The Natural History of Model Organisms: The biology of C. richardii as a tool to understand plant evolution
- Article
- Figures and data
- Abstract
- Introduction
- The variable natural history of Ceratopteris
- A transgenic model for seed-free plants
- The environmental influence on development
- Systematics and hybridization
- Plant genome structure and evolution
- Conclusion
- Data availability
- References
- Decision letter
- Author response
- Article and author information
- Metrics
Abstract
The fern Ceratopteris richardii has been studied as a model organism for over 50 years because it is easy to grow and has a short life cycle. In particular, as the first homosporous vascular plant for which genomic resources were developed, C. richardii has been an important system for studying plant evolution. However, we know relatively little about the natural history of C. richardii. In this article, we summarize what is known about this aspect of C. richardii, and discuss how learning more about its natural history could greatly increase our understanding of the evolution of land plants.
Introduction
The genus Ceratopteris has a long and complicated taxonomic history. It was first described by Linnaeus under the genus Acrostichum (Linnaeus, 1764), and the name Ceratopteris was later assigned by Brongniart (Brongniart, 1821). Since then, Ceratopteris has been placed in a number of different families, with the number of species within the genus ranging between one and twelve (Lloyd, 1974). Today it is placed within Pteridaceae, one of the largest and most diverse fern families (PPG, 2016; Figure 1).
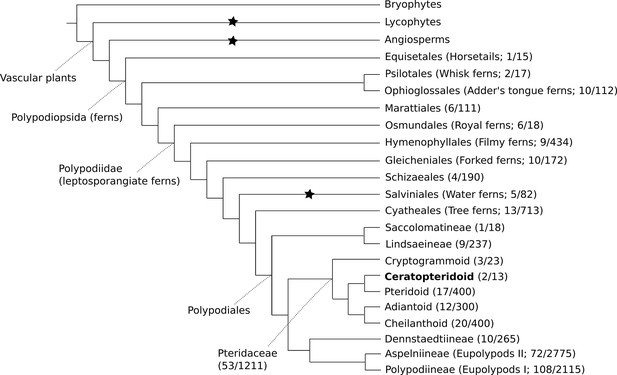
Cartoon phylogeny of land plants.
Within the Polypodiopsida (ferns), the estimated number of genera and species (genera/species) are noted for each major group. Black stars show the three extant independent evolutions of heterospory (Bateman and DiMichele, 1994). In the family Pteridaceae, the five major groups are shown, including the Ceratopteridoid clade (in bold) which includes the genera Acrostichum and Ceratopteris. Within Ceratopteris, there are about ten species found throughout the world’s tropics (see Figures 2 and 3).
There are about ten species within Ceratopteris, which can be found throughout the tropics (Figure 2; Masuyama and Watano, 2010; Zhang et al., 2020; Yu et al., 2021). The classification of these species was made difficult by their inconsistent morphologies, and molecular methods were needed to reconstruct a backbone phylogeny for the genus (Adjie et al., 2007; Kinosian et al., 2020a). Recent work has shown that cryptic and hybrid species may be quite common in Ceratopteris, warranting a more rigorous evaluation of the relationships between species in the genus (e.g., Kinosian et al., 2020b).
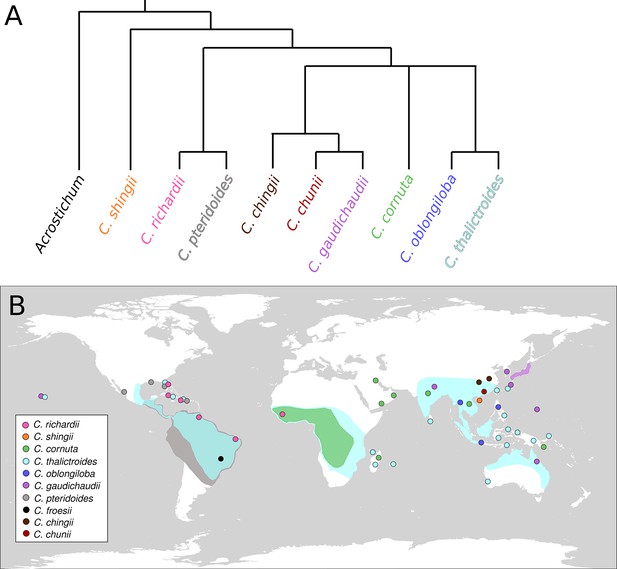
Phylogenetic and geographic representation of the genus Ceratopteris.
(A) Phylogenetic reconstruction of Ceratopteris, with the sister genus Acrostichum as the outgroup, based on Kinosian et al., 2020a; Adjie et al., 2007; Yu et al., 2021 Zhang et al., 2020. Absent from this phylogeny is the Brazilian species C. froesii, for which no genetic sequence data is available. (B) Distribution map of ten Ceratopteris species: pink, C. richardii; orange, C. shingii; green, C. cornuta; light blue, C. thalictroides; dark blue, C. oblongiloba; purple, C. gaudichaudii; grey, C. pteridoides; black, C. froesii; brown, C. chingii, and red C. chunii. Shaded areas show where a species is common, colored dots show where species occur outside their most common range, or multiple species are found in a small area. Location data from Kinosian et al., 2020a; Lloyd, 1974; Masuyama and Watano, 2010; Zhang et al., 2020; Yu et al., 2021 https://gbif.org/.
Ceratopteris richardii was first developed as a model system for ferns in the 1960s and 70s, primarily because it was easy to grow in the lab and had a short life cycle (Figure 2; Pal and Pal, 1962; Pal and Pal, 1963; Klekowski, 1970; Stein, 1971; Hickok, 1973; Hickok and Klekowski, 1973; Lloyd and Warne, 1978). Many studies used spores from a Cuban vouchered collection, now known as the Hnn strain or C-fern (Hickok, 1977). Additional strains of C. richardii and the species C. thalictroides and C. pteridoides have since been developed (Hickok and Klekowski, 1974; Nakazato et al., 2006; Muthukumar et al., 2013). In the past few decades, Ceratopteris has become an important model in the study of sex determination (Eberle et al., 1995; Ganger et al., 2019; Atallah et al., 2018; Banks, 1997), apogamy (Bui et al., 2017; Cordle et al., 2010), genome structure (Nakazato et al., 2006; Baniaga and Barker, 2019), hybridization (Hickok and Klekowski, 1974; Adjie et al., 2007), reproductive barriers (Nakazato et al., 2007), developmental biology (Hou and Hill, 2002; Conway and Di Stilio, 2020; Sun and Li, 2020; Aragón-Raygoza et al., 2020), and transgenic studies in ferns (Plackett et al., 2018; Bui et al., 2015; Cannon et al., 2018).
