Telomere length regulation by Rif1 protein from Hansenula polymorpha
Figures
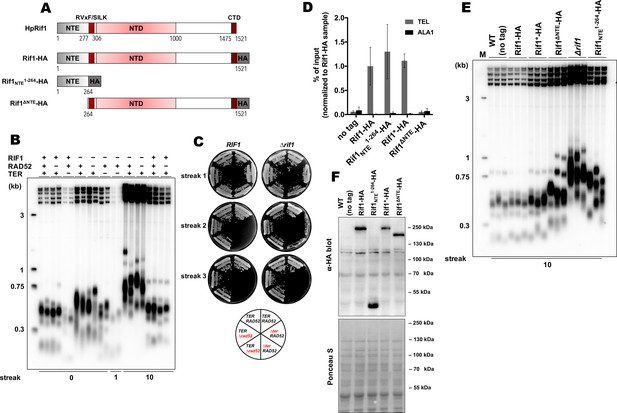
Rif1 regulates telomere length in H. polymorpha.
(A) Schematic illustration of the domain organization of the full length HpRif1 and HA-tagged Rif1 fragments expressed in H. polymorpha. (B) Southern blot analysis of terminal restriction fragments from the indicated mutant strains. Genomic DNA was isolated from the strains after the Nth streak (where N is a number under a lane; each streak is ~20 generations). ‘0’ streak – gDNA was isolated from the colonies on the transformation plate, without additional restreaks. ‘M’ – telomeric DNA containing fragments that served as markers of length (their sizes are indicated on the left of each blot). (C) Viability of the strains with the indicated genotypes was monitored during three serial restreaks on YPD agar plates, the plates were photographed after 2 days growth at 37 °C. (D) ChIP analysis. Chromatin from the indicated strains was immunoprecipitated on anti-HA magnetic beads. DNA was analyzed by qPCR with primers targeting either subtelomere region of the right end of chromosome VII (‘TEL’) or ALA1 gene locus (negative control, ‘ALA1’). The amount of DNA fragments in the IP samples as a percentage of the input DNA was calculated, the % of input of the ‘TEL’ Rif1-HA sample was set to 1. Error bars indicate SD, n = 3. ‘Rif1*-HA’ – the strain expressing the Rif1-HA protein but in a slightly different background (see Materials and methods). (E) Same as (B) but with different strains. (F) Western blot analysis of the total proteins isolated from the indicated strains using antibodies targeting HA epitope (upper panel). Ponceau S-stained membrane (lower panel) served as a loading control.
-
Figure 1—source data 1
Numerical data used to generate Figure 1.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig1-data1-v1.xlsx
-
Figure 1—source data 2
The original (raw unedited) gels/blots for Figure 1.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig1-data2-v1.zip
-
Figure 1—source data 3
The original (raw unedited) gels/blots for Figure 1.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig1-data3-v1.zip
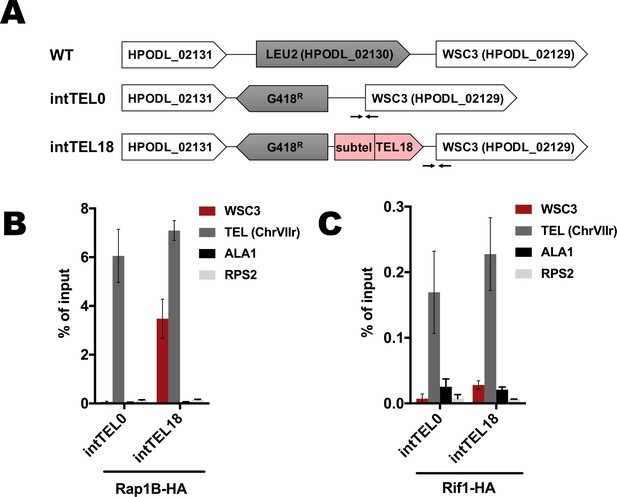
Binding of Rap1B and Rif1 at the internal telomere locus.
