Auditory mismatch responses are differentially sensitive to changes in muscarinic acetylcholine versus dopamine receptor function
Figures
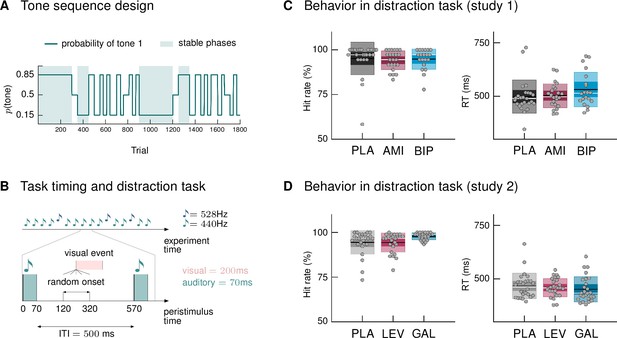
Paradigm and behavioral results.
(A) Probability structure for the tone sequence in the oddball MMN paradigm with volatility. The probability of hearing the higher tone (tone 1, with p(tone 2) = 1 – p(tone 1)) varied over the course of the tone sequence as indicated by the blue line. Tone 1 functioned as the ‹deviant› in phases where it was less likely (p = 0.15), and as the ‹standard› when it was more likely than tone 2 (p = 0.85). Stable phases (p constant for 100 or more trials) alternated with volatile phases (p changes every 25–60 trials). (B) Experimental task: Overview of timing of events. Participants passively listened to a sequence of 1800 tones while performing a visual distraction task. Visual events occurred after tone presentations at a randomly varying delay between 50 and 250 ms after tone offset, in 36 (study 1) and 90 (study 2) out of 1800 trials. ITI = Inter-stimulus interval. (C, D) Hit rates and reaction times, per drug group, for the visual distraction task, plotted using the notBoxPlot function (Campbell, 2020; https://github.com/raacampbell/notBoxPlot/). Mean values are marked by black lines, medians by white lines. The dark box around the mean reflects the 95% confidence interval around the mean, and the light outer Box 1 standard deviation. (C) There were no significant differences between drug groups in performance on the visual distraction task. (D) Participants in the galantamine group had higher hit rates in the distraction task (see main text). PLA = placebo, AMI = amisulpride, BIP = biperiden, LEV = levodopa, GAL = galantamine group.
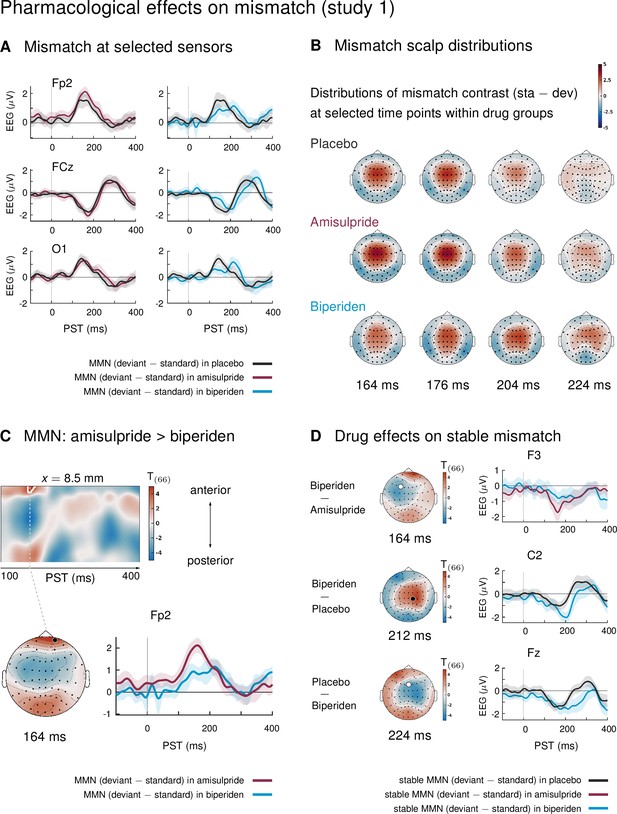
Pharmacological effects on mismatch ERPs in study 1 (interaction mismatch × drug).
(A) Difference waves (deviants – standards) at selected sensors for the different drug groups in study 1. (B) Scalp distribution of the mismatch contrast at selected time points. The mismatch response in the biperiden group peaked later and more towards right central channels than in the other groups. (C) Mismatch responses in pre-frontal sensors were significantly weaker in the biperiden group compared to the amisulpride group. Displayed are t-maps for the contrast MMN amisulpride > MMN biperiden. The first map runs across the scalp dimension y (from posterior to anterior, y-axis), and across peristimulus time (x-axis), at the spatial x-location indicated above the map. Significant t values (p < 0.05, whole-volume FWE-corrected at the peak-level) are marked by white contours. The scalp map below shows the t-map at the indicated peristimulus time point, corresponding to the peak of that cluster, across a 2D representation of the sensor layout. ERP plot shows the difference waves for a selected sensor, separately for the biperiden and amisulpride groups. The location of the chosen sensor on the scalp is marked on the scalp map by the corresponding symbol. (D) Pharmacological effects when only testing mismatch ERPs during stable phases. Logic of display as in Panel C.
