Shank promotes action potential repolarization by recruiting BK channels to calcium microdomains
Figures
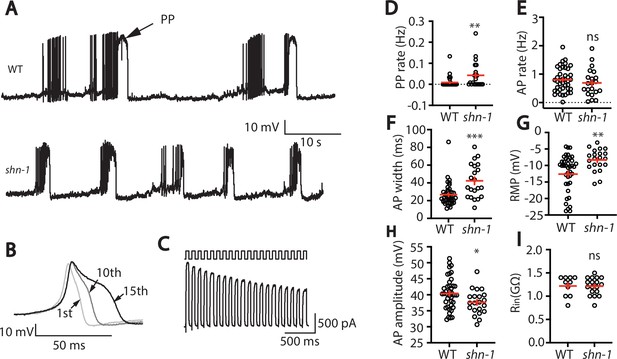
SHN-1 regulates muscle AP firing patterns.
(A) Representative traces of spontaneous muscle APs are shown for WT and shn-1(nu712 null) mutants. APs occur in bursts of ~10 APs/ burst. Plateau potentials (PPs), defined as transients lasting >150ms, are observed less frequently, often at the end of a burst. (B) APs become progressively longer during bursts. Successive APs taken from a representative burst are shown. (C) Repetitive depolarization to +30 mV leads to a progressive decrease in potassium currents. A representative recording from a WT animal is shown. This likely results from an accumulation of inactivated potassium channels during repetitive stimulation. (D–I) Mean PP rate (D), AP rate (E), AP width (F), RMP (G), AP amplitude (H), and input resistance (Rin, I) are compared in WT and shn-1 null mutants. All shn-1 data were obtained from shn-1(nu712) except for Rin (I), which were from shn-1(tm488). Values that differ significantly from wild type controls are indicated (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate SEM.
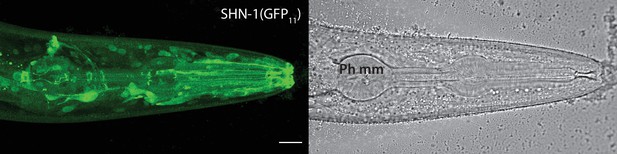
SHN-1 is expressed in many tissues.
Endogenous SHN-1 is broadly expressed, including in neurons, muscles, skin, and glia. A representative image of reconstituted fluorescence produced by shn-1(nu600 GFP11) and eft-3>GFP1-10 (left) and the corresponding bright field image (right) are shown. Pharyngeal muscles (Ph mm) are indicated in the bright field image. SHN-1(GFP11) expression in body muscles was not detected, consistent with the very low shn-1 mRNA levels reported in body muscles (Cao et al., 2017; Packer et al., 2019). Scale bar indicates 14 μm.
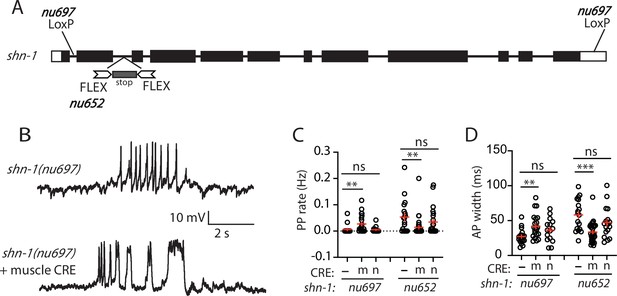
SHN-1 acts in muscles to control AP duration.
(A) A schematic of the shn-1 locus is shown. Open boxes indicate UTRs, black boxes indicate coding regions. Recombination sites mediating CRE induced deletions (LoxP) and inversions (FLEX) are indicated. The shn-1(nu697) allele allows CRE-induced shn-1 knockouts while shn-1(nu652) allows CRE-induced shn-1 rescue. In shn-1(nu652), an exon containing in frame stop codons was inserted into the second intron (in the ‘OFF’ orientation). This stop exon is bounded by FLEX sites. (B) Representative traces of spontaneous muscle APs are shown in shn-1(nu697) with and without muscle CRE expression. Mean PP rate (C) and AP width (D) are compared in the indicated shn-1 mutants without (-) and with CRE expression in muscles (m) or neurons (n). Sample sizes are as follows: shn-1(nu697) (17); shn-1(nu697) +muscle CRE (21); shn-1(nu697) +neuron CRE (15); shn-1(nu652) (18); shn-1(nu652) +muscle CRE (30); and shn-1(nu652) +neuron CRE (19). Values that differ significantly from wild-type controls are indicated (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate SEM. Representative traces for genotypes in panels C and D are shown in Figure 2—figure supplement 1.
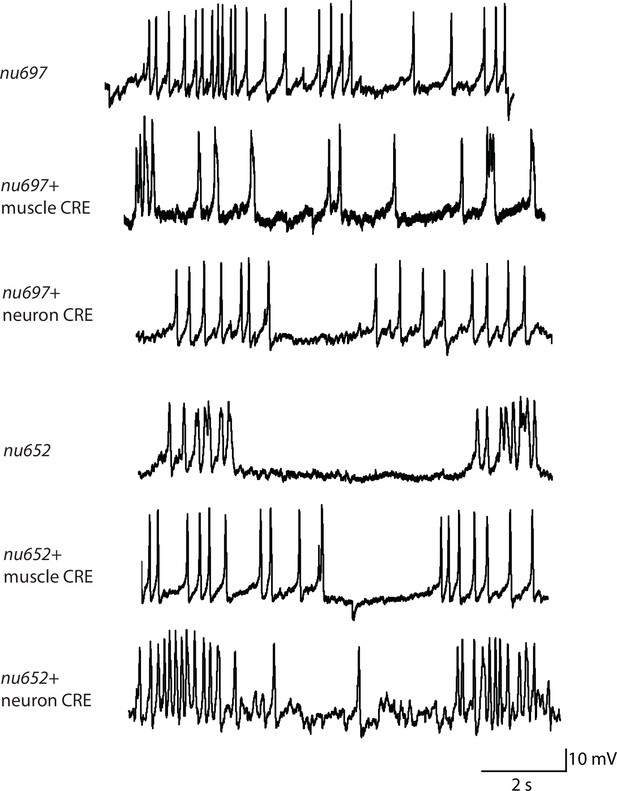
Representative traces for recordings summarized in Figure 2C and D.