Ceratopteris richardii is one of few spore-bearing model systems (e.g., Physcomitrella: Cove, 2005; Rensing et al., 2008; Selaginella: Banks et al., 2011) and the only vascular spore-bearing homosporous model species, making it a critical evolutionary lineage in comparative studies (see Box 1 for a glossary of terms used in this article). Compared to other model plants, Ceratopteris has a large genome (~11 GB) and high base chromosome number (n = 39), which has partly caused genetic resources for C. richardii to lag behind those of other plant model systems, delaying such comparative work.
Glossary
Homosporous: Plants that produce one type of spore, which germinates into a gametophyte capable of producing both eggs and sperm. This group comprises most ferns, and some lycophytes, and all non-vascular plants.
Heterosporous: Plants that produce separate spores, which produce sperm and eggs respectively. This includes all seed plants, as well as a few lineages of ferns and lycophytes.
Apomixis: A form of asexual reproduction in plants. It can proceed by ‘apogamy’, where unreduced spores germinate into gametophytes from which sporophytic tissue can grow without fertilization; the alternative is ‘apospory’, where no spore is produced and a gametophyte grows directly from the parent sporophytic tissue.
Sporophyte: The diploid generation of plants that produce spores. In mosses, this generation is dependent on the gametophyte. In ferns, lycophytes, and seed plants this generation is independent.
Gametophyte: The haploid generation in plants that produce gametes. They can be ‘archegoniate’ (having archegonia that produce eggs) or ‘antheridiate’ (having antheridia that produce sperm), or be ‘hermaphroditic’ (producing both egg and sperm). Gametophytes are free-living in mosses, ferns, and lycophytes, but dependent on the sporophyte generation in seed plants.
Sporangium: The structure in plants that create a spore. In ferns, these are found on the underside of a leaf, often grouped in small clusters called sori.
Leptosporangiate fern: One of the major lineages of ferns, in the subclass Polypodiidae. These ferns have sporangia with long stalks that produce (typically) 64 spores, all derived from a single initial cell (PPG, 2016).
Recently, however, the first draft genome sequence for C. richardii was published (Marchant, 2019a), which was the first for a homosporous fern. Ceratopteris richardii was chosen for sequencing because, although it has a large genome compared to other plants, it is relatively small for a homosporous fern (Sessa et al., 2014; Marchant et al., 2019b).
Having a reference genome for C. richardii expands its research potential and builds on decades of previous work. A homosporous plant genome provides the opportunity for exploring and comparing various aspects of plant biology such as the alternation of generations, sex determination, and reproductive modes between heterosporous and homosporous plants. In addition, a reference genome for Ceratopteris is beneficial for the development of new markers in targeted gene sequencing or whole-genome resequencing. In turn, this makes incorporating wild collections into genomic research much easier and will help us gain a more nuanced understanding of the biology, ecology, and evolutionary history of Ceratopteris.
The variable natural history of Ceratopteris
The model species Ceratopteris richardii originates in the Caribbean and Western Africa, and grows rooted or floating in shallow water (Figure 2). Indeed, all species within Ceratopteris grow in or near areas in the tropics that become inundated seasonally (Figures 2 and 3), mostly growing in fresh water, though they can tolerate salt water (Lloyd, 1974; Warne and Hickok, 1987).
Its sister genus, Acrostichum, is well-known for being able to tolerate high levels of salt as it grows in tidal and intertidal habitats (Zhang et al., 2013; Medina et al., 1990). The extent of natural salt tolerance in Ceratopteris is not fully understood, but salt-tolerant mutants of C. richardii are easy to generate in the lab (Chasan, 1992; Warne et al., 1995). Continuing to study salt tolerance in Ceratopteris may be beneficial in understanding the genetic mechanisms of this trait, or applying such findings to crop systems in the future.

Morphological diversity of four Ceratopteris species.
All photos by SPK unless credited otherwise. (A) Ceratopteris thalictroides mature plant from Townsville, Australia. (B) Detail of a sterile C. thalictroides leaf with buds. (C) Detail of a fertile C. thalictroides leaf. (D) Leaf bases and fiddleheads of C. thalictroides. (E, F) Ceratopteris pteridoides in cultivation at the Dr. Cecilia Koo Botanic Conservation Center in Taiwan. (G) Vegetative buds on C. pteridoides (photo by Christopher Haufler). (H) Ceratopteris gaudichaudii in Cairns, Australia. (I) Ceratopteris richardii in cultivation (photo by David Randall). (J) Ceratopteris richardii gametophytes (photo by Jo Ann Banks).
Through much of its range, Ceratopteris inhabits ephemeral water sources. To reproduce in this fleeting habitat C. richardii has a short life cycle of about 120 days (Stein, 1971), which is much shorter than almost all ferns’ annual or multi-year life cycles. The fern life cycle, like that of all land plants, is characterized by the alternation of generations between the diploid sporophyte and haploid gametophyte. Following fertilization, a diploid zygote is formed that is temporarily reliant on the gametophyte and becomes self-sufficient over time. In the fern life cycle, the gametophyte and sporophyte generations are completely separate at certain stages. Comparatively, in bryophytes, the sporophyte generation is entirely dependent on the gametophyte, and the opposite is true in seed plants. The fern life cycle provides an opportunity to study sporophyte and gametophyte generation separately, something that is not possible in other lineages of plants.
Because ferns have independent and free-living sporophytic and gametophytic phases, there are multiple ways in which the life cycle of Ceratopteris can proceed (Haufler et al., 2016). Fertilization can occur via gametes from different plants (sporophytic outcrossing), gametes from the same plant but different gametophytes (gametophytic outcrossing), and also gametes from the same gametophyte (gametophytic selfing). Studies show evidence of outcrossing within (Nakazato et al., 2007) and among species of Ceratopteris (i.e., hybridization, Adjie et al., 2007; Kinosian et al., 2020a; Hickok and Klekowski, 1974).