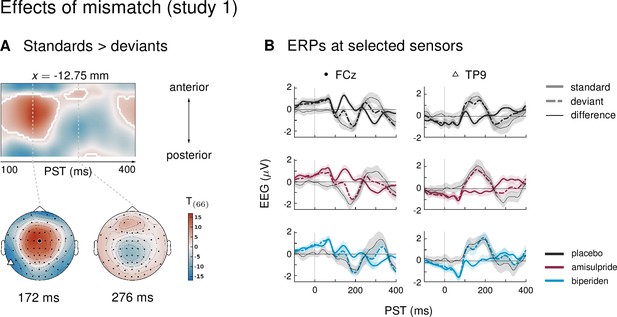
Main effect of mismatch in study 1.
(A) ERPs to deviants were significantly different from ERPs to standards in large parts of the time × sensor space, including the classical mismatch negativity in fronto-central channels between 100 and 250 ms after tone onset. Logic of display as in main Figure 2. (B) ERPs and difference waves for selected sensors, separately for the three drug groups. The location of the chosen sensors on the scalp is marked on the scalp map in panel A by the corresponding symbol.
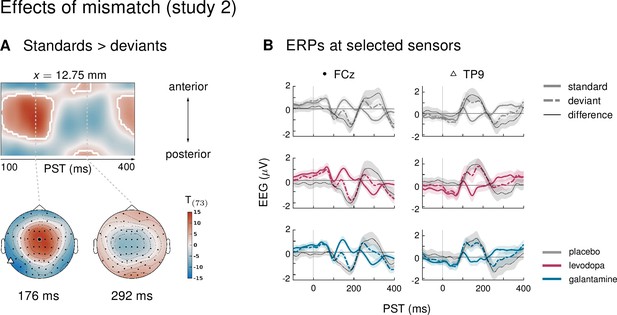
Main effect of mismatch in study 2.
(A) As in study 1, mismatch was significant in large parts of the time × sensor space, including the classical mismatch negativity. Logic of display as in main Figure 2. (B) ERPs and difference waves for selected sensors, separately for the three drug groups. Mismatch signals were highly similar across drug groups.
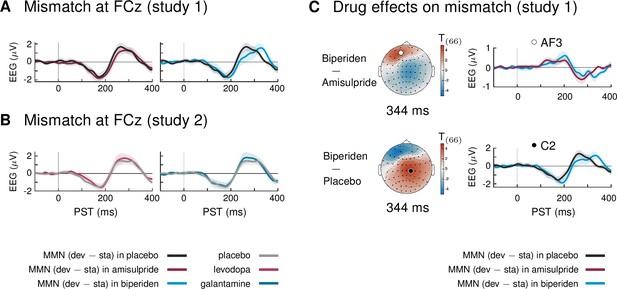
Main effect of mismatch and interaction mismatch × drug group under the new pre-processing pipeline and trial definition.
(A) Mismatch signals (deviants – standards) at sensor FCz in study 1. (B) Mismatch signals (deviants – standards) at sensor FCz in study 2. There were no significant differences between mismatch signals of the different drug groups in study 2. (C) Pharmacological effects on mismatch in study 1: the new pipeline located the dominant effect of biperiden in a later time window compared to our original pipeline. Around 344 ms, mismatch signals were significantly stronger in the biperiden group compared to placebo and amisulpride, due to a delayed peak of the P3 component.
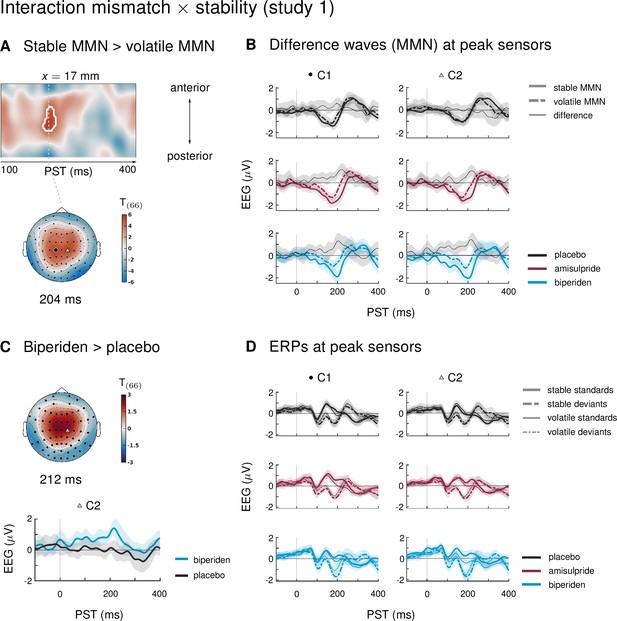
Interaction effects between mismatch and stability, and three-way interaction mismatch × stability × drug in study 1.