Representative traces of spontaneous muscle APs are shown for the indicated genotypes.
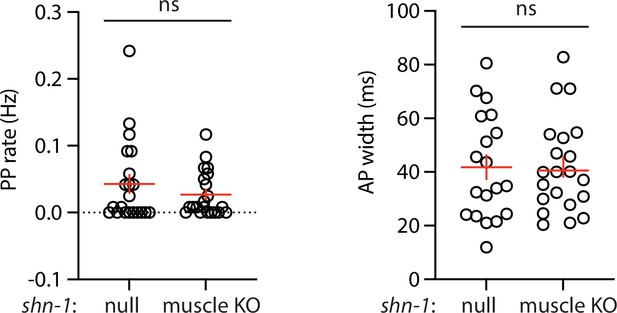
Muscle AP defects in shn-1(null) and shn-1(muscle KO) are not significantly different.
Mean PP rate and AP width are compared in the indicated shn-1 mutants. Sample sizes are as follows: shn-1(nu712 null) (21); shn-1(nu697) +muscle CRE (21). Error bars indicate SEM. No significant differences were observed.
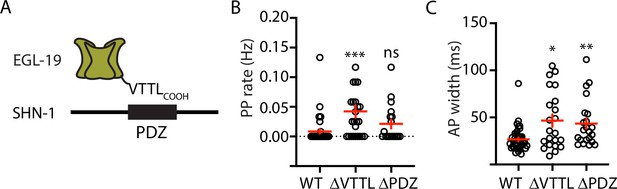
Mutations disrupting SHN-1 binding to EGL-19 increase AP duration.
(A) A schematic illustrating the binding interaction between EGL-19’s c-terminus and SHN-1’s PDZ domain is shown. (B–C) Mean PP rate and AP width are compared in the indicated genotypes. Representative traces are shown in Figure 3—figure supplement 1. Mutations deleting the SHN-1 PDZ domain (nu542 ΔPDZ) or those deleting EGL-19’s c-terminal PDZ ligand (nu496 ΔVTTL) were edited into the endogenous genes using CRISPR. These mutations significantly increased AP width, compared to WT controls. Sample sizes are as follows: WT (41), shn-1(nu542) (22), and egl-19(nu496) (22). Values that differ significantly from wild type controls are indicated (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate SEM.
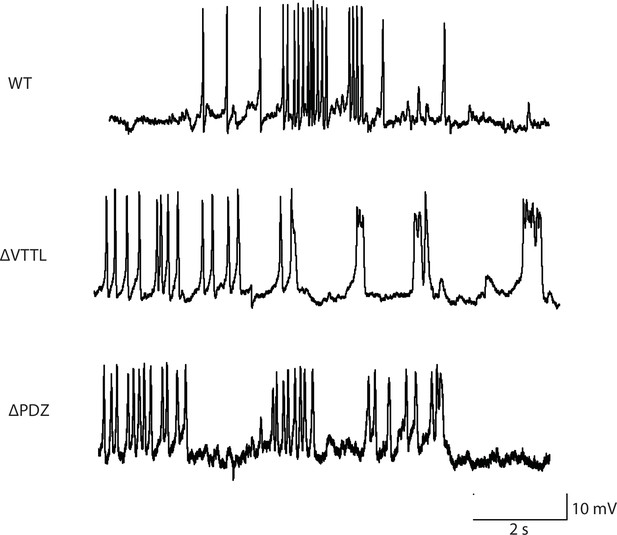
Representative traces for recordings summarized in Figure 3B and C.
Representative traces of spontaneous muscle APs are shown for the indicated genotypes.
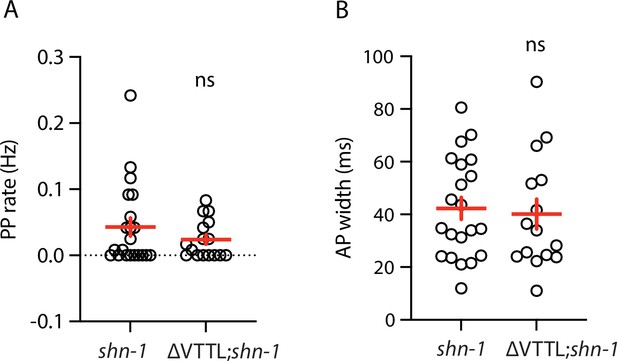
Muscle AP defects in shn-1(null) and shn-1(null); egl-19(ΔVTTL) double mutants are not significantly different.
Mean PP rate and AP width are compared in the indicated genotypes. Sample sizes are as follows: shn-1(nu712 null) (21); shn-1(nu712); egl-19(ΔVTTL) (15). Error bars indicate SEM. No significant differences were observed.
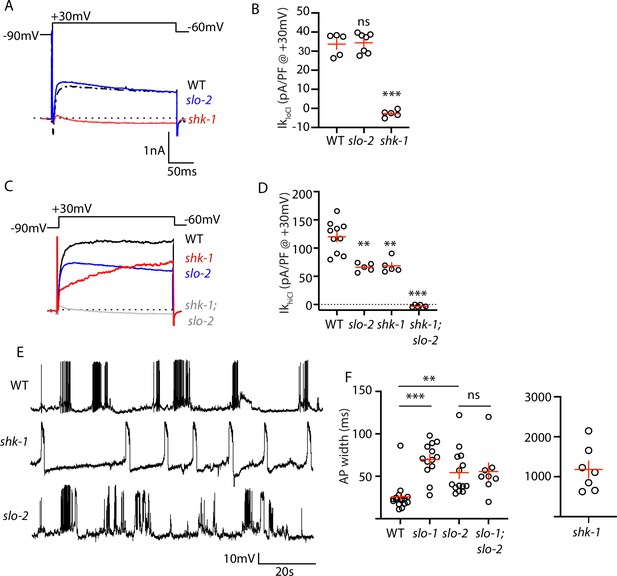
AP repolarization is mediated by SHK-1 and SLO channels.
