Key features of the genetic architecture and evolution of host-microbe interactions revealed by high-resolution genetic mapping of the mucosa-associated gut microbiome in hybrid mice
Figures
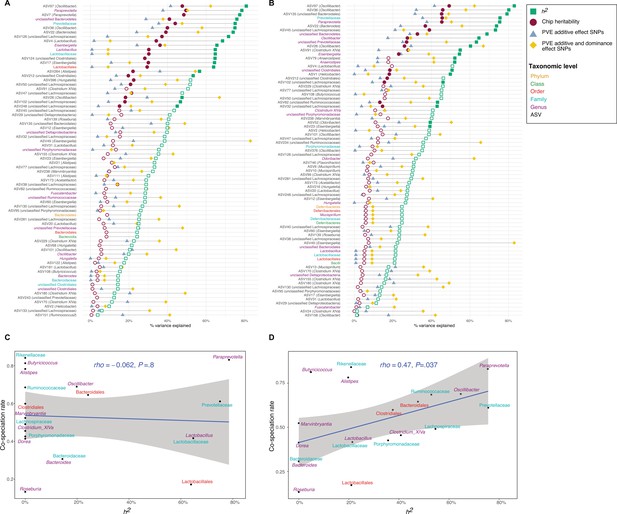
Heritability estimates and their relationship to cospeciation rates.
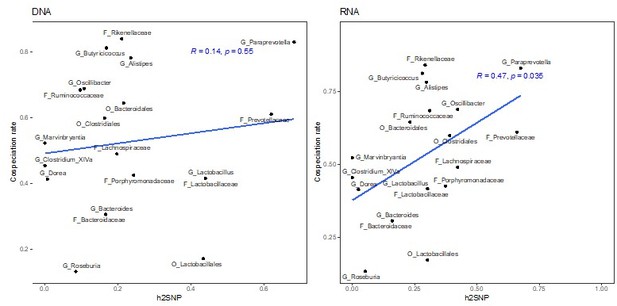
(A–B) Lollipop chart showing the values of the chip heritability (dark red circles), narrow-sense heritability (green squares), PVE by the additive effect (blue triangles), and additive and dominance effect (yellow diamonds) of all significant single nucleotide polymorphisms (SNPs) within one taxon for DNA-based traits (A) and RNA-based traits (B) Only taxa with a non-zero narrow-sense heritability estimate are shown. Only the outline of non-significant heritability estimates (p<0.05, Restricted Likelihood Ratio test) are shown. Taxa without significant hits have no PVE reported. (C–D) Relationship between the narrow-sense heritability estimates for the relative abundance of bacterial taxa and cospeciation rate for the same genus calculated by Groussin et al., 2017. DNA level (C), and RNA level (D). The blue line represents a linear regression fit to the data and the grey area the corresponding confidence interval. p-Values and the correlation coefficient are calculated using the Spearman’s correlation test. The text labels on the y-axis (A–B) and the text labels in panels (C-D) are coloured according to taxonomic level: amplicon sequence variants (ASV) in black, genus in purple, family in light blue, order in red, class in green, and phylum in yellow.
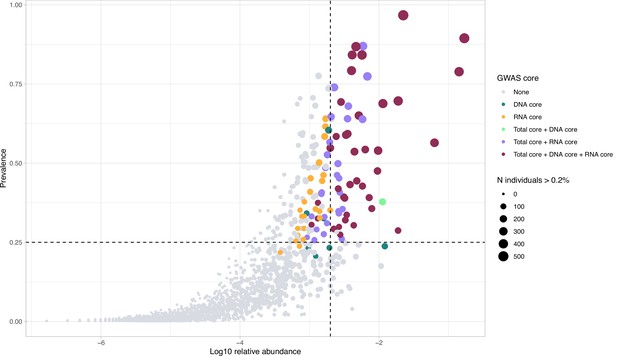
Selection of taxa for mGWAS analysis.