(A) Schematic illustration of the leu2 locus on the chromosome I in the WT, intTEL0 and intTEL18 strains. ‘TEL18’ – 18 telomeric repeats, ‘Subtel’ – 304 bp region of the subtelomeric DNA derived from HARS36 (see Materials and methods). Arrows correspond to qPCR primers targeting the WSC3 gene (located right next to the internal telomeric DNA). (B) ChIP analysis. Chromatin from the indicated strains was immunoprecipitated on anti-HA magnetic beads. DNA was analyzed by qPCR with primers targeting internal telomeric repeats/WSC3 gene locus (‘WSC3’), subtelomere region of the right end of chromosome VII (‘TEL’), ALA1 gene locus (negative control, ‘ALA1’) or RPS2 gene locus (negative control, ‘RPS2’). The amount of DNA fragments in the IP samples as a percentage of the input DNA was calculated (% of input). Error bars indicate SD, n = 3. (C) Same as (B) but with Rif1-HA-containing strains.
-
Figure 1—figure supplement 1—source data 1
Numerical data used to generate Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig1-figsupp1-data1-v1.xlsx
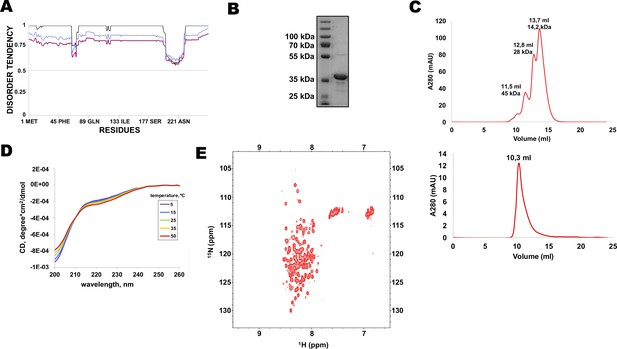
N-terminal extension of HpRif1 is intrinsically disordered.
(A) Disorder prediction by Metadisorder (Kozlowski and Bujnicki, 2012) for 1–264 fragment of HpRif1 using three different combinations of algorythms: MetaDisorder (black), MetaDisorderMD (purple), MetaDisorderMD2 (blue). All residues with the disorder probability over 0.5 are considered to be disordered. (B) An aliquot of the purified recombinant Rif1NTE1-264 was analyzed by SDS-PAGE and Coomassie staining. (C) Profiles of size-exclusion chromatography of the mixture of the standard proteins (upper profile) and the recombinant Rif1NTE1-264 (lower profile). (D) Circular dichroism spectra of the Rif1NTE1-264 recorded at 5 °C (purple), 15 °C (blue), 25 °C (green), 35 °C (yellow), and 50 °C (red). (E) Two-dimensional 1H-15N HSQC NMR spectrum of 15N-labeled Rif1NTE1-264 recorded at 25 °C.
-
Figure 2—source data 1
The original (raw unedited) gels/blots for Figure 2.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig2-data1-v1.zip
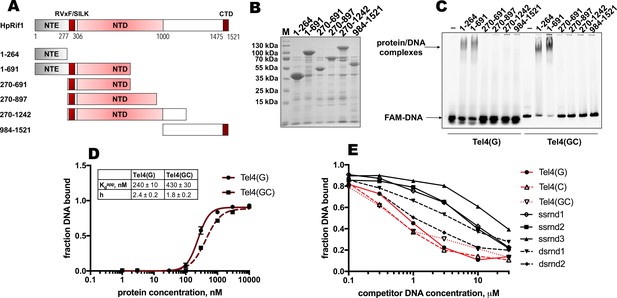
Rif1NTE1-264 binds DNA in vitro.