(A–D) Muscle voltage activated potassium currents are mediated by SHK-1 and SLO-2. Voltage activated potassium currents were recorded using pipette solutions containing low (IkloCl, A–B) and high (IkhiCl, C–D) chloride concentrations. Representative traces (A,C) and mean current density (B,D) at +30 mV are shown. IkloCl is mediated by SHK-1 whereas SHK-1 and SLO-2 equally contribute to IkhiCl. (E–F) AP durations are significantly increased in mutants lacking SHK-1, SLO-1, and SLO-2 channels. The AP widths observed in slo-1; slo-2 double mutants were not significantly different from those found in slo-2 single mutants. Representative traces (E and Figure 4—figure supplement 1) and mean AP widths (F) are shown. Alleles used in this figure were: shk-1(ok1581), slo-1(js379), and slo-2(nf100). Sample sizes are as follows: in panel B, WT (5), slo-2 (7), and shk-1 (5); in panel D, WT (10), slo-2 (5), shk-1 (5), shk-1;slo-2 (6); in panel F, WT (16), slo-1 (13), slo-2 (14), slo-1; slo-2 (8), shk-1 (7). Values that differ significantly from wild type controls are indicated (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate SEM.
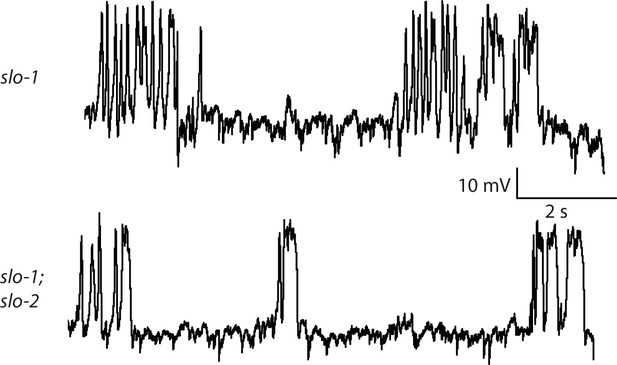
Representative traces for recordings summarized in Figure 4E.
Representative traces of spontaneous muscle APs are shown for the indicated genotypes.
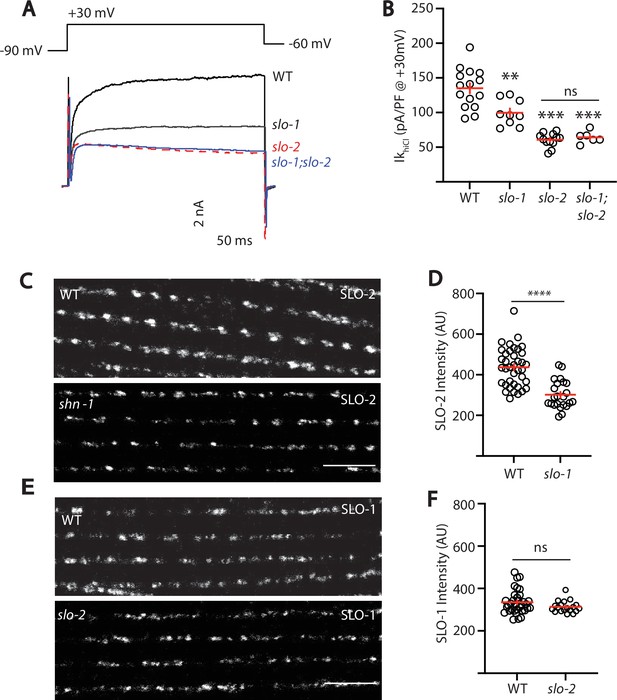
SLO-2 and SLO-1 function together in heteromeric channels.
(A–B) IkhiCl was significantly decreased in slo-1(js379) and slo-2(nf100) single mutants but was not further decreased in slo-1; slo-2 double mutants. Representative traces (A) and mean current density (B) at +30 mV are shown. Sample sizes for panel B: WT (15), slo-1 (9), slo-2 (12), slo-1;slo-2 (6). (C–F) Expression of split GFP tagged SLO-2 (C–D) and SLO-1 (E–F) was analyzed in body muscles. CRISPR alleles were constructed adding 7 copies of GFP11 to the endogenous slo-1 and slo-2 genes (Table 2) and fluorescence was reconstituted by expressing GFP1-10 in body muscles. Controls showing that the GFP11 tags had no effect on AP width, RMP, and potassium currents are shown in Figure 5—figure supplement 1. Representative images (C and E) and mean puncta intensity (D and F) are shown. SLO-2 puncta intensity was significantly decreased in slo-1(js379) mutants. SLO-1 puncta intensity was unaltered in slo-2(nf100) mutants. Sample sizes are as follows: in panel D, WT (38) and slo-1 (23); in panel F, WT (34) and slo-2 (19). Values that differ significantly from wild type controls are indicated (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate SEM. Scale bar indicates 4 μm.
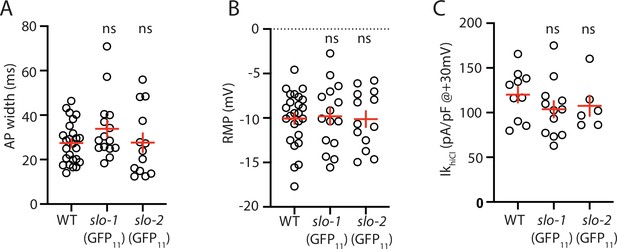
Analysis of GFP11 tagged slo-1 and slo-2 alleles.
