Structural and functional insights of the human peroxisomal ABC transporter ALDP
Figures
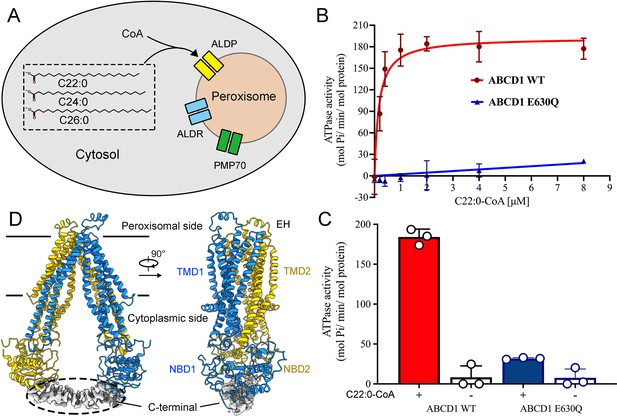
Functional and structural characterization of human adrenoleukodystrophy protein (ALDP).
(A) Schematic of very-long-chain fatty acids transport into the peroxisomes by ALDP. (B–C) ATP hydrolysis of ALDP stimulated by C22:0-CoA (data are represented as mean ± SD; n=3; three biological repeats). (D) Cryo-electron microscopy structure of ALDP. The transmembrane domains (TMDs) and nucleotide-binding domains (NBDs) are indicated.
-
Figure 1—source data 1
Cryo-electron microscopy data collection, refinement, and validation statistics.
- https://cdn.elifesciences.org/articles/75039/elife-75039-fig1-data1-v2.doc
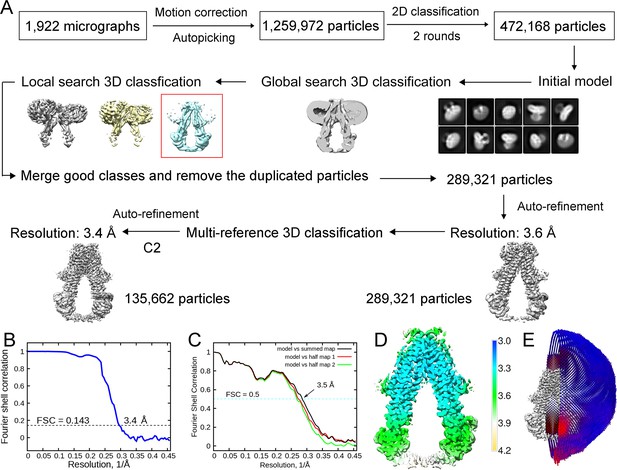
Cryo-electron microscopy (cryo-EM) analysis of adrenoleukodystrophy protein (ALDP).
(A) A flowchart description of EM data processing. (B) The Fourier Shell Correlation (FSC) curve for the cryo-EM map. The average resolution is estimated to be 3.4 Å based on the FSC value of 0.143. (C) FSC curves of the refined model versus the overall 3.4 Å map that it was refined against (black); of the model refined in the first of the two independent maps used for the FSC calculation versus that same map (red); and of the model refined in the first of the two independent maps versus the second independent map (green). The small difference between the red and green curves indicates that the refinement of the atomic coordinates did not suffer from overfitting. (D) Resolution estimation of the density map. (E) Angular distribution of the particles used for reconstruction of the ALDP. Each cylinder represents one view, and the height of the cylinder is proportional to the number of particles for that view.
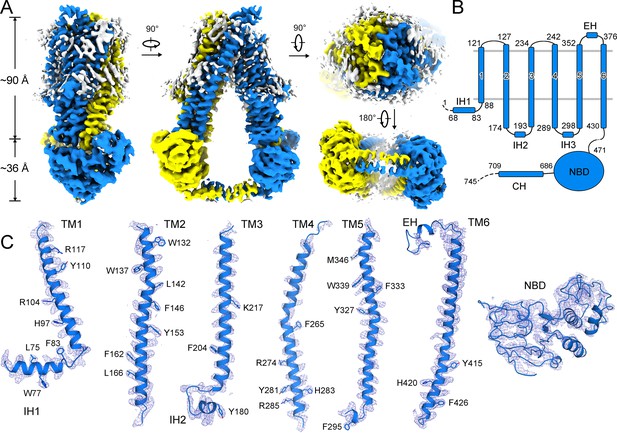
Cryo-electron microscopy (cryo-EM) density of human adrenoleukodystrophy protein (ALDP).
