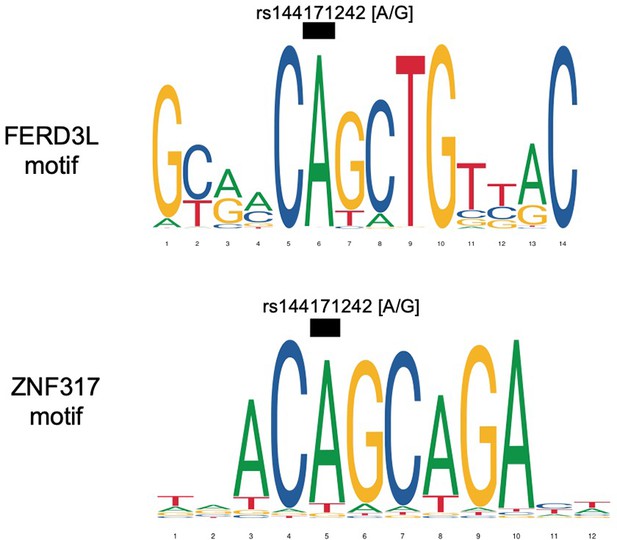
Sequence logos representing the DNA-binding motifs of transcription factors FERD3L and ZNF317.
The black boxes indicate where the risk allele [G] may disrupt binding.
Principal component analysis for the replication cohort.
Principal component analysis showing the first two principal components for a subset of cases (red) for whom genome-wide genotyping data was available (n=204), and the control (grey) cohort from 100,000 Genomes Project (100KGP) (n=4151) projected onto samples from the 1000 Genomes Project (Phase 3). Both cases and controls had confirmed European ancestry.
Manhattan plot for European sequencing-based genome-wide association study (seqGWAS).
A genome-wide single-variant association study was carried out in 88 cases and 17,993 controls for 16,938,500 variants with MAF ≥0.1%. All cases and controls had genetically determined European ancestry. Chromosomal position (GRCh38) is denoted along the x axis and strength of association using a –log10(p) scale on the y axis. Each dot represents a variant. The red line indicates the Bonferroni adjusted threshold for genome-wide significance (p<5 × 10–8). The gene in closest proximity to the lead variant at significant loci are listed.
-
Figure 6—source data 1
Comparison of diverse ancestry and European genome-wide association study (GWAS) association statistics.
The lead variants at the top four loci with p<5 × 10–7 are shown. OR, odds ratio; CI, 95% confidence interval.
- https://cdn.elifesciences.org/articles/74777/elife-74777-fig6-data1-v2.xlsx
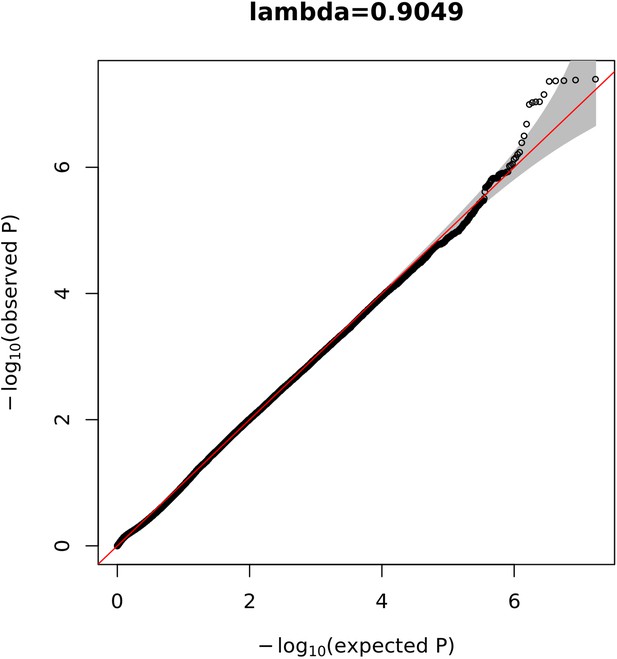
Quantile-quantile (Q-Q) plot displaying the observed vs. the expected –log10(p) for each variant tested.