APs and potassium currents were analyzed in strains containing slo-1(nu678 GFP11) and slo-2(nu725 GFP11) together with the muscle>GFP1-10 transgene. Mean AP width (A), RMP (B), and IkhiCl current density (C) were not significantly different from WT controls. Sample sizes are as follows: in panels A and B, WT (25), slo-1(nu678) (15), and slo-2(nu725) (13); in panel C, WT (10), slo-1(nu678) (12), and slo-2(nu725) (6). Error bars indicate SEM.
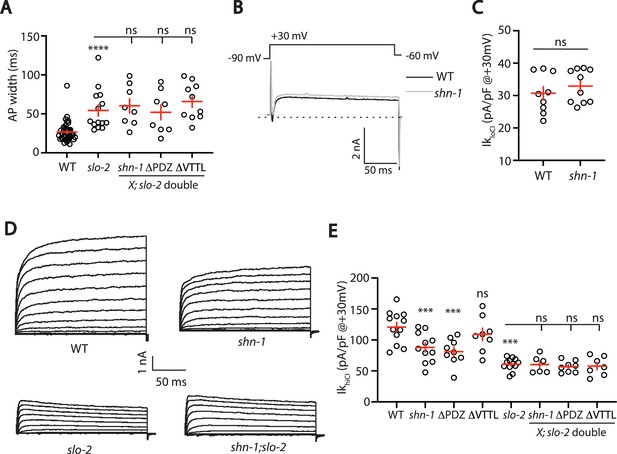
SHN-1 controls AP width by regulating SLO-2.
(A) A slo-2 null mutation blocks the effect of SHN-1 on AP width. Mean AP widths in slo-2(nf100) double mutants containing shn-1(nu712 null), shn-1(nu542 ΔPDZ), and egl-19(nu496 ΔVTTL) mutations were not significantly different from those in slo-2 single mutants. Representative traces are shown in Figure 6—figure supplement 1A. Sample sizes: WT (41), slo-2 (14), slo-2;shn-1 (8), slo-2;ΔPDZ (8), and slo-2;ΔVTTL (10). (B–C) IkloCl currents were unaltered in shn-1(nu712 null) mutants. Representative traces (B) and mean current density at +30 mV (C) are shown. Sample sizes: WT (9) and shn-1 (10). These results show that SHK-1 channel function was unaffected in shn-1 mutants. (D–E) IkhiCl currents were significantly smaller in shn-1(nu712 null) and shn-1(nu542 ΔPDZ) mutants but were unaffected in egl-19(nu496 ΔVTTL) mutants. The effect of shn-1 mutations on IkhiCl was eliminated in double mutants lacking SLO-2, indicating that the SHN-1 sensitive potassium current is mediated by SLO-2. IkhiCl currents were recorded from adult body wall muscles of the indicated genotypes at holding potentials of –60 to +60 mV. Representative traces (D and Figure 6—figure supplement 1B) and mean current density at +30 mV (E) are shown. Sample sizes in panel E: WT (12), shn-1 (11), slo-2 (12), ΔPDZ (9), ΔVTTL (8), slo-2;shn-1 (6), slo-2;ΔPDZ (8), and slo-2;ΔVTTL (7). Values that differ significantly from wild type controls are indicated (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate SEM.
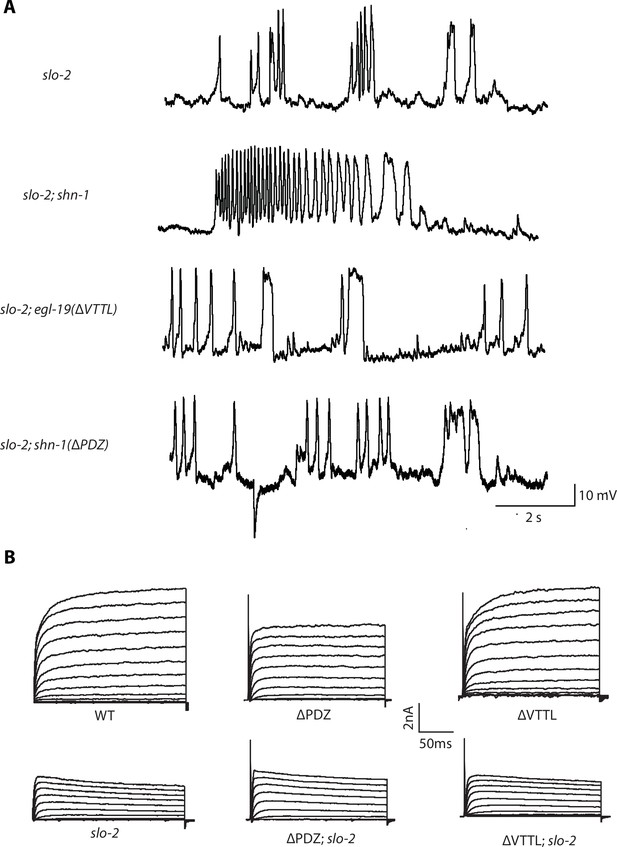
Representative traces for recordings summarized in Figure 6A and E.
Representative traces of spontaneous muscle APs (A) and IkhiCl currents at +30 mV (B) are shown for the indicated genotypes.