(A) Schematic illustration of the 6His-S-tagged Rif1 fragments expressed in E. coli and used in EMSA assays. (B) Aliquots of the Ni-NTA purified fragments were analyzed by SDS- PAGE and Coomassie staining. “M” – protein weight marker. (C) 5 μM of the indicated 6His-S-tagged HpRif1 fragments were subjected to the electrophoretic mobility shift assay, using 0.5 μM of either ss- (Tel4(G)) or ds- (Tel4(GC)) DNA oligonucleotide comprising four telomeric repeats as a probe. ‘–’ – no protein control. Positions of the free DNA and protein/DNA complexes are indicated by arrows. (D) Quantification of the titration EMSA experiment (two replicates) with increasing concentration of recombinant (tag-free) Rif1NTE1-264 (0, 1, 3, 10, 30, 100, 300, 1000, 3000, 10,000 nM) and 30 nM FAM-Tel4(G) (black circles) or FAM-Tel4(GC) (black squares) as probes (gels are shown in Figure 3—figure supplement 1A). The fits into the ‘Specific binding with Hill slope’ model are shown in dark red (FAM-Tel4(G) – solid curves; FAM-Tel4(GC) – dashed curves). The best-fit values for Kd apparent and Hill coefficient are shown. (E) Quantification of the competition EMSA experiment (the correspondent gels are shown in Figure 3—figure supplement 1B) with 30 nM FAM-Tel4(G), 1 μM (tag-free) Rif1NTE1-264 and increasing concentration of competitor DNA oligonucleotides (0.1, 0.3, 1, 3, 10, 30 μM). The sequences of the competitors are in Table 1. We note that competition with the Tel4(C) oligo may be difficult to interpret since it may first titrate out the FAM-Tel4(G) probe.
-
Figure 3—source data 1
Numerical data used to generate Figure 3.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig3-data1-v1.xlsx
-
Figure 3—source data 2
The original (raw unedited) gels/blots for Figure 3.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig3-data2-v1.zip
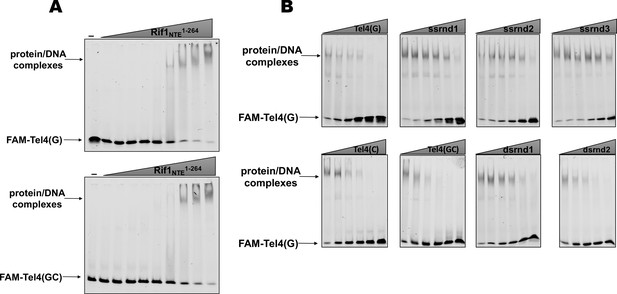
EMSA experiments.
(A) The titration EMSA experiment with increasing concentration of recombinant (tag-free) Rif1NTE1-264 (0, 1, 3, 10, 30, 100, 300, 1000, 3000, 10,000 nM) and 30 nM FAM-Tel4(G) (upper gel) or FAM-Tel4(GC) (lower gel) as probes. (B) Competition EMSA with 30 nM FAM-Tel4(G), 1 μM (tag-free) Rif1NTE1-264 and increasing concentration of the indicated competitor DNA oligonucleotides (0.1, 0.3, 1, 3, 10, 30 μM).
-
Figure 3—figure supplement 1—source data 1
The original (raw unedited) gels/blots for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig3-figsupp1-data1-v1.zip
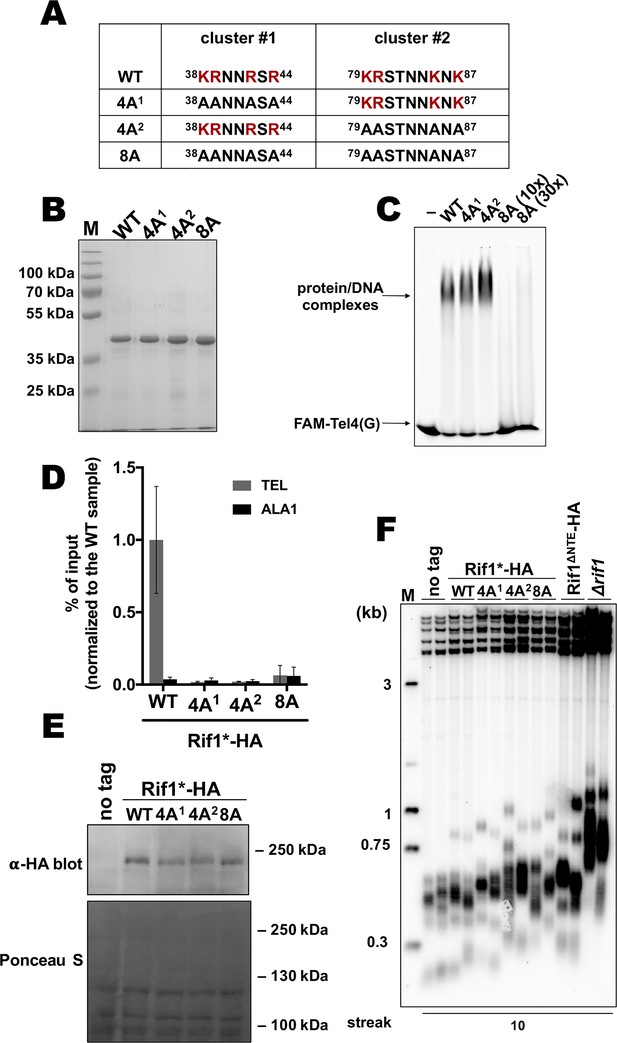
Two K/R clusters within Rif1 are important for its telomeric localization.