(A) Regions of the time × sensor space where ERPs to tones in stable phases were more positive than ERPs to tones in volatile phases. Logic of display as in Figure 2. (B) ERP difference waves (deviants – standards) at the peak sensors for the two clusters shown in panel A, separately for the three drug groups. (C) Pharmacological effect on the interaction: at right central channels, the biperiden group showed a stronger interaction effect between mismatch and stability than the placebo group (significant only within a spatio-temporal mask, see main text). Displayed is the t-map of the contrast and the difference waves (volatile – stable MMN) at sensor C2. (D) ERPs to standards and deviants at the same sensors as plotted in A and B.
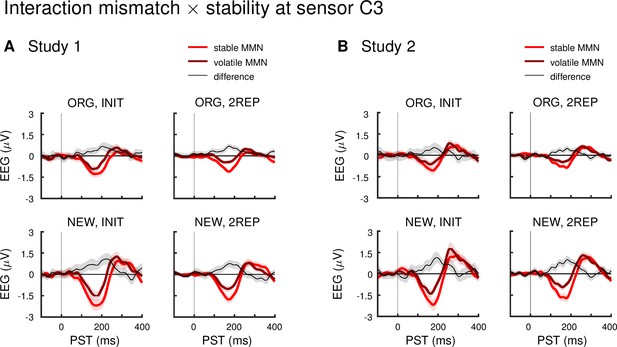
Interaction effect mismatch × stability at a left central sensor across different analysis pipelines.
Panels show mismatch difference waves (deviants – standards), averaged across all drug groups, during stable and volatile phases of the experiment, and the difference between these (volatile – stable), across variations in pre-processing (ORG vs NEW) and trial definition (INIT vs 2REP). Data are shown at an exemplar left central sensor (C3; note that the location in sensor space of the peak effect varied slightly across pipelines).
(A) Study 1: Stable mismatch responses were stronger than volatile mismatch responses around or before 200ms irrespective of the analysis pipeline used. (B) Study 2: Under all pipelines except ORG preprocessing and INIT trial definition, stable mismatch responses were stronger than volatile mismatch responses around or before 200 ms. ORG = original pre-processing pipeline as defined in the main text, NEW = adjusted pipeline as defined in section Control analyses in Materials and methods, INIT = initial trial definition: all standards and deviants following at least 5 repetitions, 2REP = new trial definition: all standards and deviants following at least 2 repetitions, for exact definitions refer to section First level general linear model and Control analyses in Materials and methods.
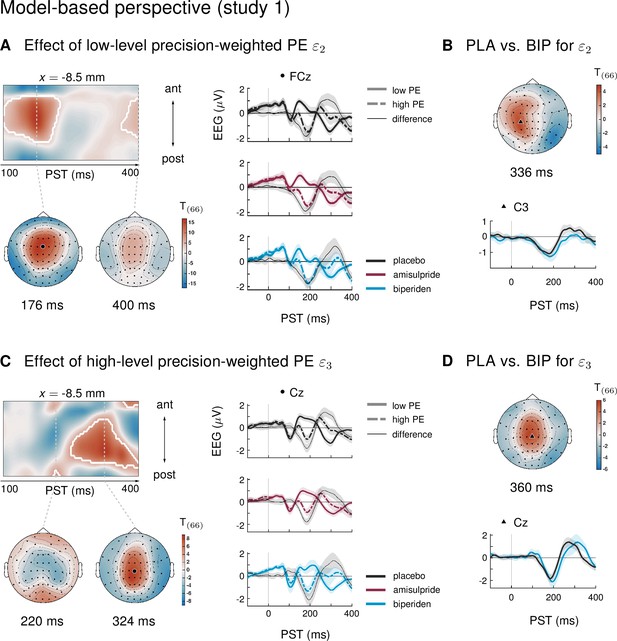
Results of the model-based single-trial analysis in study 1.
(A) The effect of the lower-level precision-weighted PE on EEG amplitudes. Left: Regions of the time × sensor space where ERPs to tones were significantly modulated by . Logic of display as in Figure 2 in the main text. Right: ERPs and difference waves for selected sensors, separately for the three drug groups. (B) Pharmacological effects on : we found a weaker modulation of ERPs by under biperiden in left central sensors. Lower plot shows the ERP difference waves (ERPs to high PE tones − ERPs to low PE tones). (C) The effect of the higher level precision-weighted PE on EEG amplitudes. (D) Biperiden shifted the late central positivity encoding the higher level PE in time.