(A) Cryo-EM map of ALDP with each monomer individually colored. The attached lipids around the transmembrane domains (TMDs) are colored in white. (B) The overall structure of ALDP. The TMDs and nucleotide-binding domains (NBDs) are indicated. ALDP contains a short helix at the peroxisomal side. (C) Representative cryo-EM densities of structural elements of ALDP.
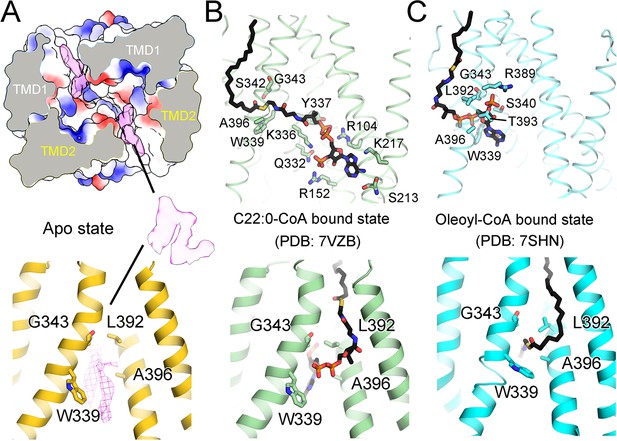
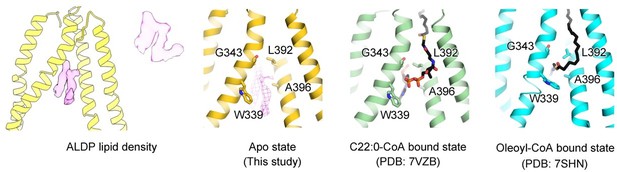
Comparison of unknown densities in the ligand-free structure with the different substrates.
(A) Two unknown densities in the ligand-free state of adrenoleukodystrophy protein (ALDP). The surface electrostatic potential is shown. (B–C) Structures of C22:0-CoA-bound and oleoyl-CoA-bound ALDP. All these residues in the lower panels are reported to be mutation sites in ALD.
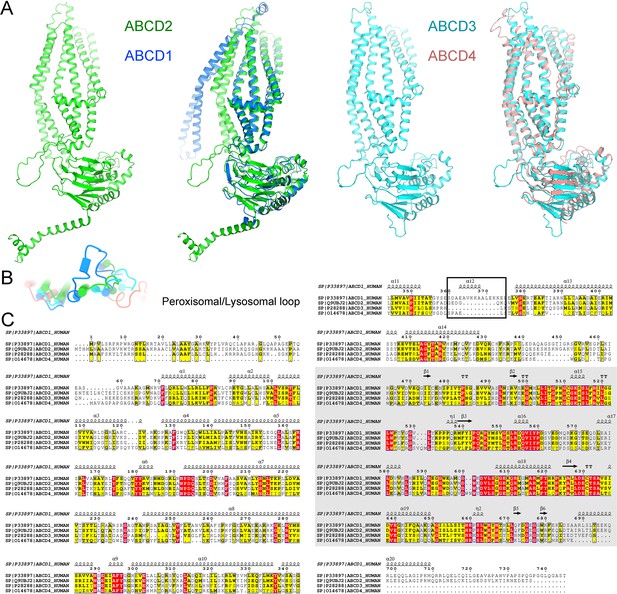
Structural comparison of all members of ATP-binding cassette sub-family D (ABCD) transporters in human.