(A) Sequences of the two clusters enriched in positively charged residues in the wild type and mutant versions of HpRif1. (B) Aliquots of the Ni- NTA purified wild type (‘WT’) and mutant 1–264 fragments of HpRif1 were analyzed by SDS-PAGE and Coomassie staining. ‘M’ – protein weight marker. (C) 10 μM of the indicated 6His-S-tagged proteins were subjected to EMSA, using 1 μM of ss- oligonucleotide comprising four telomeric repeats (FAM-Tel4(G)) as a probe. ‘–’ – no protein control. ‘30x’ – 30 μM of the 8 A mutant protein. Positions of the free DNA and protein/DNA complexes are indicated by arrows. (D) ChIP analysis of the indicated strains, same as in Figure 1D; the % of input of the ‘TEL’ Rif1*-HA WT sample was set to 1. Error bars indicate SD, n = 3. (E) Western blot analysis of the total proteins isolated from the indicated strains using antibodies targeting HA epitope (upper panel). Ponceau S-stained membrane (lower panel) served as a loading control. (F) Southern blot analysis as in Figure 1B but with different strains.
-
Figure 4—source data 1
Numerical data used to generate Figure 4.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig4-data1-v1.xlsx
-
Figure 4—source data 2
The original (raw unedited) gels/blots for Figure 4.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig4-data2-v1.zip
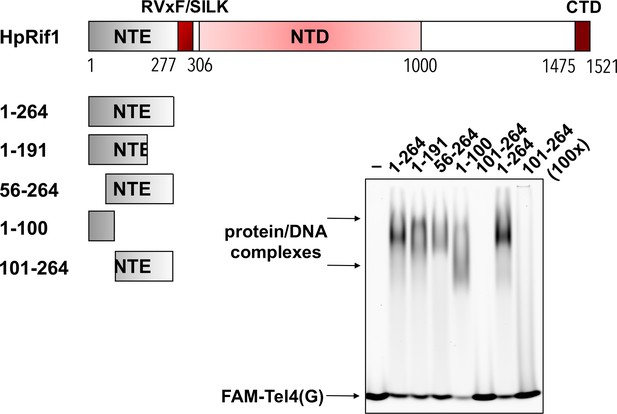
Rif1NTE truncation constructs were purified from E.
E. coli as 6His-S- fusions. 10 μM of the indicated fragments were subjected to the EMSA using 1 μM of FAM-Tel4(G) probe. ‘–’ – no protein control. ‘100 x’ – 100 μM of the protein used in the reaction.
-
Figure 4—figure supplement 1—source data 1
The original (raw unedited) gels/blots for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig4-figsupp1-data1-v1.zip
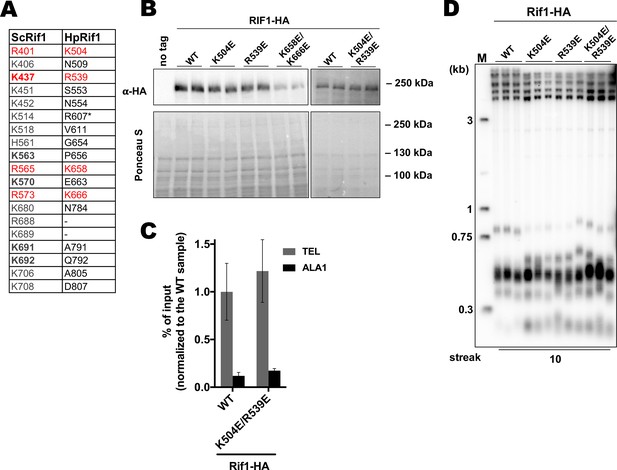
Analysis of the HpRif1 residues corresponding to the DNA-contacting residues from ScRif1 NTD.