Outcrossing and gametophytic sex expression in ferns is often controlled by pheromones known as antheridiogens. These gibberellin-related chemicals are released by early-germinating archegoniate gametophytes to promote antheridiate gametophyte development in immature gametophytes. This pheromone system confers some of the benefits of heterospory to homosporous plants, namely outcrossing (Bateman and DiMichele, 1994; Hornych et al., 2021). In the case of Ceratopteris, however, solitary gametophytes can also become bisexual and self-fertilize; therefore, theoretically, only one spore can colonize new habitats (Schedlbauer and Klekowski, 1972). This is the case for many ferns and aids Ceratopteris as a model organism because studies can be designed around this flexible life history.
In addition, asexual reproduction (apomixis) can be induced in C. richardii in the lab (Cordle et al., 2007). The variation within the life cycle of Ceratopteris makes it a powerful system in which to study reproduction in ferns, as well as an evolutionary point of reference for understanding reproduction in seed plants.
Ceratopteris was developed as a model organism for many of the same reasons as other model systems. It is easy to grow in a lab setting, has a rapid life cycle that makes experiments tangible, and is tractable for genetic transformations (Eberle et al., 1995; Hickok et al., 1995). Model organisms are often chosen for convenience, but that can make them poor representative taxa (Alfred and Baldwin, 2015).
In the case of Ceratopteris, it has several traits that are very unusual among ferns. For example, it is one of only a handful of semi-aquatic species out of around 12,000 extant ferns (PPG, 2016). The life cycle of Ceratopteris, while beneficial for lab experiments, is incredibly short for a fern (Stein, 1971). Finally, it has half or a quarter of the number of spores typical for a leptosporangiate fern: most leptosporangiate ferns produce 64 spores per sporangium, whereas species in Ceratopteris produce 32 or 16 spores per sporangium (Lloyd, 1974). This is important because a spore number of 32 or 16 is often indicative of apogamy (Grusz, 2016), but no natural apogamous taxa have been described in Ceratopteris. These characteristics, among others, make Ceratopteris a good model species but not necessarily an accurate representation of all ferns.
A transgenic model for seed-free plants
Free-living generations and a flexible life cycle make Ceratopteris an important model for evolutionary developmental studies. A reference ontogenetic framework for the Hnn strain of C. richardii (C-fern; Hickok et al., 1995) was recently published, detailing the development of the gametophyte and sporophyte, providing an important reference for future work (Conway and Di Stilio, 2020). This reference, in combination with stable transformation techniques, plus a C. richardii transcriptome (Geng et al., 2021; Atallah et al., 2018) and genome (Marchant et al., 2022; Marchant et al., 2019b) now provide the necessary suite of tools for comparative work.
In the Hnn strain of Ceratopteris richardii stable transgenic lines have been established in both the gametophyte and sporophyte generations. Transformation of the tissue in these plants has been accomplished by bombardment of sporophytic tissue by tungsten microparticles (Plackett et al., 2014; Plackett et al., 2015a), Agrobacterium infection of haploid gametophyte tissue (Bui et al., 2015), and agrobacterium infection of haploid spores (Muthukumar et al., 2013).
Transformation on gametophytes provides an important perspective on gene function in the haploid generation, something that is not as easy in seed plants. Another benefit of working with transformed gametophytes is that they can self-fertilize to produce sporophytes that are stable homozygotes. However, this can also be accomplished if transformation is done on sporophytes. Spores can be collected and screening of resulting gametophytes can then be used to produce stable homozygous transgenic lines via gametophytic selfing.
Thus, transgenic lines of C. richardii have been developed, and allow for comparative studies of gene function and evolution across land plants in both the gametophyte and sporophyte generations. The ability to have transgenic lines in both generations provides a unique perspective for studying how genes, growth conditions, or other factors affect sporophytes and gametophytes differently.
A recent study using both transgenic gametophytes and sporophytes of Ceratopteris investigated the role of the LEAFY transcription factor (LFY) in development (Plackett et al., 2018). LFY is important for cell division in moss embryos (Tanahashi et al., 2005) and angiosperm floral meristem development (Carpenter and Coen, 1990). While it is known to be important in both of these lineages, there is no functional overlap between mosses and angiosperms, so understanding the evolutionary history of LFY has been challenging.
Ceratopteris provides an evolutionary and functional midpoint with which to study the role of the LFY gene in development. Using several transgenic lines of C. richardii, Plackett et al. evaluated the role of LFY in sporophyte development. They also reported, for the first time in any land plant, that LFY function is required for C. richardii gametophyte development (Plackett et al., 2018). This suggests that LFY was important for gametophyte development for the last common ancestor of ferns and seed plants, but this function was lost in seed plants where the gametophyte is greatly reduced (Plackett et al., 2018). Incorporating other vegetative developmental systems into further studies with C. richardii (e.g., Vasco et al., 2013; Vasco et al., 2016; Hernández-Hernández et al., 2021) will help us gain a more nuanced understanding of these processes across land plants.
Similarly, there is a growing body of work on the developmental patterns associated with reproduction using Ceratopteris. It is well-established that apogamy can be induced in C. richardii (Cordle et al., 2007), but the genetic mechanism responsible was unknown until recently. In flowering plants, BABY-BOOM (BBM) genes promote somatic embryogenesis (Boutilier et al., 2002; Soriano et al., 2013). These genes are absent in non-seed plants, but an ortholog of the BBM gene AINTEGUMENTA was identified in C. richardii (Bui et al., 2017). Using Agrobacterium-mediated transformations, Bui et al., created C. richardii gametophytes with both over- and knockdown-expression of the apogamy-inducing gene, CrANT. This was the first such study conducted in a non-seed plant and provides evidence for conserved gene function in apomixis across land plant lineages (Bui et al., 2017).
Future work using transgenic lines of Ceratopteris richardii has the potential to connect gene expression and function across land plants. Work has already been done on many gene families in model bryophytes, the lycophyte Selaginella, and seed plants. As mentioned above, however, there is not always functional overlap between these lineages. Including Ceratopteris may provide such a functional or developmental link. It is important to note that ferns like Ceratopteris are an independent lineage with unique development characteristics that have continued to evolve since diverging from other land plants, as are bryophytes and seed plants (Plackett et al., 2015b; McDaniel, 2021). However, Ceratopteris does share many characteristics with bryophytes (e.g., spores, independent gametophyte generation) and seed plants (e.g., vasculature, independent sporophyte generation), which make it a key lineage to include in comparative work.