A scatter plot showing the association of average relative abundance of taxa with their prevalence in the G2 mapping population. Taxa retained for analysis are coloured according to the originating core. The size of each dot represents the number of individuals that have a median abundance higher than 0.2% of the taxon. The dashed lines represent the thresholds of the core vertical: median abundance >0.2% and horizontal prevalence of 25%.
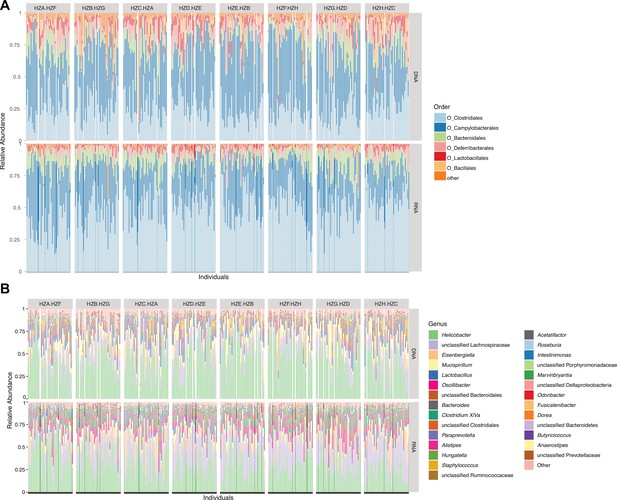
Relative abundances of core taxa.
Core orders (A) and genera (B) present in the G2 mapping population. Each vertical line represents one individual. Sub-cross (see Figure 1—figure supplement 6) is indicated at the top.
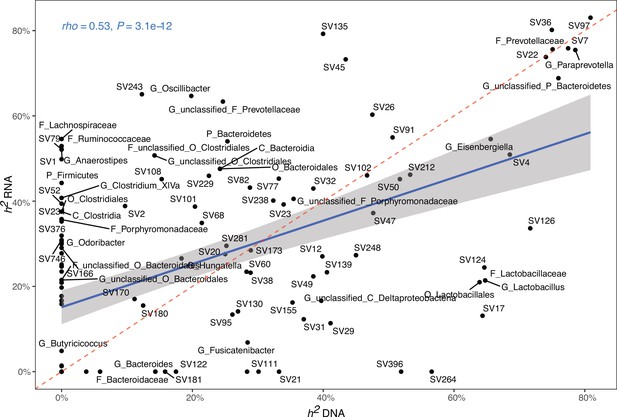
Correlation of single nucleotide polymorphisms (SNP)-based heritability estimates based on DNA (x-axis) or RNA (y-axis).
The blue line represents a linear regression fit to the data. Red dashed line represents the identity line with a slope of 1.
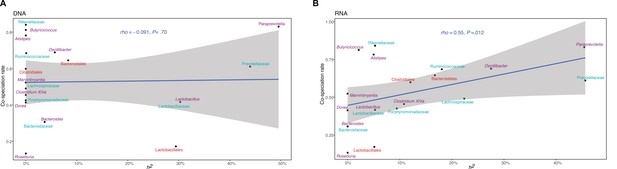
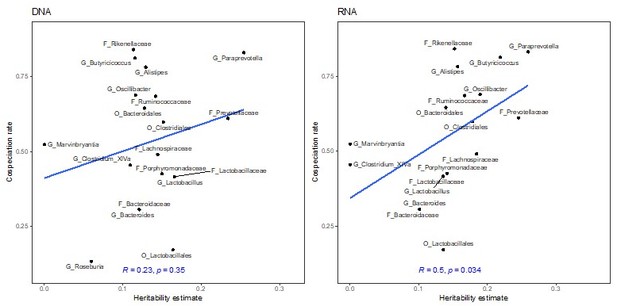
Relationship between the chip heritability estimates for the relative abundance of bacterial taxa and cospeciation rate for the same genus calculated by Groussin et al., 2017.
DNA level (A) and RNA level (B). The blue line represents a linear regression fit to the data and the grey area the corresponding confidence interval. p-Values and the correlation coefficient are calculated using the Spearman’s correlation test. The text labels in panels are coloured according to taxonomic level: amplicon sequence variants (ASV) in black, genus in purple, family in light blue, order in red, class in green, and phylum in yellow.