The grey shaded area represents the 95% confidence interval of the null distribution.
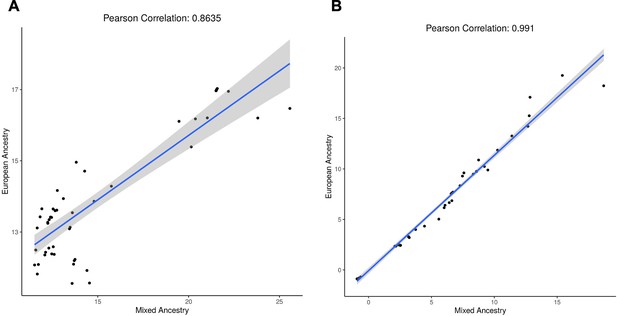
Comparison of (A) −log10(p) and (B) BETA from the diverse ancestry and European-only genome-wide association study (GWAS).
All variants with p<10–5 in both cohorts are shown. The shaded grey area represents the 95% confidence interval.
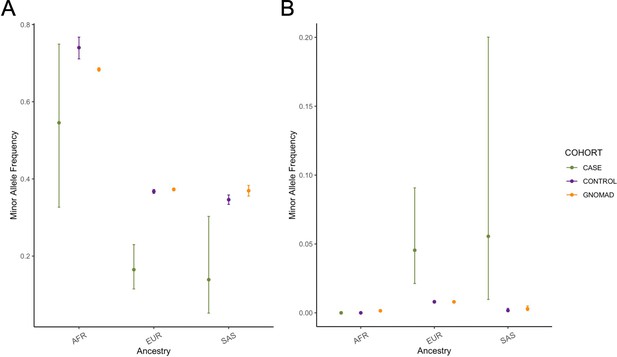
Ancestry-specific minor allele frequencies for (A) rs10774740 (T) at 12q24.21 and (B) rs144171242 (G) at 6p21.1.
Error bars represent 95% confidence intervals. The lead variant in (B) was not identified in individuals with African ancestry in this study. AFR, African ancestry (11 cases; 483 controls); EUR, European ancestry (89 cases; 17,993 controls); SAS, South Asian ancestry (18 cases; 2948 controls).
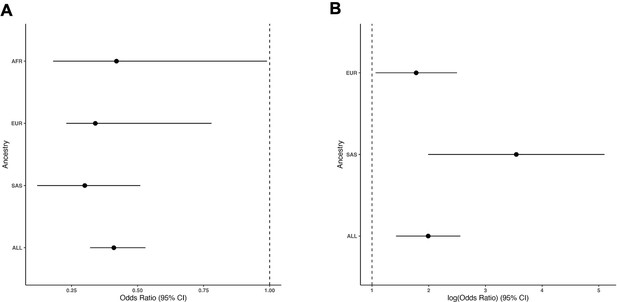
Forest plots demonstrating ancestry-specific odds ratios for (A) rs10774740 (T) and (B) rs144171242 (G).
Error bars represent 95% confidence intervals. The lead variant in (B) was not identified in individuals with African ancestry in this study. AFR, African ancestry (11 cases; 483 controls); EUR, European ancestry (89 cases; 17,993 controls); SAS, South Asian ancestry (18 cases; 2948 controls); ALL, mixed-ancestry cohort (132 cases; 23,727 controls).