Many model and non-model plants have recently established CRISPR/Cas9 gene-editing systems. Developing CRISPR in Ceratopteris is promising because C. richardii (and the Hnn strain) is diploid with a short life cycle and homozygotes can be easily created in one generation of gametophytic selfing (Shan et al., 2020). One potential avenue of study with CRISPR could be the apogamy pathway in C. richardii (Bui et al., 2017), investigating the connection between apogamy and spore number in C. richardii. This species produces 16 spores per sporangium, a number often indicative of apogamy in ferns (Grusz, 2016); C. richardii reproduces sexually but apogamy can be easily induced. If a CRISPR system could be established in Ceratopteris, one might be able to extend such technology to other members of the Pteridaceae known for apogamy (Grusz et al., 2021; Grusz, 2016; Grusz et al., 2009), or for application in other non-model ferns.
The environmental influence on development
Ceratopteris richardii is a well-known model system for studying sex determination (Banks, 1997) and the alternation of generations in homosporous plants (Eberle et al., 1995). However, there are some steps in its life cycle that are poorly understood. Almost nothing is known about how Ceratopteris gametophytes are established in the wild. It is unknown if Ceratopteris spores must germinate on soil, or if they can germinate and establish gametophytes in standing or slow-moving water.
Recently the effect of soil bacteria on sex determination in C. richardii was investigated for the first time (Ganger et al., 2019). In the presence of a soil bacterium, there were more hermaphroditic (compared to antheridiate) gametophytes as well as increased growth (Ganger et al., 2019). Ceratopteris uses an antheridiogen pheromone system to control sex determination (Scott and Hickok, 1987; Banks, 1997), and soil bacteria may be influencing sex determination in a similar way. Additional experiments would benefit our understanding of how natural conditions might affect gametophyte establishment and sex determination in Ceratopteris, outside of the known role of antheridiogens. The establishment of new plants is particularly important as climate change is a threat to the current habitat of Ceratopteris, both as sea levels rise and rainfall becomes less predictable.
In addition to the establishment of new plants, climate change may influence the morphology, ecology, and physiology of Ceratopteris. There is dramatic variation in frond morphology within Japanese C. thalictroides based on the growing season length (Masuyama, 1992); such intraspecies variety has not been systematically characterized in any other species in the genus. This is important to understand because leaves have been used by some authors as the primary method of identification in Ceratopteris (Benedict, 1909; Lloyd, 1974), despite this being one of the most variable traits. Understanding the model species C. richardii and its relatives in the wild is important for conservation efforts, as well as to understand what natural variation exists in these species that may be informative to future work.
Systematics and hybridization
Hybridization among Ceratopteris species is well-documented (Hickok and Klekowski, 1974; Nakazato et al., 2007; Hickok, 1977; Hickok, 1973), and these hybrid taxa as well as progenitor species can be morphologically cryptic (Adjie et al., 2007). Lloyd predicted the presence of multiple cryptic lineages in C. thalictroides, but detecting these taxa was not possible at the time without genetic analysis (Lloyd, 1974).
During the 1990s and early 2000 s, Masuyama and colleagues examined cryptic variation within C. thalictroides from Asia and Oceania. They used a combination of work on allozymes and cross-breeding experiments (Masuyama et al., 2002), chromosome counts (Masuyama and Watano, 2005), morphology of wild and cultivated plants (Masuyama and Adjie, 2008; Masuyama, 1992), along with plastid and nuclear markers (Adjie et al., 2007) to describe three cryptic species (Masuyama and Watano, 2010). More recently, Zhang et al. described another cryptic species of C. thalictroides endemic to Hainan Province in China. This taxon, Ceratopteris shingii, has some unique characteristics in the genus: a creeping rhizome, terrestrial growth on volcanic rock, and is sister to all other species in the genus (Zhang et al., 2020). Its phylogenetic placement and unique characteristics could provide some new hypotheses for trait evolution and an updated perspective on the life history and ecology of the genus.
In addition to the diversity of Ceratopteris in Asia, the Americas may have novel cryptic species. Natural hybrids between C. thalictroides and C. pteridoides have been described in South America (Hickok and Klekowski, 1974), as well as synthesized hybrids between several New World species (Hickok, 1973; Hickok, 1978). Kinosian et al., found several hybrid individuals and potentially a cryptic species of C. thalictroides in the Americas (Kinosian et al., 2020a). Interestingly, the same study did not find distinct wild populations of C. richardii. Future work on systematics in the group should focus on detangling cryptic species and identifying the extant range and phylogenetic placement of C. richardii.
A robust evolutionary tree is particularly important for Ceratopteris following the publication of the C. richardii genome. The taxonomy of model organisms is not always fully understood until after they become model systems (e.g., Arabidopsis, Al-Shehbaz and O’Kane, 2002; Rattus norvegicus, Musser et al., 2005 and Caenorhabditis elegans, Denver et al., 2003; De Ley, 2006), and C. richardii is no exception.
Despite having unique characteristics like a distinct deltoid leaf shape and only 16 spores per sporangium (Lloyd, 1974), C. richardii is not often identified correctly. For example, specimens identified as C. richardii from Central and South America, as well as western Africa, are each more genomically similar to other species than they are to one another (Kinosian et al., 2020b). This could be due to misidentification of collections, a poor understanding of its native range, or the extinction of C. richardii in the wild. This last possible explanation is troubling because, as we discuss above, it is important to have wild populations to best understand the potential of model organisms. Revisiting the localities of known C. richardii collections (detailed in Lloyd, 1974) should be a goal for future fieldwork. New wild collections will help elucidate the outstanding questions about the taxonomy and natural history of C. richardii, but may also provide novel populations or strains to include in lab studies.