(A) Structure similarities of the ABCD members. High structural similarities are found between adrenoleukodystrophy protein (ALDP) and ABCD2 or ABCD3 and ABCD4. The model of ABCD2 and ABCD3 is predicted by Alphafold. (B) Distinctive extracellular helix of ALDP at the peroxisomal side. (C) Sequence alignment of ABCD members. The sequences corresponding to the short peroxisomal helix are highlighted.
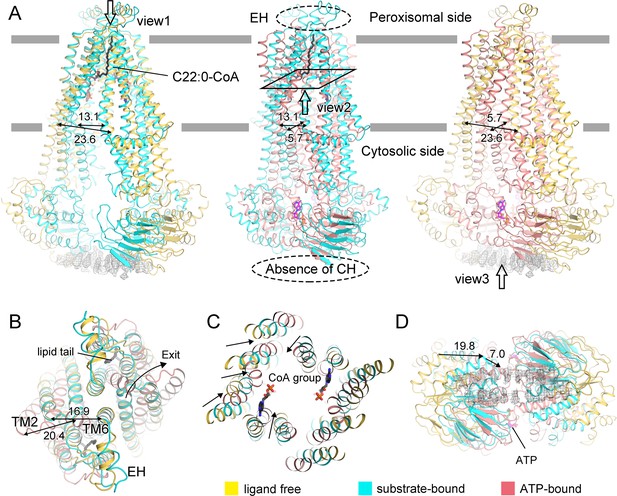
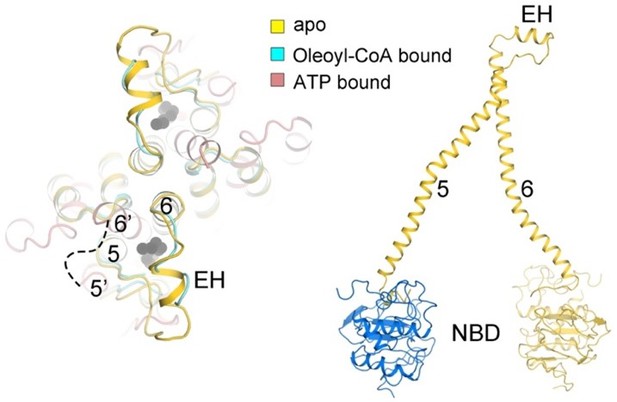
Conformational changes of adrenoleukodystrophy protein (ALDP) during the whole transport cycle.
(A) Structural alignments of ALDP in ligand-free, C22:0-CoA-bound (7VZB) (Chen et al., 2022), and ATP-bound states (7SHM) (Wang et al., 2022). The distance change (in Å) between transmembrane helix 4 (TM4) and TM6 reflects the conformational changes during lipid transport. The hollow arrows indicate three views of ALDP that are enlarged in B–D. CH: C-terminal helix; EH: extracellular helix.
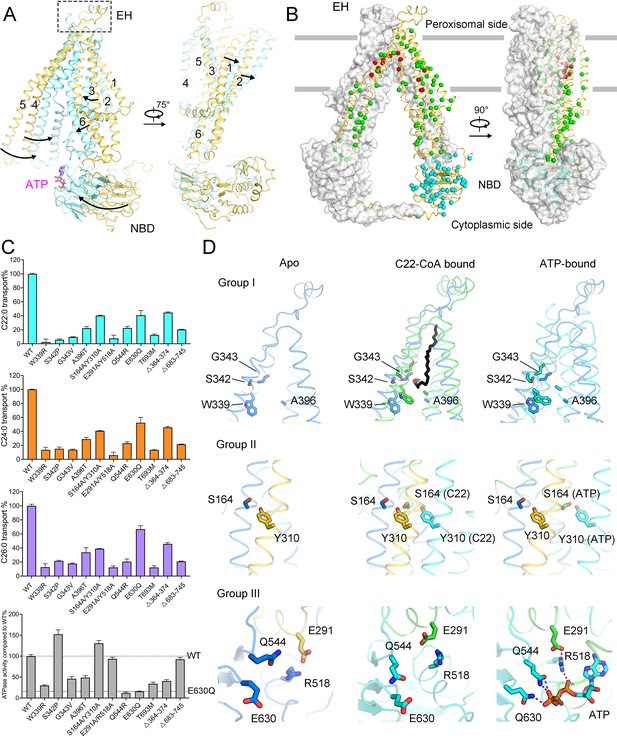
Functional analysis of adrenoleukodystrophy (ALD)-associated mutations.