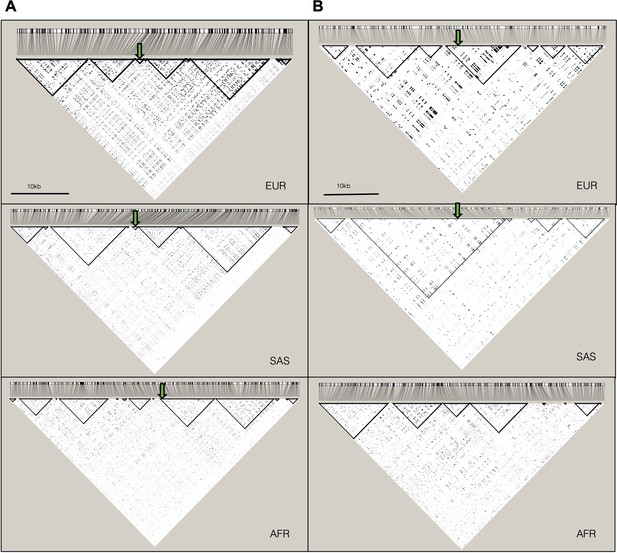
Linkage disequilibrium (LD) plots for 503 European (EUR), 489 South Asian (SAS), and 661 African (AFR) ancestry individuals from the 1000 Genomes Project (Phase 3).
Haploview (version 4.2) was used to compute pairwise LD statistics (r2) between variants for each population. The darker the shading, the higher the LD between variants. Black outlined triangles indicate haploblocks. (A) LD plot for chr12:114,663,967–114,667,916 (GRCh37) with the position of the lead variant rs10774740 represented by a green arrow; (B) LD plot for chr6:43,084,099–43,092,650 (GRCh37) with the lead variant rs144171242 represented by a green arrow. rs144171242 was not seen in the AFR population group.
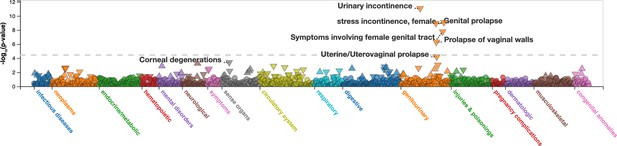
Manhattan plot of PheWAS for rs10774740 (T) at 12q24.21.
Plot downloaded from https://pheweb.org/UKB-SAIGE. The PheWAS was performed using imputed data from ~400,000 White British participants in the UK Biobank using SAIGE. The triangles indicate the direction of effect. The dashed grey line indicates a Bonferroni adjusted significance level of p< 3.6 x 10-5 (1403 phenotype codes).
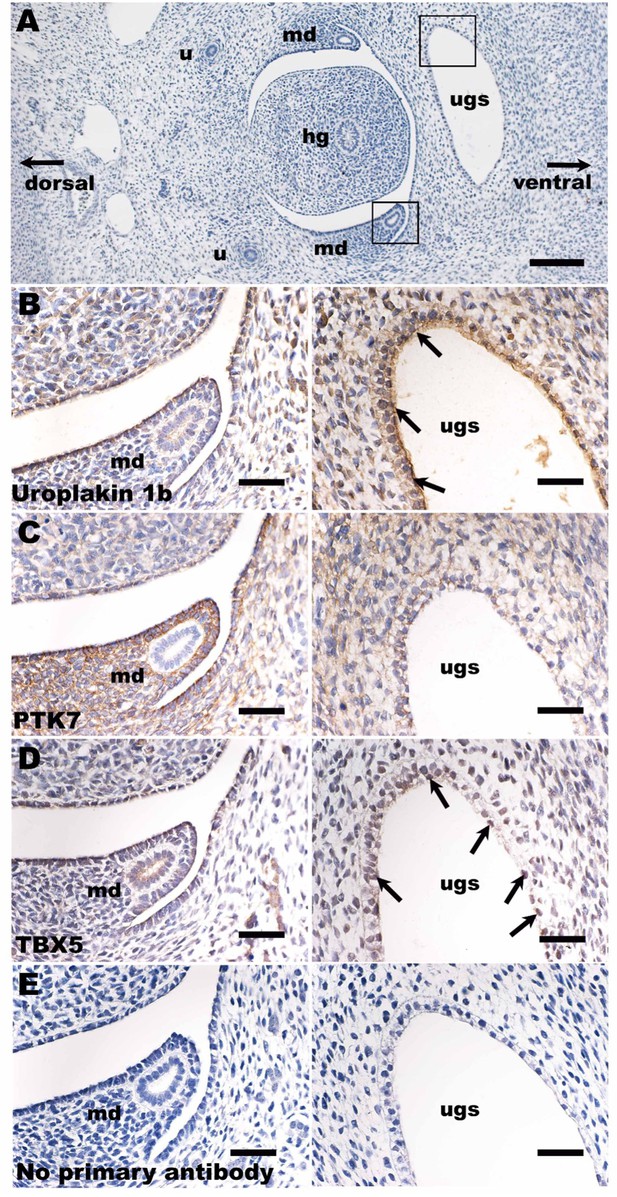
Immunohistochemistry in human embryogenesis.