Plant genome structure and evolution
On average, heterosporous plants have fewer chromosomes and smaller genomes than homosporous plants. Ceratopteris richardii is the first homosporous fern with the genomic resources to address why these differences between hetero- and homosporous genomes exist. Nakazato et al. generated a genetic linkage map for C. richardii which showed that it is likely not repeated rounds of polyploidization that leads to larger genomes in ferns, but rather small-scale gene duplications (Nakazato et al., 2006). More recently, Marchant et al. published the first draft genome assembly for C. richardii and found additional support for genetic diploidy and limited rounds of polyploidization (Marchant et al., 2019b). These data further support the theory that homosporous fern genomes are large not because of whole-genome duplication, but because they do not have the same mechanisms for genome downsizing as heterosporous plants (Clark et al., 2016; Szövényi et al., 2021).
The draft genome of Ceratopteris richardii is an important stepping stone for studying land plant evolution (Marchant et al., 2019b); a more complete genome is on the way that will be a better resource for genomic work (Marchant et al., 2022). A high-quality genome for C. richardii will also aid in the development of targeted enrichment or whole-genome resequencing. This latter advancement in sequencing resources for ferns will help us understand reticulate evolution and polyploidy in ferns, as phylogenies can be estimated with hundreds of genes.
In addition to the Ceratopteris richardii genome, there are many other fern genomes that have been recently published or will be available soon. Several heterosporous fern and lycophyte genomes have been published in the last few years, including the heterosporous ferns Azolla filiculoides and Salvinia cucullata (Li et al., 2018), and the heterosporous lycophytes Selaginella moellendorffii (Banks et al., 2011), S. lepidophylla (VanBuren et al., 2018), and Isoëtes taiwanensis (Wickell et al., 2021). In the near future, several additional homosporous fern genomes will be available, including Adiantum capillus-veneris (Polypodiales), Alsophila spinulosa (Cyatheales), Dipteris conjugata (Gleichinales), Ptisana robusta (Marattiales), Huperzia asiatica and Diphasiastrum complanata (Lycopodiales; Drs. F.-W. Li and M. Barker, personal communication). As more homosporous fern genomes become available, the preliminary work with the C. richardii genome will be tested in a more rigorous phylogenetic context, hopefully leading to a clearer picture of land plant genome evolution.
Conclusion
Although Ceratopteris richardii has been used as a model for decades, fundamental aspects of its natural history are still unknown. A few examples include basic taxonomy, origins of spore number, salt tolerance, origins of polyploids, phenotypic plasticity, and intraspecies morphological variation (see Box 2). Many of these topics are ripe for undergraduate or graduate student projects and could be integrated into existing research programs to answer fundamental aspects of fern biology.
Outstanding questions about the natural history of Ceratopteris
What is the evolutionary history of C. richardii? Kinosian et al., 2020b were unable to find a consistent genetic identity for C. richardii; is this due to poor sampling, extirpation of C. richardii from its native range, and/or misidentification of specimens?
Why do some species of Ceratopteris produce 32 spores per sporangium, and C. richardii produces only 16? This is substantially less than the typical leptosporangiate fern which produces 64 spores per sporangium.
What is the genetic population structure of Ceratopteris species? Plants are typically locally abundant but regionally rare; is this due to environmental conditions, spore dispersal, or other factors? How does it affect genetic diversity across a landscape?
How are Ceratopteris gametophytes established in the wild?
What is the biogeographic history of the genus? How might that be influencing current species distributions and hybridization?
How does the habit (aquatic vs. semi-aquatic) of different Ceratopteris species influence population structure, breeding system, or genetic structure and function?
Additionally, C. richardii is a useful tool for teaching students at all grade levels about plant biology (https://www.c-fern.org/). As detailed by Marchant, the C-fern can be incorporated into curriculum topics ranging from basic plant biology to evolution and development to bioinformatics. In the lab, field, or classroom, the recently published C. richardii genome provides a new window into the study of this fern (Marchant et al., 2019b). As more fern genomic resources become available, having Ceratopteris as a well-established model system will only become more important to test novel hypotheses about land plant evolution.
Data availability
Source data for Figure 2 (Range map of Ceratopteris) can be found in the file cer_locations.csv in https://github.com/sylviakinosian/ceratopteris-map (copy archived at swh:1:rev:02f4523dc32b20cb18b17e226eb6f2ffb60cb05a) (previously published in Kinosian et al., 2020a, https://doi.org/10.1016/j.ympev.2020.106938), and in Ceratopteris Brongn. in GBIF Secretariat (2021). GBIF Backbone Taxonomy. Checklist dataset https://doi.org/10.15468/39omei (accessed via GBIF.org on 2021-10-4).
References
-
Independent origins of tetraploid cryptic species in the fern Ceratopteris thalictroidesJournal of Plant Research 120:129–138.https://doi.org/10.1007/s10265-006-0032-5
-
Taxonomy and phylogeny of Arabidopsis brassicaceaeThe Arabidopsis Book 1:e0001.https://doi.org/10.1199/tab.0001
-
Sex determination in the fern CeratopterisTrends in Plant Science 2:175–180.https://doi.org/10.1016/S1360-1385(97)85223-5
-
The genus Ceratopteris: a preliminary revisionBulletin of the Torrey Botanical Club 36:463.https://doi.org/10.2307/2479023
-
Description d’un nouveau genre de fougere, nomme CeratopterisBull. Sci. Soc. Philom. Paris Sér 3:184–187.