(A) Conformational changes of adrenoleukodystrophy protein (ALDP) from ligand-free to ATP-bound state. (B) Structural mapping of clinical-derived pathogenic mutations of ALDP. The Cα atoms of the indicated residues are shown as spheres. The residues involved in substrate binding are shown as red spheres. Other mutation sites on transmembrane domains are presented in green. Mutations on nucleotide-binding domain (NBD) and the single-mutation T693M on C-terminal helix are colored cyan, respectively. (C) Spatial location of selected ALD mutants and rationally designed mutations. (D) Transport of cytosolic C22:0-CoA, C24:0-CoA, C26:0-CoA, and ATP hydrolysis by wild-type (WT) ALDP and mutations (data are represented as mean ± SD; n = 3; three biological repeats). EH: extracellular helix.
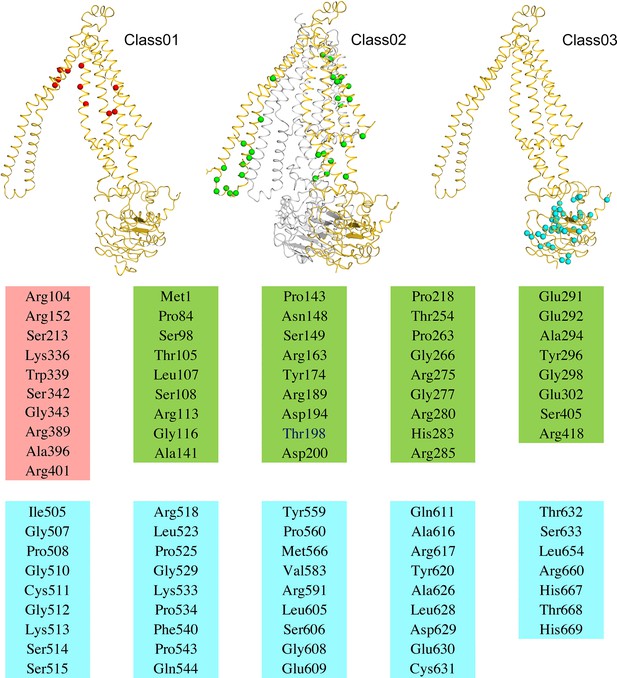
Structural mapping of residues that harbor recurring adrenoleukodystrophy-associated mutants.
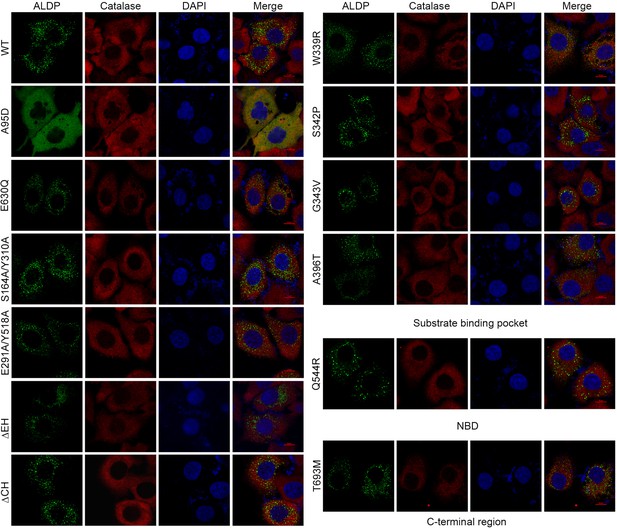
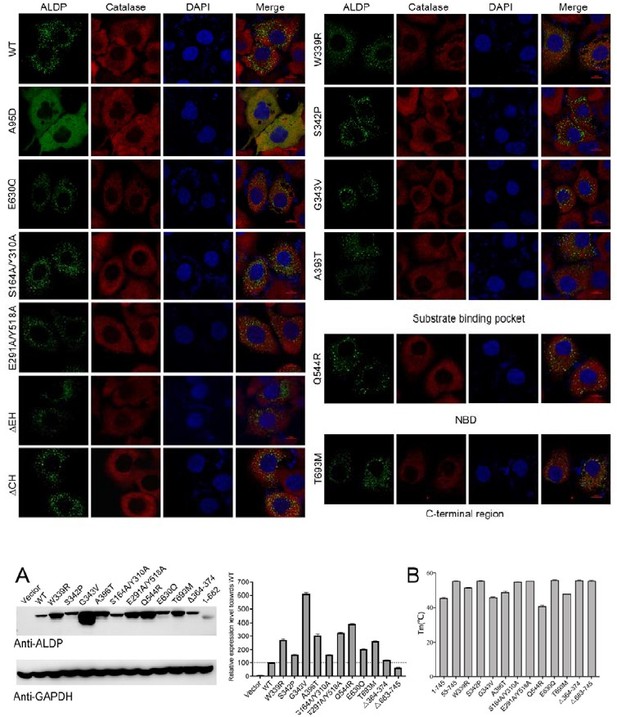
Cellular localization of representative mutations.