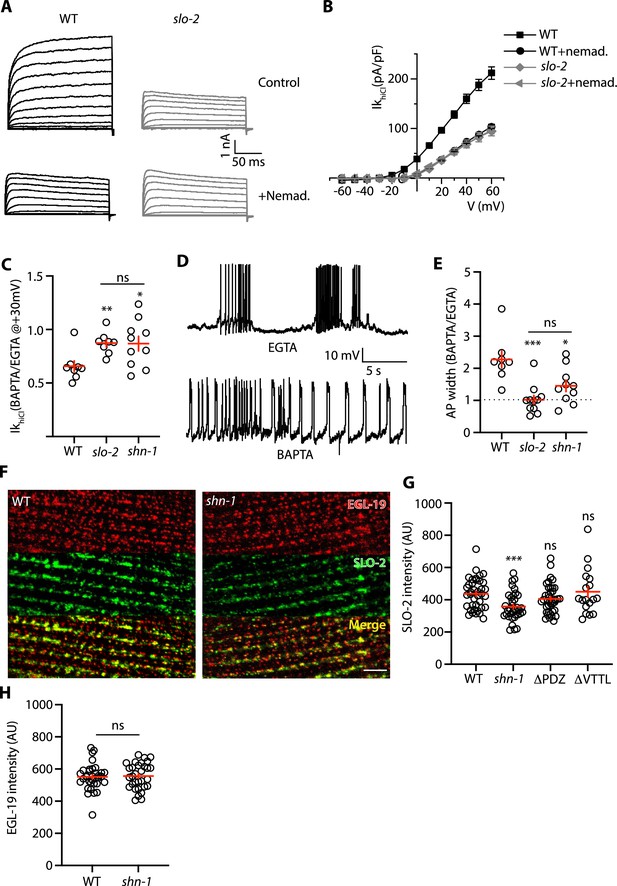
SHN-1 promotes EGL-19 to SLO-2 microdomain coupling.
(A–B) SLO-2 activation is functionally coupled to EGL-19. IkhiCl was significantly reduced by nemadipine (an EGL-19 antagonist). This inhibitory effect of nemadipine on IkhiCl was eliminated in slo-2(nf100) mutants, indicating that the nemadipine sensitive current is mediated by SLO-2. IkhiCl currents were recorded from adult body wall muscles of the indicated genotypes at holding potentials of –60 to +60 mV. Representative IkhiCl traces (A) and mean current density as a function of membrane potential (B) are shown. (C) SLO-2 activation requires microdomain coupling to EGL-19. IkhiCl currents recorded in BAPTA are significantly smaller than those in EGTA. The inhibitory effect of BAPTA was reduced in shn-1(nu712 null) mutants and was eliminated in slo-2(nf100) mutants, indicating that the BAPTA sensitive current is mediated by SLO-2. The ratio of IkhiCl current density at +30 mV recorded in BAPTA to the mean current density recorded in EGTA is plotted for the indicated genotypes. Representative traces are shown in Figure 7—figure supplement 1A. Sample sizes for panel C: WT (8), slo-2 (8), and shn-1 (10). (D–E) AP repolarization is mediated by microdomain activation of SLO-2. AP widths recorded in solutions containing BAPTA are wider than those recorded in EGTA. The effect of BAPTA on AP widths was reduced in shn-1(nu712 null) mutants and was eliminated in slo-2(nf100) mutants, indicating that BAPTA’s effect is mediated by SLO-2. Representative traces of WT muscle APs recorded in EGTA and BAPTA are shown (D). The ratio of AP widths recorded in BAPTA to the mean AP widths recorded in EGTA is plotted for the indicated genotypes (E). Representative traces for panel E are shown in Figure 7—figure supplement 1B. Sample sizes for panel E: WT (8), slo-2 (11), and shn-1 (10). (F–H) SLO-2(nu725 GFP11) is partially co-localized with EGL-19(nu722 Cherry11) in body muscles. GFP11 and Cherry11 fluorescence were reconstituted by expressing GFP1-10 and Cherry1-10 in body muscles. SLO-2 puncta intensity was significantly reduced in shn-1(nu712 null) mutants but was unaffected in shn-1(nu542 ΔPDZ) and egl-19(nu496 ΔVTTL) mutants. Representative images (F) and mean puncta intensity for SLO-2 (G) and EGL-19 (H) are shown. Sample sizes for panel G: slo-2(GFP11) single mutants (38), and double mutants containing the shn-1 (35), ΔPDZ (39), and ΔVTTL (18) mutations. Sample sizes for panel H: egl-19(Cherry11) single mutants (34) and shn-1; egl-19(Cherry11) double mutants (31). Values that differ significantly from wild type controls are indicated (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate SEM. Scale bar indicates 4 μm.
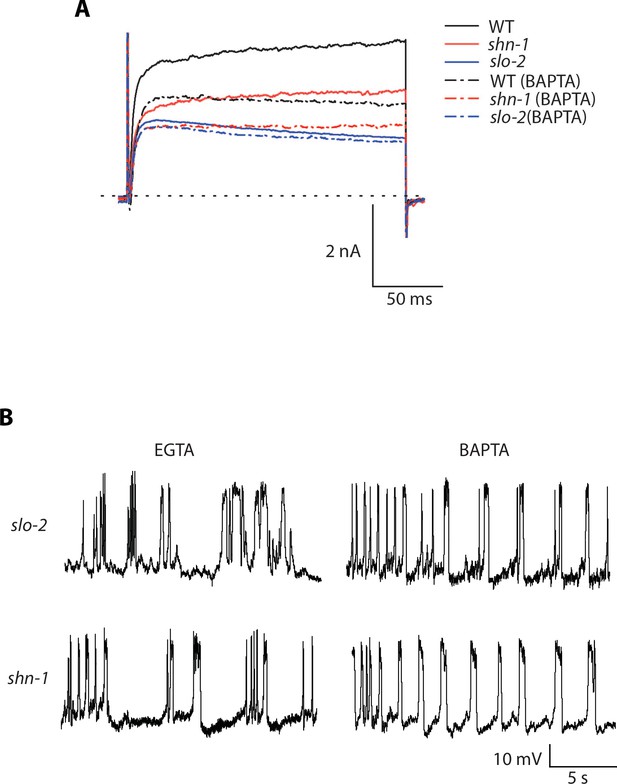
Representative traces for recordings summarized in Figure 7C and E.
Representative traces of IkhiCl currents at +30 mV (A) and spontaneous muscle APs (B) are shown for the indicated genotypes and recording conditions.