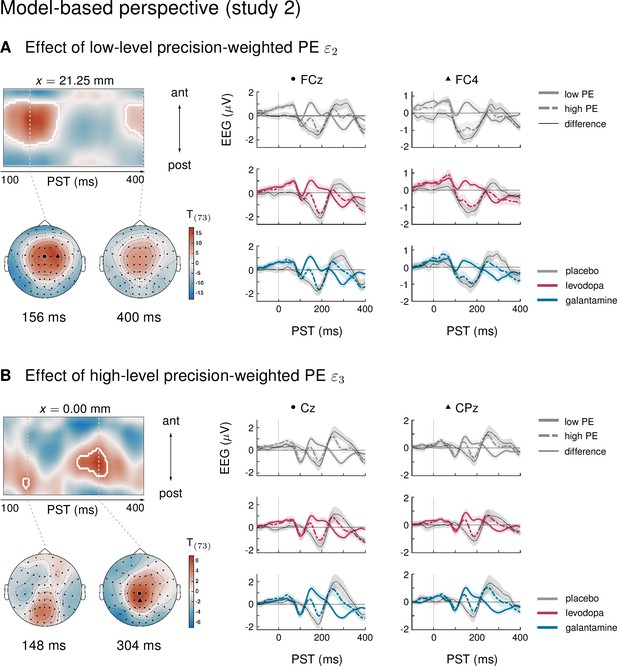
Results of the model-based single-trial analysis in study 2.
(A) The effect of the lower-level precision-weighted PE on EEG amplitudes. Left: Regions of the time × sensor space where ERPs to tones were significantly modulated by . Logic of display as in Figure 2 in the main text. Right: ERPs and difference waves for selected sensors, separately for the three drug groups. (B) The effect of the higher-level precision-weighted PE on EEG amplitudes. There were no significant differences between drug groups in either prediction error.
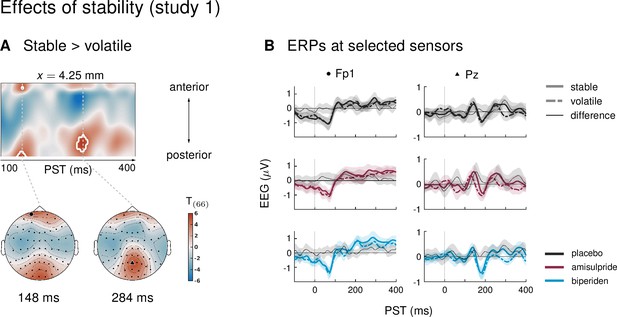
Main effect of stability in study 1.
(A) Regions of the time ×sensor space where ERPs to tones in stable phases were more positive than ERPs to tones in volatile phases. Logic of display as in Figure 2 in the main text. We found early (at 148ms in pre-frontal sensors and at 144ms in occipital sensors) and late effects (at 284ms in parietal sensors) of stability on the ERPs. (B) ERPs and difference waves for selected sensors, separately for the three drug groups. In all clusters, ERPs to tones in stable phases were more positive than ERPs to tones in volatile phases.
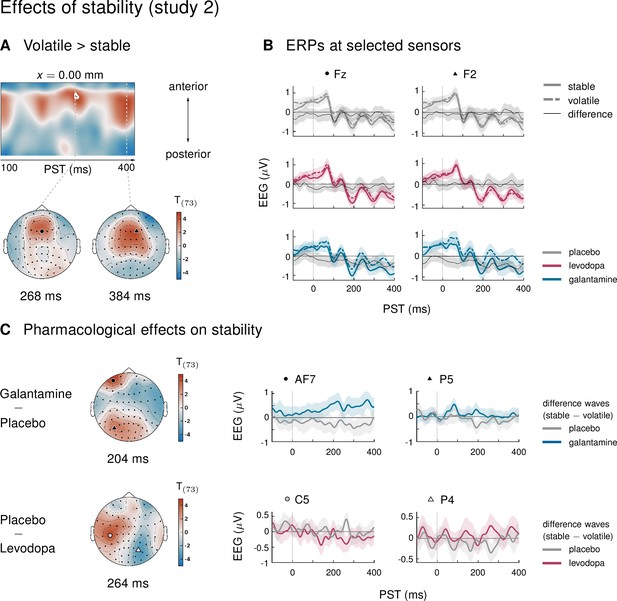
Main effect of stability and interaction stability ×drug group in study 2.