-
BookGeneration of transgenic spores of the fern Ceratopteris richardii to analyze Ca2+ transport dynamics during gravity-directed polarizationIn: Fernández Helena, editors. In Current Advances in Fern Research. Cham: Springer International Publishing. pp. 285–303.https://doi.org/10.1007/978-3-319-75103-0_14
-
Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majusGenes & Development 4:1483–1493.https://doi.org/10.1101/gad.4.9.1483
-
Ceratopteris: a model plant for the 90sThe Plant Cell 4:113–115.https://doi.org/10.1105/tpc.4.2.113
-
Genome evolution of ferns: evidence for relative stasis of genome size across the fern phylogenyThe New Phytologist 210:1072–1082.https://doi.org/10.1111/nph.13833
-
Apogamy induction in Ceratopteris richardiiInternational Journal of Plant Sciences 168:361–369.https://doi.org/10.1086/511049
-
BookLaboratory-induced apogamy and apospory in Ceratopteris richardiiIn: Kumar A, Fernández H, Revilla MA, editors. In Working with Ferns: Issues and Applications. New York, NY: Springer New. pp. 25–36.https://doi.org/10.1007/978-1-4419-7162-3
-
The moss Physcomitrella patensAnnual Review of Genetics 39:339–358.https://doi.org/10.1146/annurev.genet.39.073003.110214
-
A quick tour of nematode diversity and the backbone of nematode phylogenyWormBook: The Online Review of C. Elegans Biology 1:1–8.https://doi.org/10.1895/wormbook.1.41.1
-
Phylogenetics in Caenorhabditis elegans: an analysis of divergence and outcrossingMolecular Biology and Evolution 20:393–400.https://doi.org/10.1093/molbev/msg044
-
Ceratopteris: a model system for studying sex-determining mechanisms in plantsInternational Journal of Plant Sciences 156:359–366.https://doi.org/10.1086/297257
-
Deciphering the origins of apomictic polyploids in the Cheilanthes yavapensis complex (PteridaceaeAmerican Journal of Botany 96:1636–1645.https://doi.org/10.3732/ajb.0900019
-
A current perspective on apomixis in fernsJournal of Systematics and Evolution 54:656–665.https://doi.org/10.1111/jse.12228
-
A drought-driven model for the evolution of obligate apomixis in ferns: evidence from pellaeids (Pteridaceae)American Journal of Botany 108:263–283.https://doi.org/10.1002/ajb2.1611
-
R2R3-MYB gene evolution in plants, incorporating ferns into the storyInternational Journal of Plant Sciences 182:1–8.https://doi.org/10.1086/710579
-
Karyotype evolution in Ceratopteris. An analysis of the synthesized hybrid C. pteridoides x C. richardiiAmerican Journal of Botany 60:1010–1022.
-
Inchoate speciation in Ceratopteris: an analysis of the synthesized hybrid C. richardii x C. pteroidesEvolution; International Journal of Organic Evolution 28:439–446.https://doi.org/10.1111/j.1558-5646.1974.tb00765.x
-
Cytological relationships between three diploid species of the fern genus CeratopterisCanadian Journal of Botany 55:1660–1667.https://doi.org/10.1139/b77-194
-
The biology of the fern Ceratopteris and its use as a model systemInternational Journal of Plant Sciences 156:332–345.https://doi.org/10.1086/297255
-
Heteroblastic root development in Ceratopteris richardii (Parkeriaceae)International Journal of Plant Sciences 163:341–351.https://doi.org/10.1086/339156
-
Cryptic diversity in the model fern genus Ceratopteris (Pteridaceae)Molecular Phylogenetics and Evolution 152:106938.https://doi.org/10.1016/j.ympev.2020.106938
-
There and back again: reticulate evolution in CeratopterisAmerican Fern Journal 110:193–210.https://doi.org/10.1640/0002-8444-110.4.193
-
Reproductive biology of the Pteridophyta IV. An experimental study of mating systems in Ceratopteris thalictroides (L.) BrongnBotanical Journal of the Linnean Society 63:153–169.https://doi.org/10.1111/j.1095-8339.1970.tb02547.x
-
Ferns with benefits: incorporating Ceratopteris into the genomics eraAmerican Fern Journal 109:183.https://doi.org/10.1640/0002-8444-109.3.183
-
Cryptic species in the fern Ceratopteris thalictroides (L.) Brongn. (Parkeriaceae). II. Cytological characteristics of three cryptic speciesActa Phytotaxonomica et Geobotanica 56:231–240.https://doi.org/10.18942/apg.KJ00004623254
-
Three forms of Ceratopteris thalictroides in GuamAmerican Fern Journal 98:104–107.https://doi.org/10.1640/0002-8444(2008)98[104:TFOCTI]2.0.CO;2
-
Bryophytes are not early diverging land plantsThe New Phytologist 230:1300–1304.https://doi.org/10.1111/nph.17241
-
Soil salinity, sun exposure, and growth of Acrostichum aureum, the mangrove fernBotanical Gazette 151:41–49.https://doi.org/10.1086/337803
-
BookMammal species of the world: A taxonomic and geographic referenceIn: Wilson DE, Reeder DM, editors. The Johns. Oxford university Press. pp. 824–830.https://doi.org/10.1644/06-MAMM-R-422.1
-
Ferns: the missing link in shoot evolution and developmentFrontiers in Plant Science 6:972.https://doi.org/10.3389/fpls.2015.00972
-
A community-derived classification for extant lycophytes and fernsJournal of Systematics and Evolution 54:563–603.https://doi.org/10.1111/jse.12229
-
The Physcomitrella genome reveals evolutionary insights into the conquest of land by plantsScience (New York, N.Y.) 319:64–69.https://doi.org/10.1126/science.1150646
-
Antheridogen activity in the fern Ceratopteris thalictroides (L.) BrongnBotanical Journal of the Linnean Society 65:399–413.https://doi.org/10.1111/j.1095-8339.1972.tb02280.x
-
Genetic analysis of antheridiogen sensitivity in Ceratopteris richardiiAmerican Journal of Botany 74:1872–1877.https://doi.org/10.1002/j.1537-2197.1987.tb08789.x
-
Considerations in adapting CRISPR/Cas9 in nongenetic model plant systemsApplications in Plant Sciences 8:e11314.https://doi.org/10.1002/aps3.11314
-
Charting the genomic landscape of seed-free plantsNature Plants 7:554–565.https://doi.org/10.1038/s41477-021-00888-z
-
Diversification of gene function: homologs of the floral regulator FLO/LFY control the first zygotic cell division in the moss Physcomitrella patensDevelopment (Cambridge, England) 132:1727–1736.https://doi.org/10.1242/dev.01709
-
The evolution, morphology, and development of fern leavesFrontiers in Plant Science 4:345.https://doi.org/10.3389/fpls.2013.00345
-
Challenging the paradigms of leaf evolution: Class III HD-Zips in ferns and lycophytesThe New Phytologist 212:745–758.https://doi.org/10.1111/nph.14075
-
The analysis of genetically and physiologically complex traits using Ceratopteris: a case study of NaCl-tolerant mutantsInternational Journal of Plant Sciences 156:374–384.https://doi.org/10.1086/297259
-
Underwater CAM photosynthesis elucidated by Isoetes genomeNature Communications 12:6348.https://doi.org/10.1038/s41467-021-26644-7
-
Ceratopteris shingii, a new species of Ceratopteris with creeping rhizomes from Hainan, ChinaPhytotaxa 449:23–30.