Cytoplasmic localization of adrenoleukodystrophy protein (ALDP) mutants. Green, ALDP-GFP (Green fluorescent protein). Red, catalase, a peroxisome marker. Blue, DAPI (4',6-diamidino-2-phenylindole dihydrochloride).
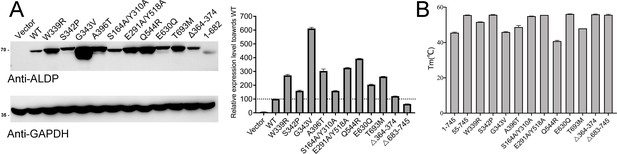
Expression level and thermo-stability of representative mutations.
(A) Expression level of adrenoleukodystrophy protein (ALDP) mutants. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as loading control (data are represented as mean ± SD; n = 3; three statistical repeats). (B) The Tm values of selected mutations (data are represented as mean ± SD; n = 2; two technical repeats).
-
Figure 3—figure supplement 3—source data 1
Western blot for Figure 3—figure supplement 3A.
- https://cdn.elifesciences.org/articles/75039/elife-75039-fig3-figsupp3-data1-v2.docx
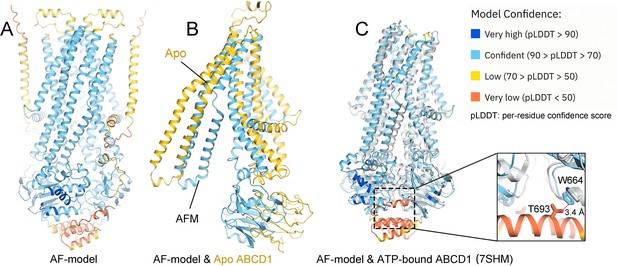
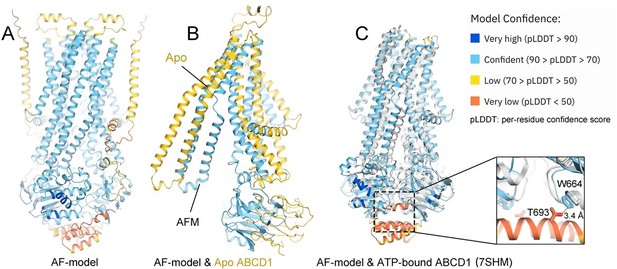
Structural comparison between cryo-electron microscopy (EM) structure and the predicted one by Alphafold (AF).
(A) A predicted model of adrenoleukodystrophy protein (ALDP) by AF (AF-ALDP). (B) Comparison of AF-model with the cryo-EM structure in apo state. (C) Structural comparison between the AF-model and the cryo-EM model in ATP-bound state (colored white).