(A) Overview of transverse section of a normal human embryo 7 weeks after fertilization. The section has been stained with haematoxylin (blue nuclei). Boxes around the urogenital sinus and the mesonephric duct mark similar areas depicted under high power in (B–E). In (B–D), sections were reacted with primary antibodies, as indicated; in (E) the primary antibody was omitted. (B–E) were counterstained with haematoxylin. In (B–E), the left-hand frame shows the region around the mesonephric duct, while the right-hand frame shows one lateral horn of the urogenital sinus. (B) Uroplakin 1b immunostaining revealed positive signal (brown) in the apical aspect of epithelia lining the urogenital sinus (arrows, right frame), the precursor of the urinary bladder and proximal urethra. Uroplakin 1b was also detected in the flat monolayer of mesothelial cells (left frame) that line the body cavity above the mesonephric duct. (C) There were strong PTK7 signals (brown cytoplasmic staining) in stromal-like cells around the mesonephric duct (left frame), whereas the epithelia of the duct itself were negative. PTK7 was also detected in a reticular pattern in epithelia lining the urogenital sinus (right frame) and in stromal cells near the sinus. (D) A subset of epithelial cells lining the urogenital sinus (right frame) immunostained for TBX5 (brown nuclei; some are arrowed). The mesothelial cells near the mesonephric duct (left frame) were also positive for TBX5. (E) This negative control section had the primary antibody omitted; no specific (brown) signal was noted. Bar is 400 μm in (A), and bars are 100 μm in (B–E). ugs, urogenital sinus; md, mesonephric duct; hg, hindgut; u, ureter.
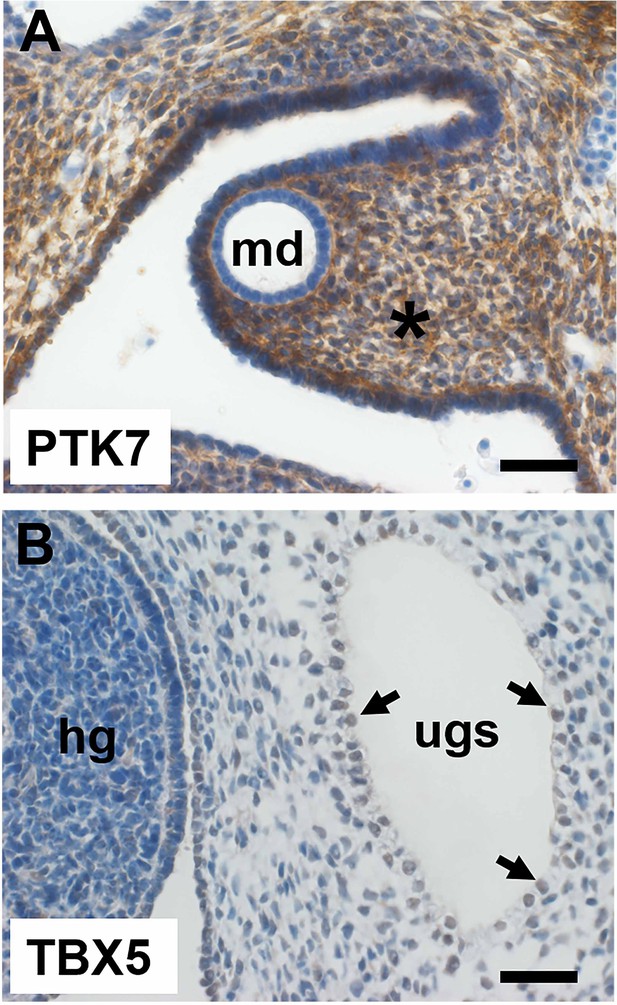
Immunohistochemistry of a second 7-week human embryo counterstained with haematoxylin.