(A) List of the ScRif1NTD residues potentially involved in the DNA binding (‘ScRif1’) and corresponding residues in the HpRif1 (‘HpRif1’) (alignment in the Supplementary file 1). ScRif1 residues which were confirmed to be important for telomeric function are in bold. Conserved residues are marked by red. * – different HpRif1 residues correspond to ScRif1 K514 according to alignments with different algorithms. (B) Anti-HA western blot analysis (upper panel). Ponceau S-stained membrane (lower panel) served as a loading control. (C) ChIP analysis of the indicated strains, same as in Figure 1D. (D) Southern blot analysis as in Figure 1B but with different strains.
-
Figure 4—figure supplement 2—source data 1
Numerical data used to generate Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig4-figsupp2-data1-v1.xlsx
-
Figure 4—figure supplement 2—source data 2
The original (raw unedited) gels/blots for Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig4-figsupp2-data2-v1.zip
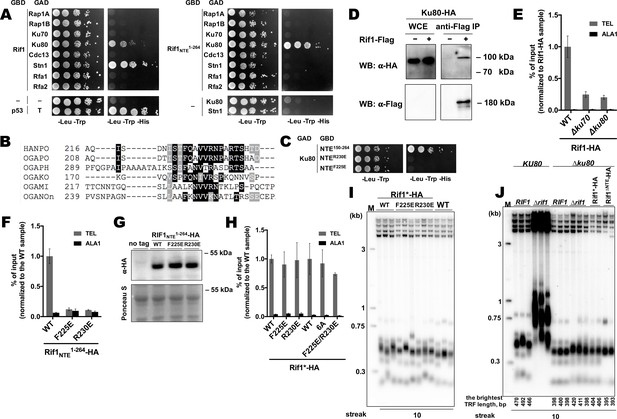
Rif1 interacts with Ku80 in H.polymoprha.
(A) Y2H analysis. AH109 colonies expressing pairs of the indicated proteins (fused to either Gal4-BD (GBD) or Gal4-AD (GAD)); cultures with A600 ~0.5 and four 10-fold serial dilutions were plated on the SC medium lacking amino acids as indicated, and incubated at 30°С for 4 days. “T” – SV40 large T antigen. (B) A fragment of the alignment of the NTE regions from H. polymoprha DL-1 (HANPO) and five of its closest relatives. Full alignment is in Supplementary file 2. (C) Y2H analysis as in (A), 'NTER230’ and ‘NTEF225E’ – 150–264 fragments of Rif1 with the R230E and F225E mutations, respectively. (D) Co-IP analysis. IP on the anti-Flag resin. The amount of tagged proteins in whole cell extracts (WCE) and the IP samples (IP) was monitored by Western blot (WB) using anti-Flag and anti-HA antibodies. The IP experiment was performed in the presence of benzonase nuclease. (E, F) ChIP analysis of the indicated strains, same as in Figure 1D; the % of input of the ‘TEL’ Rif1-HA WT sample was set to 1 (E); the % of input of the ‘TEL’ Rif1NTE1-264-HA WT sample was set to 1 (F). (G) Western blot analysis. Same as in Figure 1F, but with different strains. (H) ChIP analysis of the indicated strains, same as in Figure 1D; Error bars indicate SD, n = 3. ‘6 A’ mutation: 225FQAVVR230/225AAAAAA230. (I, J) Southern blot analysis as in Figure 1B (J) Mean lengths of the brightest TFR bands are: RIF1KU80 476 bp, RIF1∆ku80 399 bp. WT telomere length reported to be ~160 bp (~20 telomeric repeats, Sohn et al., 1999), therefore telomere length is reduced by ~50% in the knockout strain.