(A) Regions of the time ×sensor space where ERPs to tones in volatile phases were more positive than ERPs to tones in stable phases. Logic of display as in Figure 2 in the main text. (B) ERPs and difference waves for selected sensors, separately for the three drug groups. In this cluster, ERPs to tones in volatile phases were more positive than ERPs to tones in stable phases. This effect was driven by the galantamine group. (C) Pharmacological effects on stability: we found significant differences in the impact of stability on ERPs in two clusters. Plots show the ERP difference waves (ERPs to tones in stable phases − ERPs to tones in volatile phases) in different drug conditions.
Tables
Significant clusters of activation for main effect of mismatch (standards versus deviants) and pharmacological effects on mismatch in study 1.
The table lists the peak coordinates (x, y, and z for time), peak t values, corresponding Z values, whole-volume FWE-corrected p-values at the peak level, and cluster size (kE). The last column lists the minimal and maximal time points of the cluster, i.e., the significant time window tsig.
| Study 1: Mismatch | cluster | [ms] | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A standards > deviants | 1 | –13 | 2 | 172 | 16.59 | Inf | 0.000 | 8,217 | 100–232 |
| 2 | 0 | 13 | 400 | 8.24 | 6.81 | 0.000 | 1,037 | 364–400 | |
| 3 | –13 | 50 | 276 | 6.66 | 5.81 | 0.000 | 284 | 240–300 | |
| 4 | -4 | –95 | 304 | 5.38 | 4.88 | 0.002 | 135 | 284–328 | |
| 5 | –60 | –57 | 268 | 4.76 | 4.40 | 0.015 | 166 | 240–280 | |
| –60 | –36 | 260 | 4.73 | 4.38 | 0.016 | ||||
| –60 | –46 | 264 | 4.73 | 4.37 | 0.016 | ||||
| B deviants > standards | 1 | –47 | –68 | 176 | 14.42 | Inf | 0.000 | 6,293 | 100–328 |
| 64 | –62 | 200 | 13.55 | Inf | 0.000 | ||||
| 42 | –78 | 164 | 11.83 | Inf | 0.000 | ||||
| 2 | 17 | 72 | 168 | 13.93 | Inf | 0.000 | 1,592 | 100–236 | |
| 3 | 34 | –46 | 364 | 6.10 | 5.41 | 0.000 | 336 | 352–400 | |
| 47 | –52 | 400 | 5.27 | 4.80 | 0.003 | ||||
| 4 | –51 | –30 | 400 | 5.79 | 5.19 | 0.000 | 286 | 376–400 | |
| 5 | 4 | 72 | 400 | 4.56 | 4.24 | 0.027 | 5 | 400–400 | |
| 6 | 26 | 67 | 400 | 4.46 | 4.15 | 0.036 | 1 | 400–400 | |
| C AMI > BIP | 1 | 8 | 67 | 164 | 4.83 | 4.45 | 0.012 | 31 | 160–172 |
Significant clusters of activation for main effect of mismatch (standards versus deviants) in study 2.
Columns are organized as in Table 1.
| Study 2: Mismatch | cluster | [ms] | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A standards > deviants | 1 | 13 | -9 | 176 | 14.13 | Inf | 0.000 | 7,583 | 100–216 |
| 4 | 18 | 160 | 13.76 | Inf | 0.000 | ||||
| 42 | –25 | 124 | 10.85 | Inf | 0.000 | ||||
| 2 | 0 | -9 | 396 | 8.66 | 7.16 | 0.000 | 1,338 | 364–400 | |
| 3 | 4 | –95 | 292 | 7.34 | 6.33 | 0.000 | 713 | 244–332 | |
| 26 | –89 | 280 | 6.66 | 5.87 | 0.000 | ||||
| 47 | –62 | 252 | 5.71 | 5.18 | 0.001 | ||||
| 4 | 4 | 61 | 288 | 6.14 | 5.50 | 0.000 | 320 | 256–304 | |
| –4 | 56 | 268 | 6.07 | 5.44 | 0.000 | ||||
| 5 | –47 | –68 | 256 | 5.93 | 5.34 | 0.000 | 364 | 232–284 | |
| –60 | –57 | 260 | 5.65 | 5.13 | 0.001 | ||||
| B deviants > standards | 1 | –42 | –73 | 172 | 13.97 | Inf | 0.000 | 5,637 | 100–216 |
| 55 | –68 | 196 | 10.87 | Inf | 0.000 | ||||
| –42 | –73 | 124 | 10.38 | Inf | 0.000 | ||||
| 2 | -8 | –30 | 256 | 7.41 | 6.38 | 0.000 | 2,876 | 232–328 | |
| –26 | –14 | 288 | 6.83 | 5.99 | 0.000 | ||||
| 8 | –9 | 304 | 6.67 | 5.87 | 0.000 | ||||
| 3 | 38 | –68 | 400 | 6.65 | 5.86 | 0.000 | 302 | 372–400 | |
| 4 | 4 | 72 | 388 | 6.03 | 5.41 | 0.000 | 168 | 368–400 | |
| 5 | 68 | 18 | 192 | 5.36 | 4.91 | 0.002 | 20 | 168–204 | |
| 6 | –60 | -9 | 400 | 5.26 | 4.83 | 0.003 | 153 | 388–400 | |
| –34 | –62 | 396 | 5.04 | 4.65 | 0.005 |
Results of the ROI analysis.