(A) View of the mesonephric duct (the epithelial tube with md in its lumen). Note the prominent signal (brown) for PTK7 in the stromal cells surrounding the duct. (B) View of the urogenital sinus (ugs) with a subset of nuclei (three shown by arrows) in its monolayer epithelium that stain (light brown) for the transcription factor TBX5. The hindgut (hg) is nearby. Bars are 100 μm.
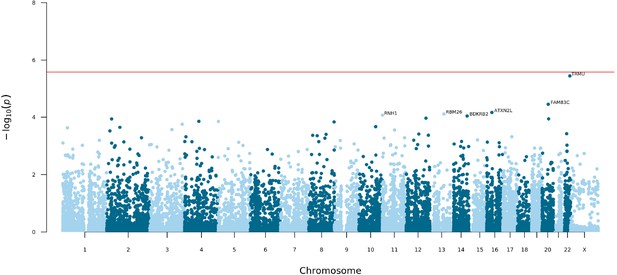
Manhattan plot of exome-wide rare coding variant analysis.
Chromosomal position (GRCh38) is shown on the x axis and strength of association using a –log10(p) scale on the y axis. Each dot represents a gene. The red line indicates the Bonferroni adjusted threshold for exome-wide significance (p=2.58 × 10–6). Genes with p<10–4 are labelled.
-
Figure 9—source data 1
Exome-wide rare coding variant analysis by gene.
p-Values generated using SAIGE-GENE and the SKAT-O rare variant test.
- https://cdn.elifesciences.org/articles/74777/elife-74777-fig9-data1-v2.txt
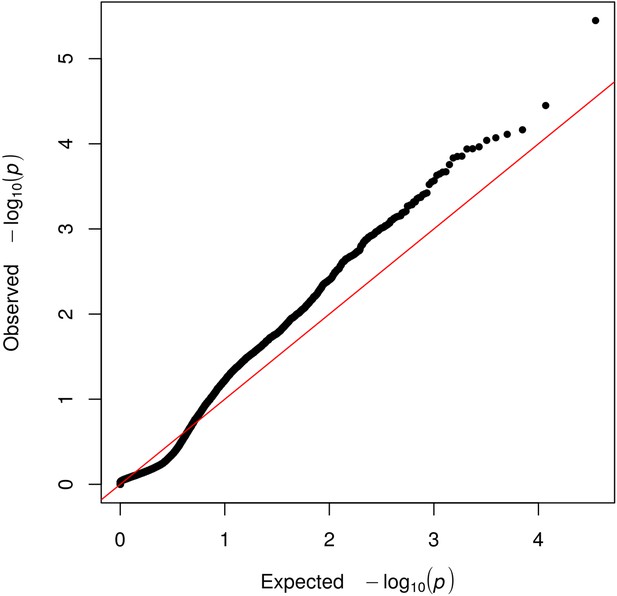
Quantile-quantile (Q-Q) plot displaying the observed vs. the expected –log10(p) for each gene tested.
The grey shaded area represents the 95% confidence interval of the null distribution.