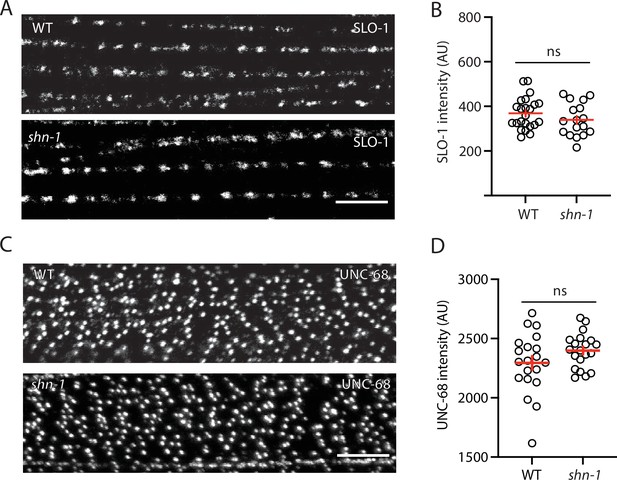
SLO-1 and UNC-68/RYR puncta intensity is unaltered in shn-1 mutant muscles.
SLO-1(nu678 GFP11) and UNC-68(nu664 GFP11) puncta intensity in body muscles is compared in WT and shn-1(null) mutants. GFP11 fluorescence was reconstituted by expressing GFP1-10 in body muscles. Representative images (A and C) and mean puncta intensity (B and D) are shown. Sample sizes: slo-1(GFP11) single mutants (23); shn-1(nu712 null);slo-1(GFP11) double mutants (17); unc-68(GFP11) single mutants (20); and shn-1(tm488 null);unc-68(GFP11) double mutants (20). No significant differences were observed. Error bars indicate SEM. Scale bars indicate 4 μm.
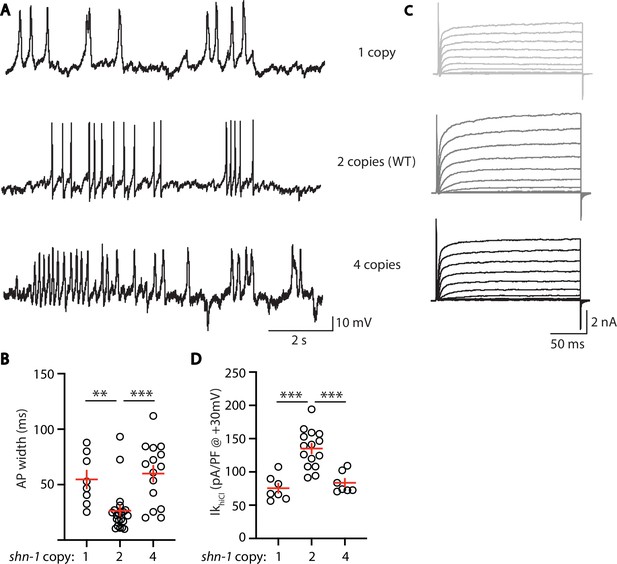
AP width and SLO-2 current are sensitive to shn-1 gene dosage.
The effect of shn-1 gene dosage on AP widths (A–B) and IkhiCl current (C–D) was analyzed. IkhiCl was significantly decreased while AP duration was significantly increased in animals containing 1 and 4 copies of shn-1 compared to WT controls (i.e. 2 copies). The following genotypes were analyzed: 1 copy of shn-1 [shn-1(nu712)/ + heterozygotes], 2 copies of shn-1 (WT) and 4 copies of shn-1 (nuSi26 homozygotes in wild-type). IkhiCl currents were recorded from adult body wall muscles of the indicated genotypes at holding potentials of –60 to +60 mV. Representative traces (A,C), mean AP width (B), and mean IkhiCl current density at +30 mV (D) are shown. Sample sizes: for panel B, 1 copy (8), 2 copies (21), and four copies (15); for panel D, 1 copy (8), 2 copies (15), and four copies (7). Significant differences are indicated (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate SEM.
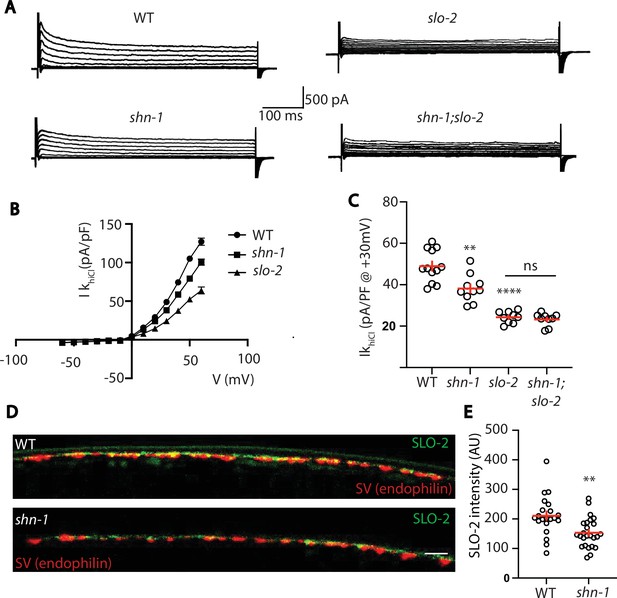
SHN-1 controls SLO-2 currents in motor neurons.
(A–B) IkhiCl currents in cholinergic motor neurons were significantly decreased in shn-1(nu712 null) mutants. IkhiCl currents were recorded from adult cholinergic motor neurons of the indicated genotypes at holding potentials of –60 to +60 mV. Representative traces (A), mean current density as a function of membrane potential (B), and mean current density at +30 mV (C) are shown. Sample sizes for panels B and C: WT (12), shn-1 (10), slo-2 (9), and shn-1; slo-2 (9). (D–E) SLO-2 puncta intensity in motor neuron axons was significantly decreased in shn-1(nu712 null) mutants. Representative images of SLO-2(nu725 GFP11) and a synaptic vesicle marker [UNC-57/Endophilin(mCherry)] in dorsal cord axons of DA/DB motor neurons are shown (D). GFP11 fluorescence was reconstituted with GFP1-10 expressed in DA/DB motor neurons (using the unc-129 promoter). Mean SLO-2 puncta intensity in axons is plotted (E). Sample sizes for panel E: WT (21) and shn-1 (25). Values that differ significantly from wild type controls are indicated (ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001). Error bars indicate SEM. Scale bar indicates 2 μm.