-
Figure 5—source data 1
Numerical data used to generate Figure 5.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig5-data1-v1.xlsx
-
Figure 5—source data 2
The original (raw unedited) gels/blots for Figure 5.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig5-data2-v1.zip
-
Figure 5—source data 3
The original (raw unedited) gels/blots for Figure 5.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig5-data3-v1.zip
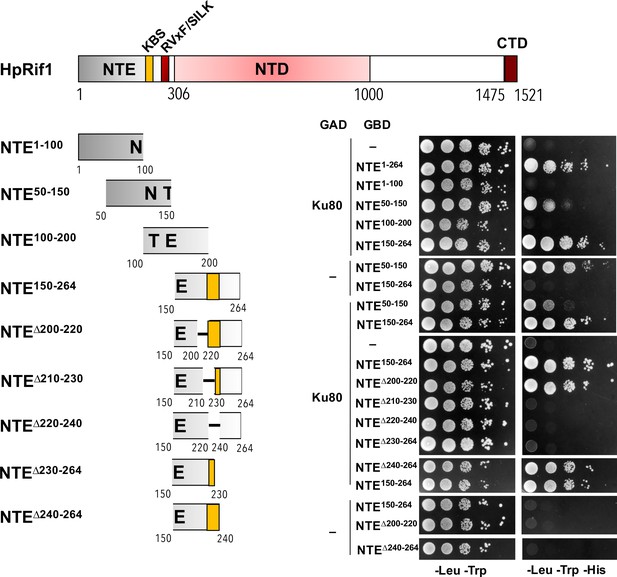
Y2H analysis.
AH109 colonies expressing pairs of the indicated proteins (fused to either Gal4-BD [GBD] or Gal4-AD [GAD]); cultures with A600 ~0.5 and four 10-fold serial dilutions were plated on the SC medium lacking amino acids as indicated, and incubated at 30°С for 4 days. KBS – Ku80 binding site.
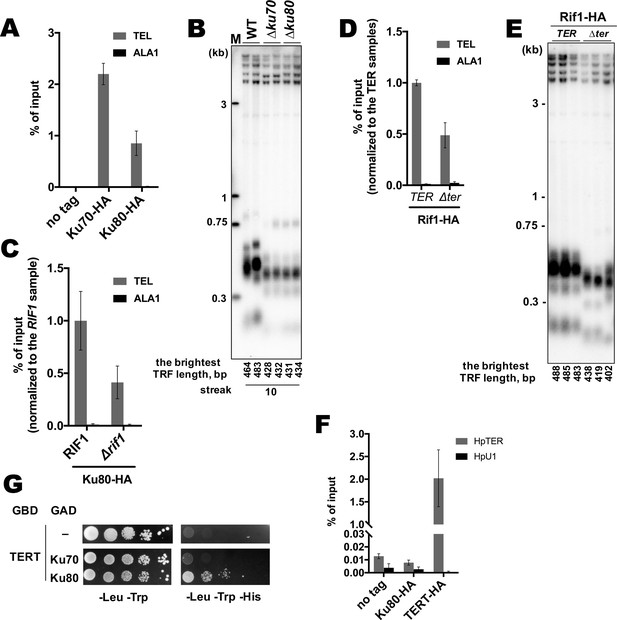
Additional experiments on the Rif1-Ku interaction.
(A) Anti-HA ChIP analysis of the indicated strains, same as in Figure 1D. The amount of DNA fragments in the IP samples is represented as a percentage of the input DNA (mean ± SD, n = 3). (B) Southern blot analysis as in Figure 1B but with different strains. Mean lengths of the brightest TFR bands are: WT 474 bp, ∆ku70 430 bp, ∆ku80 433 bp. WT telomere length reported to be ~160 bp (~20 telomeric repeats, Sohn et al., 1999), therefore telomere length is reduced by ~25% in each knockout strain. (C, D) Anti-HA ChIP analysis same as (A) (E) Southern blot analysis (as in Figure 1B) of the strains analyzed by ChIP in (D). Mean length of the brightest TFR bands are: TER 485 bp, ∆ter 420 bp (telomere length is reduced by ~40% in the ∆ter strains compared to TER). (F) RNA co-precipitated from the indicated strains on anti-HA agarose was analyzed by qRT-PCR targeting HpTER or snU1 RNA (HpU1). The amount of RNA fragments in the IP samples is represented as a percentage of the input RNA (mean ± SD, n = 3). (G) Y2H analysis. cultures with A600 ~0.5 and four 10-fold serial dilutions were plated on the SC medium lacking amino acids as indicated, and incubated at 30°С for 4 days.