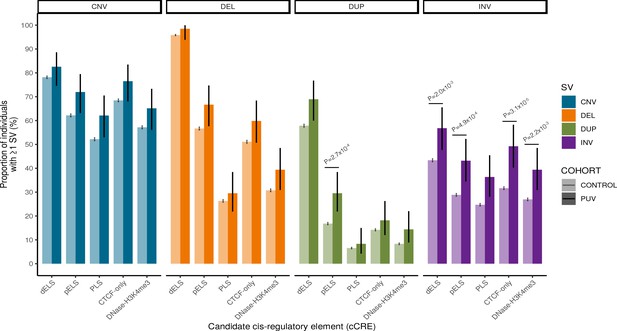
Rare structural variant burden analysis.
The proportion of individuals with ≥1 rare autosomal structural variant intersecting with an ENCODE candidate cis-regulatory element (cCRE) in cases and controls was enumerated using a two-sided Fisher’s exact test. Note that inversions affecting cCRE are enriched in PUV. Vertical black bars indicate 95% confidence intervals. Unadjusted p-values shown are significant after correction for multiple testing (p<2.5 × 10–3). CNV, copy number variant; DEL, deletion; DUP, duplication; INV, inversion; PUV, posterior urethral valves; dELS, distal enhancer-like signature; pELS, proximal enhancer-like signature; PLS, promoter-like signature; cCRE, candidate cis-regulatory element.
-
Figure 10—source data 1
Structural variant cCRE analysis.
The burden of rare autosomal structural variants intersecting with each cis-regulatory element type was compared between 132 cases and 23,727 controls. cCRE, candidate cis-regulatory element; CNV, copy number variant; DEL, deletion; DUP, duplication; INV, inversion; dELS, distal enhancer-like signature; pELS, proximal enhancer-like signature; PLS, promoter-like signature; OR, odds ratio; CI, 95% confidence interval.
- https://cdn.elifesciences.org/articles/74777/elife-74777-fig10-data1-v2.xlsx
Tables
Clinical characteristics and genetic ancestry of the discovery cohort.
| PUV (n=132) | Controls (n=23,727) | ||
|---|---|---|---|
| Median age (range) | 13 (2–66) | ||
| Males (%) | 132 (100) | 10,425 (43.9) | |
| PCA determined ancestry | |||
| EUR (%) | 89 (67.4) | 19,418 (81.8) | |
| SAS (%) | 18 (13.6) | 2847 (12.0) | |
| AFR (%) | 11 (8.3) | 449 (1.9) | |
| AMR (%) | 0 (0) | 7 (0.03) | |
| Admixed (%) | 14 (10.6) | 1006 (4.2) | |
| Additional renal/urinary phenotypes | |||
| Hydronephrosis (%) | 56 (42.4) | ||
| Bladder abnormality (%) | 32 (24.2) | ||
| Hydroureter (%) | 30 (22.7) | ||
| VUR (%) | 27 (20.5) | ||
| Renal dysplasia (%) | 16 (12.1) | ||
| Hypertension (%) | 11 (8.3) | ||
| Renal agenesis (%) | 8 (6.1) | ||
| Recurrent UTIs (%) | 5 (3.8) | ||
| Renal hypoplasia (%) | 4 (3.0) | ||
| Renal duplication (%) | 2 (1.5) | ||
| Extrarenal manifestations (%) | 35 (26.5) | ||
| Cardiac anomaly (%) | 4 (3.0) | ||
| Neurodevelopmental disorder (%) | 7 (5.3) | ||
| Family history (%) | 5 (3.8) | ||
| End-stage renal disease (%) | 23 (17.4) | ||
| Median age ESRD (range) | 14 (0–39) |
-
PUV, posterior urethral valves; PCA, principal component analysis; EUR, European; SAS, South Asian; AFR, African; AMR, Latino/Admixed American; VUR, vesico-ureteral reflux; UTI, urinary tract infection; ESRD, end-stage renal disease.