Table lists mean (std) values of the peak amplitudes and latencies separately for each drug group in the two studies. F (p) values refer to the effect of the factor drug group in a 3 × 3 ANOVA (drug × sensor). Last row lists the significant post-hoc comparisons between pairs of drug groups. Lat. = Latency.
| Study 1 | Study 2 | |||||||
|---|---|---|---|---|---|---|---|---|
| PLA | AMI | BIP | F2,208(p) | PLA | LEV | GAL | F (p) | |
| Peaks (µV) | –1.83 (0.1) | –1.95 (0.1) | –2.04 (0.1) | 1.1 (0.33) | –1.75 (0.1) | –2.0 (0.1) | –1.76 (0.1) | 1.73 (0.18) |
| Lat. (ms) | 168.8 (3.2) | 173.7 (3.2) | 181.9 (3.4) | 4.06 (0.02) | 165.1 (2.75) | 173.2 (2.75) | 163.7 (2.75) | 3.5 (0.03) |
| Post-hoc t | Lat.: BIP > PLA | p = 0.013 | Lat.: LEV >GAL | p = 0.038 | ||||
Significant clusters of activation for interaction effects (mismatch × stability) on ERPs in study 1.
Columns are organized as in Table 1.
| Study 1: mismatch × stability | cluster | [ms] | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A stable MMN > volatile MMN | 1 | 17 | –19 | 204 | 5.73 | 5.14 | 0.001 | 591 | 180–220 |
| –17 | –25 | 196 | 5.68 | 5.10 | 0.001 | ||||
| B volatile MMN > stable MMN | 1 | 42 | –78 | 200 | 5.63 | 5.07 | 0.001 | 119 | 188–236 |
| 55 | –68 | 200 | 5.17 | 4.72 | 0.005 | ||||
| 64 | –62 | 200 | 5.04 | 4.62 | 0.007 | ||||
| 2 | –60 | –57 | 208 | 5.33 | 4.85 | 0.003 | 29 | 200–220 |
Significant clusters of activation for pharmacological effects on stable mismatch in study 1.
Columns are organized as in Table 1.
| Study 1:Stable Mismatch | cluster | [ms] | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A PLA >BIP | 1 | –26 | 56 | 204 | 4.56 | 4.24 | 0.027 | 8 | 204–212 |
| 2 | –26 | 61 | 224 | 4.36 | 4.07 | 0.049 | 1 | 224–224 | |
| B BIP > PLA | 1 | 21 | –19 | 212 | 4.92 | 4.52 | 0.009 | 78 | 200–220 |
| C AMI > BIP | 1 | 17 | 72 | 164 | 4.74 | 4.38 | 0.016 | 11 | 160–168 |
Parameter settings for the HGF.
The first two rows show the priors under which the surprise-minimizing parameters were estimated, the last row displays the result of the estimation and thus the values used in the simulation of belief trajectories. All perceptual parameters and starting values of beliefs were fixed to their prior means (indicated by zero prior variance) except for the tonic learning rate on the second level, ω2.
| Parameter | µ2(0) | µ3(0) | σ2(0) | σ3(0) | κ1 | κ2 | ω2 | ω3 |
|---|---|---|---|---|---|---|---|---|
| Prior mean | 0 | 1 | 0.25 | 1 | 1 | 1 | -3 | –10 |
| Prior variance | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 0 |
| Posterior mean | 0 | 1 | 0.25 | 1 | 1 | 1 | –3.03 | –10 |
Results of the model-based single-trial analysis in study 1.
Columns are organized as in Table 1 of the main text.