Decision letter
-
Helena Pérez ValleReviewing Editor; eLife, United Kingdom
-
Peter RodgersSenior Editor; eLife, United Kingdom
In the interests of transparency, eLife publishes the most substantive revision requests and the accompanying author responses.
Decision letter after peer review:
Thank you for submitting your article "The Natural History of Model Organisms: The biology of the C-fern as a tool for understanding plant evolution" to eLife for consideration as a Feature Article. Your article has been reviewed by three peer reviewers, and the evaluation has been overseen by a member of the eLife Features Team (Helena Perez Valle). The following individuals involved in review of your submission have agreed to reveal their identity: Chi Lien Cheng and Verónica di Stilio.
The reviewers and editors have discussed the reviews and we have drafted this decision letter to help you prepare a revised submission.
Summary:
This short article on the fern model Ceratopteris richardii pulls together information about this fern's natural history, as well as reviewing its history as a model system. The use of understandable language and the framing of this species within the gametophyte-sporophyte relationship and the homospory-heterospory trends in plant evolution is welcome and refreshing. The article should be useful for those interested in learning more about ecological and genetic diversity in the genus, which could be leveraged in understanding genotype – phenotype relationships in this species.
Essential revisions:
1. Currently, the text is lacking in references on the breakthrough development of transgenic techniques in this model. Please discuss how the following references are a key aspect of this organism rising to model status:
– Plackett AR, Huang L, Sanders HL, Langdale JA. High-efficiency stable transformation of the model fern species Ceratopteris richardii via microparticle bombardment. Plant Physiol. 2014 May;165(1):3-14. doi: 10.1104/pp.113.231357. Epub 2014 Mar 12. PMID: 24623851; PMCID: PMC4012588.
– Plackett AR, Rabbinowitsch EH, Langdale JA. Protocol: genetic transformation of the fern Ceratopteris richardii through microparticle bombardment. Plant Methods. 2015 Jul 3;11:37. doi: 10.1186/s13007-015-0080-8. PMID: 26146510; PMCID: PMC4490597.
– Muthukumar B, Joyce BL, Elless MP, Stewart CN Jr. Stable transformation of ferns using spores as targets: Pteris vittata and Ceratopteris thalictroides. Plant Physiol. 2013 Oct;163(2):648-58. doi: 10.1104/pp.113.224675. Epub 2013 Aug 9. PMID: 23933990; PMCID: PMC3793046.
– Bui, L.T., Cordle, A.R., Irish, E.E. et al., Transient and stable transformation of Ceratopteris richardii gametophytes. BMC Res Notes 8, 214 (2015). https://doi.org/10.1186/s13104-015-1193-x
2. Please add a summary of the handful of functional studies that have been conducted since 2015 to illustrate the use of the transgenic techniques mentioned in the point above, mentioning how C. richardii's natural history could be used to complement the knowledge that is emerging from these studies.
3. Please emphasize the potential use of C. richardii as a model to study plant evo-devo, based on ferns bridging the gap between the bryophyte systems (moss and Marchantia) and the angiosperms. For an example see the following review:
– Plackett AR, Di Stilio VS, Langdale JA. Ferns: the missing link in shoot evolution and development. Front Plant Sci. 2015 Nov 6;6:972. doi: 10.3389/fpls.2015.00972. PMID: 26594222; PMCID: PMC4635223.
4. Please update the photograph in Figure 1 to include a fern that is in a good state, preferably growing in the wild, the current plant depicted does not have the full set of leaves and does not look to be in a healthy state. If possible, please update the figure to also include photographs of several mutants. Please remember that any photographs used in the manuscript must be available to be published under a CC BY 4.0 license. Please contact me [h.perezvalle@elifesciences.org] if you have any queries about this.
5. If possible, please update Figure 2 to include photographs representing the diverse morphologies found in the different natural species, and update the figure caption to include details as to what questions each is most amenable for. Please remember that any photographs used in the manuscript must be available to be published under a CC BY 4.0 license.
6. Please include a figure with a simplified fern phylogeny to help a broader readership to comprehend the text better, with more details surrounding the genus Ceratopteris in the figure caption.
7. Please include further references regarding factors influencing sex determination, including light and hormones. In line 154, references from the Banks lab would be appropriate.
8. Please update the article to reflect that C. richardii is a semi-aquatic (not aquatic) species of fern, and if possible please discuss any work studying differences between aquatic and semi aquatic species of Ceratopteris.
9. The term "C-Fern" was trademarked by Leslie Hickock, so please use Ceratopteris richardii in the article (including the title), but please include a brief mention of the C-fern cultivar and its use in research.
https://doi.org/10.7554/eLife.75019.sa1Author response
Essential revisions:
1. Currently, the text is lacking in references on the breakthrough development of transgenic techniques in this model. Please discuss how the following references are a key aspect of this organism rising to model status:
– Plackett AR, Huang L, Sanders HL, Langdale JA. High-efficiency stable transformation of the model fern species Ceratopteris richardii via microparticle bombardment. Plant Physiol. 2014 May;165(1):3-14. doi: 10.1104/pp.113.231357. Epub 2014 Mar 12. PMID: 24623851; PMCID: PMC4012588.
– Plackett AR, Rabbinowitsch EH, Langdale JA. Protocol: genetic transformation of the fern Ceratopteris richardii through microparticle bombardment. Plant Methods. 2015 Jul 3;11:37. doi: 10.1186/s13007-015-0080-8. PMID: 26146510; PMCID: PMC4490597.
– Muthukumar B, Joyce BL, Elless MP, Stewart CN Jr. Stable transformation of ferns using spores as targets: Pteris vittata and Ceratopteris thalictroides. Plant Physiol. 2013 Oct;163(2):648-58. doi: 10.1104/pp.113.224675. Epub 2013 Aug 9. PMID: 23933990; PMCID: PMC3793046.