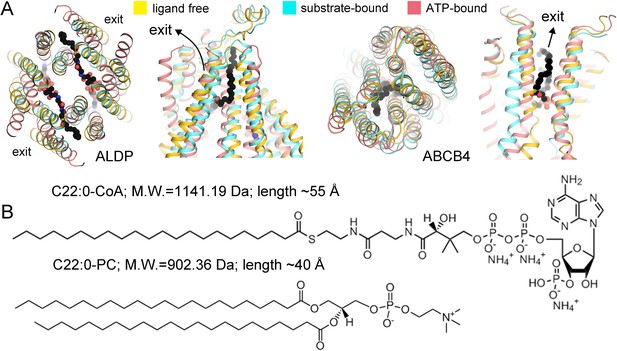
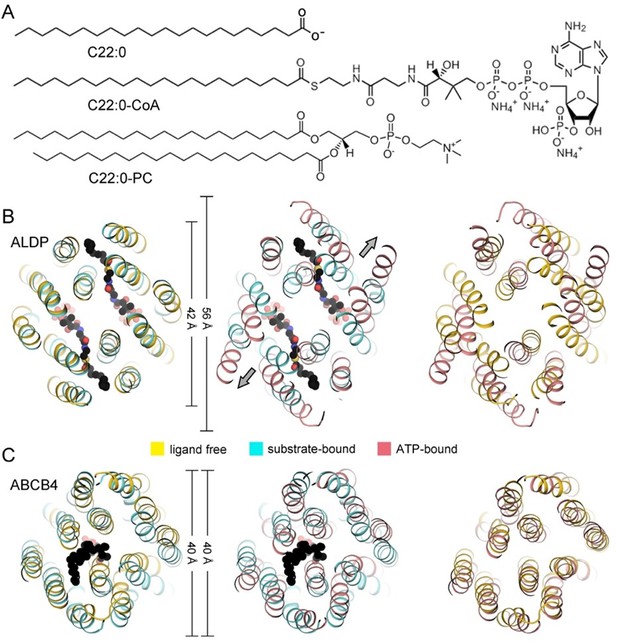
Comparison between adrenoleukodystrophy protein (ALDP) and phosphatidylcholine transporter ABCB4.
(A) Superimposition of the transmembrane domains (TMDs) of ALDP and ABCB4 in three major states of a working cycle. PDB (Protein Data Bank) code of ALDP: 7VR1 (ligand-free state, this study), 7VZB (C22:0-CoA-bound state) (Chen et al., 2022), 7SHM (ATP-bound state) (Wang et al., 2022) PDB of ABCB4: 7NIU (ligand-free state) (Nosol et al., 2021), 7NIV (PC bound state) (Nosol et al., 2021), and 6S7P (ATP-bound state) (Olsen et al., 2020). (B) The substrate of ALDP displays a much larger head group and a flexible hydrophobic tail. The molecular weight and size are labeled.
Tables
| State | Confirmation | Peroxisomal helix | c-terminal coiled domain | Reference |
|---|---|---|---|---|
| Apo | Cytosol-facing | Presence | Presence | This study |
| Apo (chimera) | Cytosol-facing | Absence | Presence | Chen et al |
| C22-CoA bound (chimera) | Intermediate | Presence | Absence | Chen et al |
| ATP bound (Chimera) | Peroxisomal-facing | Absence | Absence | Chen et al |
| Oleoyl-CoA bound | Cytosol-facing | Presence | Presence | Wang et al |
| ATP bound | Peroxisomal-facing | Absence | Absence | Wang et al |
| C26 bound | Cytosol-facing | Presence | Presence | Xiong et al |
| C26-CoA & ATP bound | Cytosol-facing | Presence | Presence | Xiong et al |
| ATP bound | Peroxisomal-facing | Absence | Absence | Xiong et al |
| ATP-γS bound state 1 | Peroxisomal-facing | Absence | Absence | Le et al |
| ATP-γS bound state 2 | Cytosol-facing | Presence | Absence | Le et al |