| Study 1: Model | cluster | [ms] | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Low-level PE: pos. | 1 | –60 | –57 | 176 | 14.19 | Inf | 0.000 | 5583 | 108–228 |
| –17 | 72 | 144 | 12.43 | Inf | 0.000 | ||||
| –8 | 72 | 144 | 12.41 | Inf | 0.000 | ||||
| 2 | 47 | –36 | 360 | 6.53 | 5.71 | 0.000 | 1353 | 284–400 | |
| 42 | –41 | 336 | 6.51 | 5.70 | 0.000 | ||||
| 47 | –41 | 400 | 5.71 | 5.13 | 0.001 | ||||
| 3 | –55 | –25 | 400 | 6.06 | 5.38 | 0.000 | 423 | 360–400 | |
| 4 | 68 | 18 | 192 | 4.79 | 4.42 | 0.013 | 4 | 184–196 | |
| 5 | –42 | –30 | 312 | 4.58 | 4.25 | 0.025 | 12 | 312–316 | |
| Low-level PE: neg. | 1 | -8 | -3 | 176 | 16.40 | Inf | 0.000 | 7361 | 100–232 |
| 4 | 29 | 168 | 15.84 | Inf | 0.000 | ||||
| 2 | –13 | –14 | 400 | 6.51 | 5.70 | 0.000 | 663 | 364–400 | |
| Low-level PE: PLA >BIP | 1 | –26 | –30 | 328 | 4.46 | 4.15 | 0.036 | 8 | 328–332 |
| Low-level PE: BIP > PLA | 1 | 42 | –62 | 336 | 4.91 | 4.51 | 0.009 | 47 | 328–344 |
| 2 | 38 | –62 | 392 | 4.43 | 4.13 | 0.040 | 10 | 388–396 | |
| High-level PE: pos. | 1 | -8 | –36 | 324 | 8.48 | 6.95 | 0.000 | 3679 | 248–400 |
| –13 | –46 | 364 | 7.77 | 6.52 | 0.000 | ||||
| –26 | –68 | 400 | 5.60 | 5.04 | 0.001 | ||||
| 2 | 4 | 72 | 392 | 6.53 | 5.71 | 0.000 | 208 | 364–400 | |
| 3 | 8 | –95 | 220 | 5.21 | 4.75 | 0.003 | 79 | 212–228 | |
| 4 | 4 | 61 | 212 | 4.46 | 4.15 | 0.036 | 11 | 208–212 | |
| 21 | 61 | 212 | 4.41 | 4.11 | 0.042 | ||||
| High-level PE: neg. | 1 | 64 | –52 | 328 | 6.79 | 5.89 | 0.000 | 2483 | 272–400 |
| 34 | 13 | 380 | 6.10 | 5.41 | 0.000 | ||||
| 60 | –36 | 284 | 6.06 | 5.38 | 0.000 | ||||
| 2 | –55 | –46 | 308 | 4.85 | 4.47 | 0.011 | 165 | 296–320 | |
| 3 | 0 | 50 | 276 | 4.77 | 4.40 | 0.014 | 20 | 260–280 | |
| 4 | 30 | –30 | 216 | 4.76 | 4.40 | 0.014 | 97 | 208–224 | |
| 4 | 2 | 216 | 4.59 | 4.26 | 0.024 | ||||
| 5 | 13 | –95 | 320 | 4.49 | 4.18 | 0.033 | 4 | 316–324 | |
| 6 | –21 | 40 | 256 | 4.41 | 4.11 | 0.042 | 2 | 252–256 | |
| High-level PE: BIP > PLA | 1 | -8 | –14 | 360 | 5.35 | 4.86 | 0.002 | 327 | 336–376 |
Results of the model-based single-trial analysis in study 2.
Columns are organized as in Table 1 of the main text.
| Study 2: Model | cluster | [ms] | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Low-level PE: pos. | 1 | –47 | –68 | 168 | 14.03 | Inf | 0.000 | 6196 | 100–224 |
| 0 | 72 | 164 | 13.51 | Inf | 0.000 | ||||
| 47 | –73 | 192 | 12.61 | Inf | 0.000 | ||||
| 2 | –34 | –68 | 400 | 7.21 | 6.24 | 0.000 | 432 | 368–400 | |
| 3 | –17 | –25 | 256 | 6.85 | 6.00 | 0.000 | 1386 | 240–300 | |
| 4 | 0 | 72 | 400 | 6.51 | 5.76 | 0.000 | 225 | 364–400 | |
| 5 | 38 | –73 | 400 | 6.31 | 5.62 | 0.000 | 147 | 384–400 | |
| 6 | 68 | 18 | 176 | 4.51 | 4.22 | 0.032 | 5 | 168–184 | |
| Low-level PE: neg. | 1 | 21 | 8 | 156 | 17.50 | Inf | 0.000 | 8067 | 100–220 |
| 13 | 8 | 176 | 17.07 | Inf | 0.000 | ||||
| 34 | –3 | 124 | 11.88 | Inf | 0.000 | ||||
| 2 | -8 | –14 | 400 | 9.01 | 7.37 | 0.000 | 1582 | 360–400 | |
| 3 | –42 | –73 | 264 | 5.44 | 4.97 | 0.002 | 221 | 252–296 | |
| –26 | –89 | 264 | 5.29 | 4.85 | 0.003 | ||||
| –60 | –57 | 268 | 5.27 | 4.84 | 0.003 | ||||
| 4 | 42 | 50 | 324 | 5.34 | 4.89 | 0.002 | 74 | 296–332 | |
| 34 | 61 | 300 | 4.74 | 4.41 | 0.016 | ||||
| 60 | 29 | 324 | 4.68 | 4.36 | 0.019 | ||||
| 5 | 64 | –62 | 284 | 5.01 | 4.63 | 0.007 | 19 | 280–292 | |
| 6 | 34 | 61 | 264 | 4.40 | 4.13 | 0.044 | 2 | 264–264 | |
| High-level PE: pos. | 1 | 0 | –25 | 304 | 6.57 | 5.80 | 0.000 | 619 | 260–320 |
| 0 | –25 | 272 | 5.38 | 4.92 | 0.002 | ||||
| 2 | -4 | –68 | 148 | 4.90 | 4.54 | 0.010 | 41 | 140–156 | |
| High-level PE: neg. | 1 | –42 | –73 | 304 | 5.66 | 5.13 | 0.001 | 111 | 288–316 |
| 2 | -4 | 56 | 268 | 5.26 | 4.82 | 0.003 | 121 | 248–296 | |
| 3 | 42 | –78 | 104 | 4.52 | 4.23 | 0.034 | 6 | 104–108 | |
| 4 | –60 | –57 | 308 | 4.48 | 4.20 | 0.038 | 2 | 304–308 |
Significant clusters for main effects of stability and interactions stability × drug group across the two studies.