-
Figure 5—figure supplement 2—source data 1
Numerical data used to generate Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig5-figsupp2-data1-v1.xlsx
-
Figure 5—figure supplement 2—source data 2
The original (raw unedited) gels/blots for Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig5-figsupp2-data2-v1.zip
-
Figure 5—figure supplement 2—source data 3
The original (raw unedited) gels/blots for Figure 5—figure supplement 2.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig5-figsupp2-data3-v1.zip
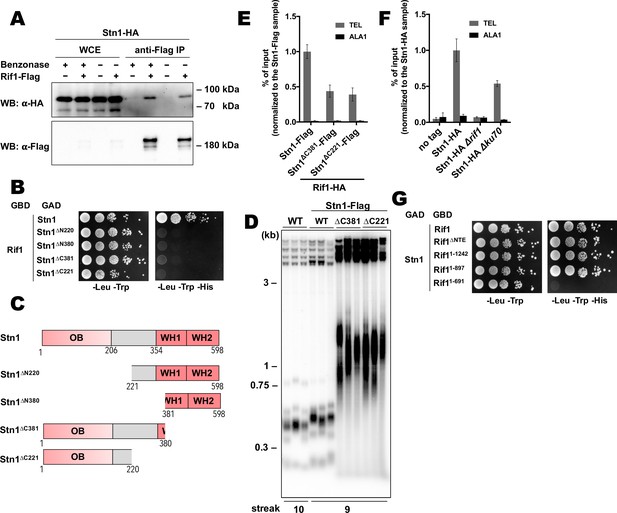
Rif1 recruits Stn1 at H.polymorpha telomeres.
(A) Co-IP analysis. Same as in Figure 5I, but with different strains; the IP experiment was performed either with or without benzonase treatment as indicated. (B) Y2H analysis. Same as in in Figure 5A, but with different protein pairs. (C) Schematic illustration of the domain organization of the full-length HpStn1 and its truncation variants used in this study. (D) Southern blot analysis as in Figure 1B but with different strains. (E, F) ChIP analysis of the indicated strains, same as in Figure 1D (IP on the anti-HA beads); the % of input of the ‘TEL’ Rif1-HA/Stn1-Flag sample was set to 1. Error bars indicate SD, n = 3. (E) Or the % of input of the ‘TEL’ Stn1-HA sample was set to 1 (F). (G) Y2H analysis. Same as in in Figure 5A, but with different protein pairs.
-
Figure 6—source data 1
Numerical data used to generate Figure 6.
- https://cdn.elifesciences.org/articles/75010/elife-75010-fig6-data1-v1.xlsx
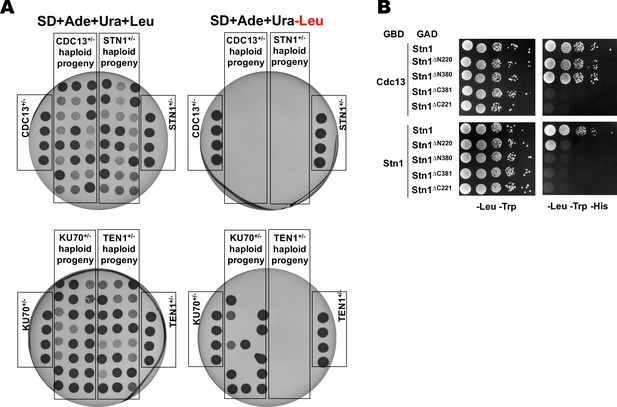
Cdc13, Stn1 and Ten1 are essential for viability in H. polymorpha.