Tables
Comparison of shn-1 null alleles.
| Genotype: | PP rate (Hz): | AP width (ms): | AP Amp. (mV): | RMP (mV): |
|---|---|---|---|---|
| WT | 0.01 ± 0.00 | 26.73 ± 1.98 | 40.42 ± 0.77 | –12.61 ± 0.96 |
| shn-1(nu712) | 0.04 ± 0.01** | 42.28 ± 4.17*** | 37.63 ± 0.83* | –8.33 ± 0.72** |
| shn-1(nu652) | 0.05 ± 0.02*** | 58.06 ± 5.27*** | 34.78 ± 0.92*** | –7.19 ± 0.99*** |
| shn-1(tm488) | 0.05 ± 0.01*** | 49.06 ± 7.07*** | 41.36 ± 1.64 | –12.06 ± 1.54 |
-
Mean, SEM, and significant differences from WT controls are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (C. elegans) | N2 Bristol | https://cgc.umn.edu/ | N2 | Wild-type reference |
| Strain, strain background (C. elegans) | slo-1(js379) | Wang et al., 2001 | NM1968 | |
| Strain, strain background (C. elegans) | slo-2(nf100) | Santi et al., 2003 | LY100 | |
| Strain, strain background (C. elegans) | slo-1(js379);slo-2(nf100) | This paper | KP10046 | |
| Strain, strain background (C. elegans) | shk-1(ok1581) | Liu et al., 2011 | RB1392 | |
| Strain, strain background (C. elegans) | shk-1(ok1581);slo-2(nf100) | This paper | KP10879 | |
| Strain, strain background (C. elegans) | shn-1(tm488) | Oh et al., 2011 | KP7032 | |
| Strain, strain background (C. elegans) | shn-1(nu712) | This paper | KP10151 | |
| Strain, strain background (C. elegans) | shn-1(nu697) | This paper | KP10082 | |
| Strain, strain background (C. elegans) | nuSi26 | Pym et al., 2017 | KP7493 | Pmyo-3::shn-1A MOSSci |
| Strain, strain background (C. elegans) | nuSi572 | This paper | KP10696 | Muscle CRE |
| Strain, strain background (C. elegans) | nuSi502 | This paper | KP10497 | Pan neuron CRE |
| Strain, strain background (C. elegans) | shn-1(nu697);nuSi572 | This paper | KP10767 | |
| Strain, strain background (C. elegans) | shn-1(nu697);nuSi502 | This paper | KP10768 | |
| Strain, strain background (C. elegans) | nuSi205 | This paper | KP9234 | Ubiquitous GFP1-10 |
| Strain, strain background (C. elegans) | shn-1(nu604 GFP11) | This paper | KP8587 | |
| Strain, strain background (C. elegans) | shn-1(nu652); nuSi205 | This paper | KP9548 | |
| Strain, strain background (C. elegans) | nuSi470 | This paper | KP10393 | Muscle CRE |
| Strain, strain background (C. elegans) | shn-1(nu652);nuSi470 | This paper | KP10437 | |
| Strain, strain background (C. elegans) | shn-1(nu604);nusi205 | This paper | KP9232 | |
| Strain, strain background (C. elegans) | shn-1(nu542ΔPDZ) | This paper | KP9898 | |
| Strain, strain background (C. elegans) | egl-19(nu496ΔVTTL) | Pym et al., 2017 | KP7992 | |
| Strain, strain background (C. elegans) | egl-19(nu496);shn-1(tm488) | Pym et al., 2017 | KP8046 | |
| Strain, strain background (C. elegans) | nuSi144 | This paper | KP9814 | muscle GFP1-10 |
| Strain, strain background (C. elegans) | slo-2(nu725 GFP11) | This paper | KP10285 | |
| Strain, strain background (C. elegans) | slo-2(nu725);nuSi144 | This paper | KP10031 | |
| Strain, strain background (C. elegans) | shn-1(nu712); slo-2(nu725);nuSi144 | This paper | KP10894 | |
| Strain, strain background (C. elegans) | shn-1(nu542); slo-2(nu725);nuSi144 | This paper | KP10890 | |
| Strain, strain background (C. elegans) | egl-19(nu496); slo-2(nu725);nuSi144 | This paper | KP10891 | |
| Strain, strain background (C. elegans) | slo-1(nu678 GFP11) | This paper | KP9826 | |
| Strain, strain background (C. elegans) | slo-1(nu678);nuSi144 | This paper | KP10030 | |
| Strain, strain background (C. elegans) | shn-1(nu712); slo-1(nu678);nuSi144 | This paper | KP10892 | |
| Strain, strain background (C. elegans) | shn-1(nu712);slo-2(nf100) | This paper | KP10880 | |
| Strain, strain background (C. elegans) | shn-1(nu542);slo-2(nf100) | This paper | KP10881 | |
| Strain, strain background (C. elegans) | egl-19(nu496);slo-2(nf100) | This paper | KP10882 | |
| Strain, strain background (C. elegans) | nuSi458 | This paper | KP10374 | muscle Cherry1-10 SL2 GFP1-10 |
| Strain, strain background (C. elegans) | egl-19(nu722 Cherry11) | This paper | KP10230 | |
| Strain, strain background (C. elegans) | slo-2(nu725);egl-19(nu722);nusi458 | This paper | KP10816 | |
| Strain, strain background (C. elegans) | shn-1(nu712); slo-2(nu725);egl-19(nu722);nusi458 | This paper | KP10816 | |
| Strain, strain background (C. elegans) | shn-1(nu712);vsIs48 | This paper | KP10883 | vsIs48 is Punc-17::GFP |
| Strain, strain background (C. elegans) | slo-2(nf100);vsIs48 | This paper | KP10884 | |