Columns are organized as in Table 1 of the main text.
| Effects of stability | cluster | [ms] | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A study 1: stable >volatile | 1 | 4 | –62 | 284 | 5.32 | 4.83 | 0.003 | 110 | 272–292 |
| 2 | 4 | –95 | 144 | 5.04 | 4.62 | 0.007 | 45 | 136–156 | |
| 3 | 0 | 61 | 148 | 4.76 | 4.39 | 0.017 | 14 | 144–148 | |
| B study 2: volatile >stable | 1 | 0 | 40 | 268 | 4.51 | 4.22 | 0.035 | 6 | 268–272 |
| 2 | 4 | 24 | 384 | 4.42 | 4.15 | 0.046 | 4 | 384–384 | |
| C study 2: PLA >LEV | 1 | –38 | –14 | 264 | 4.58 | 4.28 | 0.028 | 4 | 264–264 |
| D study 2: GAL > PLA | 1 | –30 | 61 | 204 | 4.73 | 4.40 | 0.018 | 7 | 200–208 |
Genetic effects and pharmaco-genetic interactions in study 1.
Columns are organized as in Table 1 of the main text. Mismatch ERPs in volatile periods were differentially modulated by CHAT polymorphism in the placebo versus the biperiden group, whereas mismatch ERPs overall and in stable periods were modulated by COMT polymorphism in the amisulpride group.
| Study 1: Genetic effects | cluster | [ms] | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A volatile mismatch: CHAT: PLA >BIP | 1 | –34 | –52 | 380 | 5.77 | 5.15 | 0.001 | 302 | 356–392 |
| B volatile mismatch: PLA: pos. effect of CHAT | 1 | –34 | –46 | 384 | 4.57 | 4.23 | 0.032 | 6 | 380–384 |
| C volatile mismatch: BIP: neg. effect of CHAT | 1 | –51 | –25 | 384 | 4.49 | 4.17 | 0.040 | 6 | 380–384 |
| D mismatch: AMI: pos. effect of COMT | 1 | 0 | –73 | 304 | 4.51 | 4.18 | 0.033 | 18 | 300–308 |
| E mismatch: AMI: neg. effect of COMT | 1 | 42 | –19 | 300 | 4.79 | 4.41 | 0.014 | 33 | 292–304 |
| F stable mismatch: AMI: neg. effect of COMT | 1 | 42 | –14 | 300 | 4.44 | 4.12 | 0.042 | 6 | 300–304 |
Genetic effects and pharmaco-genetic interactions in study 2.
Columns are organized as in Table 1 of the main text. Volatile mismatch ERPs in the placebo group were positively modulated by CHAT polymorphism, and negatively modulated by COMT polymorphism.
| Study 2: Genetic effects | cluster | [ms] | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A volatile mismatch: PLA: pos. effect of CHAT | 1 | –13 | –78 | 104 | 4.42 | 4.13 | 0.046 | 4 | 104–108 |
| B volatile mismatch: PLA: neg. effect of COMT | 1 | 4 | 45 | 192 | 5.31 | 4.84 | 0.003 | 53 | 180–200 |
Additional files
-
Supplementary file 1
Significant clusters of activation for a reduced sample size in study 1 (Table S1) and study 2 (Table S2).
Tables show the results for the main contrasts reported in the main text when excluding data sets due to lack of behavioral data or low performance in the visual distraction task.
- https://cdn.elifesciences.org/articles/74835/elife-74835-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/74835/elife-74835-transrepform1-v2.pdf