(A) Results of the analysis of random spores derived from the indicated heterozygous strains. After ether treatment and growth on YPD plate, 24 colonies derived from each heterozygous strain were randomly picked, resuspended in water, plated onto the indicated selective plates, grown for 4 days at 37°C and photographed. Each plate also contained the parental heterozygous strains (phenotype: Ade+Ura+Leu+). None of the spores from strains CDC13+/-; STN1+/- and TEN1+/- displayed the Leu+ phenotype, whereas 50% (12/24) of the spores from the KU70+/- had the Leu+ phenotype (as expected for the non-essential KU70 gene). (B) Y2H analysis. AH109 colonies expressing pairs of the indicated proteins (fused to either Gal4-BD (GBD) or Gal4-AD (GAD)); cultures with A600 ~0.5 and four 10-fold serial dilutions were plated on the SC medium lacking amino acids as indicated, and incubated at 30°С for 4 days.
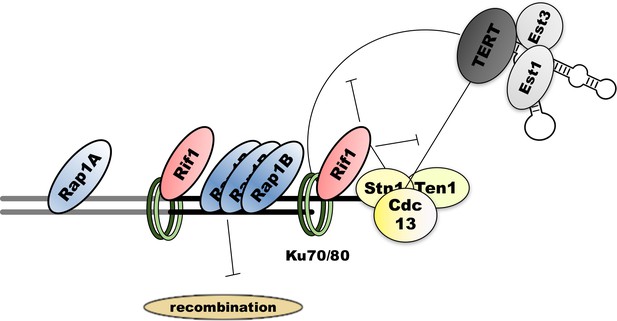
Schematic representation of the putative protein-protein interactions at H.polymorpha telomeres.
Double thick line represents subtelomeric (gray) and telomeric (black) DNA. Thin lines denote interactions, blunt arrows – inhibitory effect.
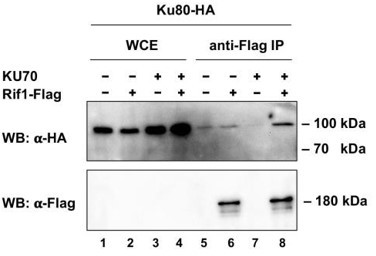
Co-IP analysis.
Rif1-Flag protein was immunoprecipitated from the indicated strains on the anti-Flag resin. The amount of tagged proteins in whole cell extracts (WCE) and IP samples (IP) was monitored by Western blot (WB) using anti-Flag and anti-HA antibodies. The IP experiment was performed in the presence of benzonase nuclease.
Tables
Oligonucleotides used in the EMSA experiments.
| Oligo name | Sequence (5'–3') |
|---|---|
| Tel4(G) | GGGTGGCGGGGTGGCGGGGTGGCGGGGTGGCG |
| Tel4(C) | CGCCACCCCGCCACCCCGCCACCCCGCCACCC |
| Tel4(GC) | annealed from Tel4(G) and Tel4(C) |
| ssrnd1 | ACGACTCACTGTAGATACGACTCACTGTAGAT |
| ssrnd2 | ATCTACAGTGAGTCGTATCTACAGTGAGTCGT |
| ssrnd3 | AAATCTAGACATGAAAAAAAAAATGTTAGTAATCGAAATCTC |
| dsrnd1 | annealed from ssrnd1 and ssrnd2 |
| dsrnd2antisense | GAGATTTCGATTACTAACATTTTTTTTTTCATGTCTAGATTT |
| dsrnd2 | annealed from ssrnd3 and dsrnd2antisense |
Additional files
-
Supplementary file 1
Multiple alignment of Rif1 homologues from budding yeasts.
- https://cdn.elifesciences.org/articles/75010/elife-75010-supp1-v1.docx
-
Supplementary file 2
Multiple alignment of Rif1NTE regions from H. polymoprha DL-1 and five of its closest relatives.
- https://cdn.elifesciences.org/articles/75010/elife-75010-supp2-v1.docx
-
Supplementary file 3
H. polymorpha strains used in this study.
- https://cdn.elifesciences.org/articles/75010/elife-75010-supp3-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/75010/elife-75010-transrepform1-v1.docx