| Strain, strain background (C. elegans) | shn-1(nu712);slo-2(nf100);vsIs48 | This paper | KP10885 | |
| Strain, strain background (C. elegans) | slo-2(nu725);nuSi144 | This paper | KP10886 | |
| Strain, strain background (C. elegans) | shn-1(nu712);slo-2(nu725);nusi144 | This paper | KP10887 | |
| Strain, strain background (C. elegans) | slo-1(nu678); slo-2(nf100); nuSi144 | This paper | KP10895 | |
| Strain, strain background (C. elegans) | slo-2(nu725); slo-1(js379);nuSi144 | This paper | KP10896 | |
| Strain, strain background (C. elegans) | slo-2(nu725);nuSi250 | This paper | KP10897 | nuSi250 is Punc-129 GFP1-10 |
| Strain, strain background (C. elegans) | shn-1(nu712); slo-2(nu725 GFP11);nuSi250 | This paper | KP10898 | |
| Strain, strain background (C. elegans) | egl-19(nu496); shn-1(nu712) | This paper | KP10906 | |
| Strain, strain background (C. elegans) | unc-68(nu664); nuSi144 | This paper | KP9802 | |
| Strain, strain background (C. elegans) | unc-68(nu664); shn-1(tm488); nuSi144 | This paper | KP10040 | |
| Strain, strain background (C. elegans) | shn-1(nu604,nu652); nuSi502 | This paper | KP10907 | |
| Strain, strain background (E. coli) | OP50 | Brenner, 1974 | OP50 | Worm food |
| Sequence-based reagent | egl-19 residue 2 | This paper | N/A | TTACCTGACATGATGGACAC |
| Sequence-based reagent | shn-1 ∆PDZ 5' | This paper | N/A | gtgattccacgtggtgtcaa |
| Sequence-based reagent | shn-1 ∆PDZ 3' | This paper | N/A | gtagctgatatgagtagggg |
| Sequence-based reagent | shn-1 intron one loxP insertion | This paper | N/A | tcaatttcagAAGTTCCTTG |
| Sequence-based reagent | shn-1 3' UTR loxP insertion | This paper | N/A | gaaaaggcatagaatcagtg |
| Sequence-based reagent | shn-1 intron two insertion for STOP cassette | This paper | N/A | ggggaaagatatgcatctga |
| Sequence-based reagent | shn-1 residue 946 insertion | This paper | N/A | CACATCTTCTCGAACGTCAC |
| Sequence-based reagent | slo-1 residue 1,121 insertion | This paper | N/A | cccggctcgtactccagtcc |
| Sequence-based reagent | slo-2 residue 1,092 insertion | This paper | N/A | ctgcgtcttagaccccttct |
| Recombinant DNA reagent | Pmyo-3::gfp1-10 | This paper | KP#3,315 | muscle GFP1-10 |
| Recombinant DNA reagent | Peft-3::gfp1-10 | This paper | KP#4,524 | ubiquitous GFP1-10 |
| Recombinant DNA reagent | Punc-129::gfp1-10 | This paper | KP#4,525 | GFP1-10 in DA/B neurons |
| Recombinant DNA reagent | Ppat-10 Cherry1-10 SL2 GFP 1-10 | This paper | KP#4,526 | Cherry1-10 and GFP1-10 in muscles |
| Recombinant DNA reagent | Pmyo-3::CRE | This paper | KP#4,527 | muscle CRE |
| Recombinant DNA reagent | Psbt-1::CRE | This paper | KP#4,528 | Pan neuron CRE |
| Recombinant DNA reagent | Peft-3::CRE | This paper | KP#4,529 | germline CRE |
| Chemical compound, drug | Nemadipine-A | Abcam | ab145991 | N/A |
| Software, algorithm | MATLAB R2018a | MATLAB | N/A | N/A |
| Software, algorithm | Fiji | https://fiji.sc/ | N/A | N/A |
| Software, algorithm | ClampFit | Molecular Devices | N/A | N/A |
| Software, algorithm | Prism 9 | GraphPad | N/A | N/A |
| Software, algorithm | Origin 2019 | OriginLab | N/A | N/A |
| Software, algorithm | Adobe illustrator 2020 | Adobe | N/A | N/A |
Alleles used in this study.
| Allele: | Description: | Reference: | |
|---|---|---|---|
| shn-1(tm488) | 1537 nt deletion, frameshift at codon 118 | Oh et al., 2011 | |
| shn-1(nu697) | LoxP sites in intron 1 and 3'UTR | This study | |
| shn-1(nu712) | derived by germline CRE recombination of nu697 | This study | |
| shn-1(nu652) | stop cassette (flanked by FLEX sites) in intron two in "OFF" orientation | This study | |
| shn-1(nu600 GFP11) | seven copies GFP11 inserted at codon 945 of SHN-1A | This study | |
| shn-1(nu542ΔPDZ) | deletes aa 446–532 of SHN-1A | This study | |
| egl-19(nu722 Cherry11) | six copies sfCherry11 inserted at codon 2 | This study | |
| egl-19(nu496ΔVTTL) | WT C-term PAENSSRQHDSRGGSQEDLLLVTTL replaced with PMIHAEDHKKSYF | Pym et al., 2017 | |
| unc-68(nu664 GFP11) | seven copies GFP11 inserted at codon 3,705 of UNC-68A | Piggott et al., 2021 | |
| slo-2(nu725 GFP11) | seven copies GFP11 inserted at codon 1,092 of SLO-2A | This study | |
| slo-1(nu678 GFP11) | seven copies GFP11 inserted at codon 1,130 of SLO-1A | This study | |
| slo-1(js379) | Q251stop | Wang et al., 2001 | |
| slo-2(nf100) | in frame deletion of aa 450–569 | Santi et al., 2003 | |
| shk-1(ok1581) | P253stop | Liu et al., 2011 | |