– Bui, L.T., Cordle, A.R., Irish, E.E. et al., Transient and stable transformation of Ceratopteris richardii gametophytes. BMC Res Notes 8, 214 (2015). https://doi.org/10.1186/s13104-015-1193-x
Thank you for pointing out the lack of discussion on transgenic techniques in C. richardii.
We agree that this is an important part of this model, and have added a section titled, “Ceratopteris as a transgenic model for non-seed plants” that describes the various transformation approaches and how the life history of Ceratopteris enables the simple establishment of transgenic lines.
“Free-living generations and a flexible life cycle make Ceratopteris an important model for evolutionary developmental studies. […] The ability to have transgenic lines in both generations provides a unique perspective for studying how genes, growth conditions, or other factors affect sporophytes and gametophytes differently.”
2. Please add a summary of the handful of functional studies that have been conducted since 2015 to illustrate the use of the transgenic techniques mentioned in the point above, mentioning how C. richardii's natural history could be used to complement the knowledge that is emerging from these studies.
3. Please emphasize the potential use of C. richardii as a model to study plant evo-devo, based on ferns bridging the gap between the bryophyte systems (moss and Marchantia) and the angiosperms. For an example see the following review:
– Plackett AR, Di Stilio VS, Langdale JA. Ferns: the missing link in shoot evolution and development. Front Plant Sci. 2015 Nov 6;6:972. doi: 10.3389/fpls.2015.00972. PMID: 26594222; PMCID: PMC4635223.
Thank you for these suggestions; we will address the two comments above here. We have added a discussion of transgenic techniques and how the natural history and phylogenetic placement of Ceratopteris makes it an important model for these studies:
“A recent study using both transgenic gametophytes and sporophytes of Ceratopteris investigated the role of the LEAFY transcription factor (LFY) in development (Plackett et al., 2018). LFY is important for cell division in moss embryos (Tanahashi et al., 2005) and angiosperm floral meristem development (Carpenter and Coen 1990). […] If a CRISPR system could be established in Ceratopteris, one might be able to extend such technology to other members of the Pteridaceae known for apogamy (Grusz et al., 2021; Grusz 2016; Grusz, Windham, and Pryer 2009), or for application in other non-model ferns.”
4. Please update the photograph in Figure 1 to include a fern that is in a good state, preferably growing in the wild, the current plant depicted does not have the full set of leaves and does not look to be in a healthy state. If possible, please update the figure to also include photographs of several mutants. Please remember that any photographs used in the manuscript must be available to be published under a CC BY 4.0 license. Please contact me [h.perezvalle@elifesciences.org] if you have any queries about this.
5. If possible, please update Figure 2 to include photographs representing the diverse morphologies found in the different natural species, and update the figure caption to include details as to what questions each is most amenable for. Please remember that any photographs used in the manuscript must be available to be published under a CC BY 4.0 license.
Thank you for these suggestions, we will address the two comments above here. We have added a new figure (Figure 3) and included photographs of C. thalictroides, C. pteridoides, and C. gaudichaudii, in addition to a C. richardii sporophyte and gametophyte. We have included a new picture of C. richardii growing in cultivation; we are not aware of any recent (in the last 20 years) wild collections of C. richardii but would be very interested to see any.
6. Please include a figure with a simplified fern phylogeny to help a broader readership to comprehend the text better, with more details surrounding the genus Ceratopteris in the figure caption.
We have included a cartoon phylogeny (Figure 1) of land plants to place Ceratopteris in phylogenetic context. We have also updated Figure 2 to include a phylogenetic reconstruction of the current estimation of species relationships within the genus Ceratopteris. This was included in Figure 2 along with the distribution map of the genus to help the reader better contextualize the species range and relationships together.
7. Please include further references regarding factors influencing sex determination, including light and hormones. In line 154, references from the Banks lab would be appropriate.
Thank you for this suggestion, we have provided additional references throughout the manuscript where appropriate.
8. Please update the article to reflect that C. richardii is a semi-aquatic (not aquatic) species of fern, and if possible please discuss any work studying differences between aquatic and semi aquatic species of Ceratopteris.
We appreciate the reviewers pointing this out. We have made this distinction more clear throughout the manuscript. Little work has been done to investigate differences between the semi-aquatic and aquatic species of Ceratopteris. The two species most commonly found growing as fully aquatic plants are C. pteridoides (Americas) and C. chingii (endemic to China); the remaining species in the genus are mostly semi-aquatic, with the exception of C. cornuta which can sometimes be found growing unrooted in still or slow-moving water. The aquatic and semi-aquatic species can hybridize (e.g., C. richardii and C. pteridoides) and form morphologically intermediate offspring (Hickok and Klekowski, 1974). Additionally, a few gene expression studies have used C. pteridoides, but do not directly compare it to C. richardii or another semi-aquatic species. Researching potential differences between the semi-aquatic and aquatic species of Ceratopteris could be a great new avenue for work on fern ecology, gene function, or hybridization. We have added a short note to this extent in Box 2, on line 364:
“How does the habit (aquatic vs. semi-aquatic) of different Ceratopteris species influence population structure, breeding system, or genetic structure and function?”
9. The term "C-Fern" was trademarked by Leslie Hickock, so please use Ceratopteris richardii in the article (including the title), but please include a brief mention of the C-fern cultivar and its use in research.
Thank you for pointing this out. We have altered the title to be “The biology of Ceratopteris richardii as a tool for understanding plant evolution” and changed the usage of “C-fern” throughout the article to be Ceratopteris richardii or C. richardii. We have also added a mention of the C-fern cultivar on lines 58-9:
“Many studies used spores from a Cuban vouchered collection, now known as the Hnn strain or C-fern (Hickok 1977).”
https://doi.org/10.7554/eLife.75019.sa2Article and author information
Author details
Funding
National Science Foundation (DEB-1911459)
- Paul G Wolf
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank Jacob Suissa for the thoughtful comments on the manuscript. We are grateful to Christopher Haufler, David Randall, and Jo Ann Banks for providing the photograph of Ceratopteris richardii in cultivation.
Publication history
- Received:
- Accepted:
- Accepted Manuscript published:
- Accepted Manuscript updated:
- Version of Record published:
Copyright
© 2022, Kinosian and Wolf
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,969
- views
-
- 334
- downloads
-
- 15
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 15
- citations for umbrella DOI https://doi.org/10.7554/eLife.75019