Association statistics for significant genome-wide loci.
| P value | OR (95% CI) | Case EAF | Control EAF | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lead variant | CHR:POS | Effect Allele | Closest gene | Discovery | Replication | Discovery | Replication | Discovery | Replication | Discovery | Replication |
| rs10774740 | chr12:114228397 | T | TBX5 | 7.81x10–12 | 5.17x10–3 | 0.4 | 0.8 | 0.19 | 0.31 | 0.37 | 0.36 |
| (0.31–0.52) | (0.68–0.93) | ||||||||||
| rs144171242 | chr6:43120356 | G | PTK7 | 2.02x10–8 | 7.21x10–3 | 7.2 | 2.18 | 0.05 | 0.018 | 0.01 | 0.008 |
| (4.08–12.70) | (1.22–3.90) | ||||||||||
-
Table 2—source data 1
Replication study.
The lead variants at the top four loci with p<5 × 10–7 were genotyped in an independent European cohort of 395 posterior urethral valves (PUV) cases and 4151 controls. p-Values in the replication cohort were calculated using a one-sided Cochran Armitage Trend test. OR, odds ratio; CI, 95% confidence interval.
- https://cdn.elifesciences.org/articles/74777/elife-74777-table2-data1-v2.xlsx
Structural variant burden analysis.
The burden of rare autosomal structural variants intersecting with (a) at least one exon or (b) a cis-regulatory element was compared between 132 cases and 23,727 controls. cCRE, candidate cis-regulatory element; PUV, posterior urethral valves; CNV, copy number variant; DEL, deletion; DUP, duplication; INV, inversion; OR, odds ratio; CI, 95% confidence interval, IQR, interquartile range.
| EXON | cCRE | ||||
|---|---|---|---|---|---|
| PUV | Controls | PUV | Controls | ||
| CNV | n (%) | 109 (82.6) | 17,961 (75.7) | 111 (84.1) | 18,773 (79.1) |
| OR (CI) | 1.52 (0.96–2.50) | 1.39 (0.87–2.34) | |||
| Fisher’s exact p | 0.07 | 0.2 | |||
| Median size (kb) (IQR) | 104 (183) | 94 (158) | 80 (165) | 80 (128) | |
| p (Wilcoxon) | 0.75 | 0.75 | |||
| DEL | n (%) | 117 (88.6) | 19,987 (84.2) | 132 (100) | 23,031 (97.1) |
| OR (CI) | 1.46 (0.85–2.69) | Inf | |||
| Fisher’s exact p | 0.19 | 0.04 | |||
| Median size (kb) (IQR) | 1.4 (4.2) | 1.8 (4.6) | 1.5 (4.1) | 1.9 (4.5) | |
| p (Wilcoxon) | 0.18 | 4.1×10–4 | |||
| DUP | n (%) | 59 (44.7) | 8,476 (35.7) | 104 (78.8) | 16,004 (67.5) |
| OR (CI) | 1.45 (1.01–2.08) | 1.79 (1.17–2.83) | |||
| Fisher’s exact p | 0.04 | 5.0×10–3 | |||
| Median size (kb) (IQR) | 3.7 (5.7) | 3.0 (5.6) | 2.0 (5.3) | 2.3 (5.3) | |
| p (Wilcoxon) | 0.16 | 0.49 | |||
| INV | n (%) | 66 (50.0) | 8,736 (36.8) | 81 (61.4) | 11,171 (47.1) |
| OR (CI) | 1.72 (1.20–2.45) | 1.79 (1.24–2.59) | |||
| Fisher’s exact p | 2.1×10–3 | 1.2×10–3 | |||
| Median size (kb) (IQR) | 253 (1931) | 261 (1642) | 129 (459) | 94 (779) | |
| p (Wilcoxon) | 0.44 | 0.12 |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/74777/elife-74777-transrepform1-v2.docx
-
Source code 1
Ancestry matching.
- https://cdn.elifesciences.org/articles/74777/elife-74777-code1-v2.zip
-
Source code 2
SV burden code.
- https://cdn.elifesciences.org/articles/74777/elife-74777-code2-v2.zip