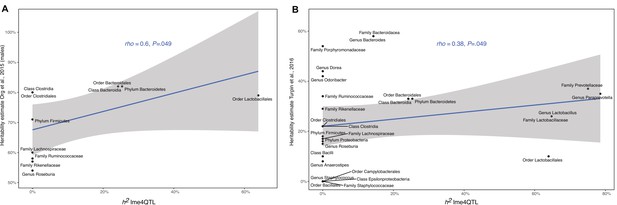
Correlation between heritability estimates.
Estimates calculated with lme4QTL and heritability estimates reported in Org et al., 2015 (males) (A) and heritability estimates reported in Turpin et al., 2016 (B). Only taxa present in both studies are shown (n=11 and n=28, respectively). The blue line represents a linear regression fit to the data and the grey area the corresponding confidence interval. p-Values and the correlation coefficient are calculated using the Spearman’s correlation test.
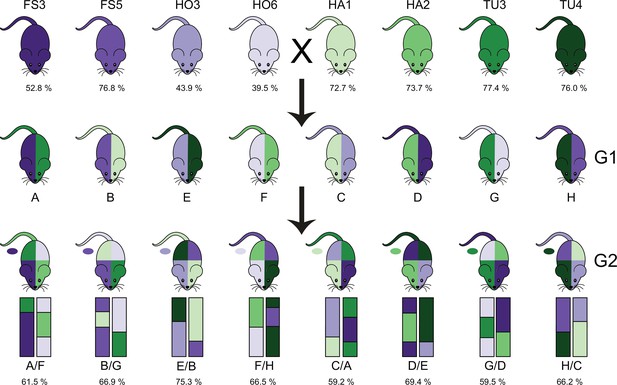
Overview of the intercross design.
G0 mice are from eight partially inbred lines derived from mice wild-caught in four hybrid zone sites. Hybrid index—the percentage of musculus alleles—is reported as the mean for the G0 mice from each line (top), or mean of 40 G2s from each sub-cross (bottom). We performed eight G1 crosses with one line with hybrid index ~50% (purple shades) and one line with hybrid index >50% (green shades); colour on the left side of mouse diagram indicates dam line and right side indicates sire line. Next, G1 mice were crossed in eight combinations such that each G2 mouse had one grandparent from each of the four breeding stocks, indicated by colours of mouse diagram, and representative chromosomes below. Tail colour indicates Y chromosome strain, and oval indicates mitochondrial strain.
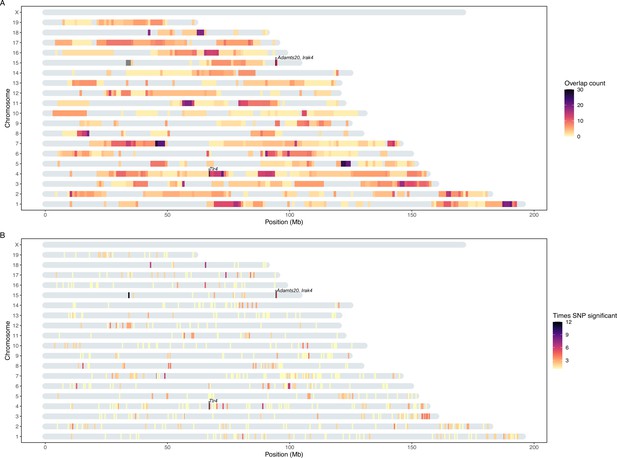
Heatmap of significant host loci from association mapping of bacterial abundances.
Karotype plot showing the number of significant loci found using a study-wide threshold, where (A) plots the significance intervals and (B) the significant single nucleotide polymorphisms (SNP) markers on the chromosomes. The position of the SNPs on panel (B) has been amplified by 0.5 Mb to visualise it. The position of the genes closest to SNPs with the lowest p-values (Tlr4, and Irak4 and Adamsts20) are indicated.
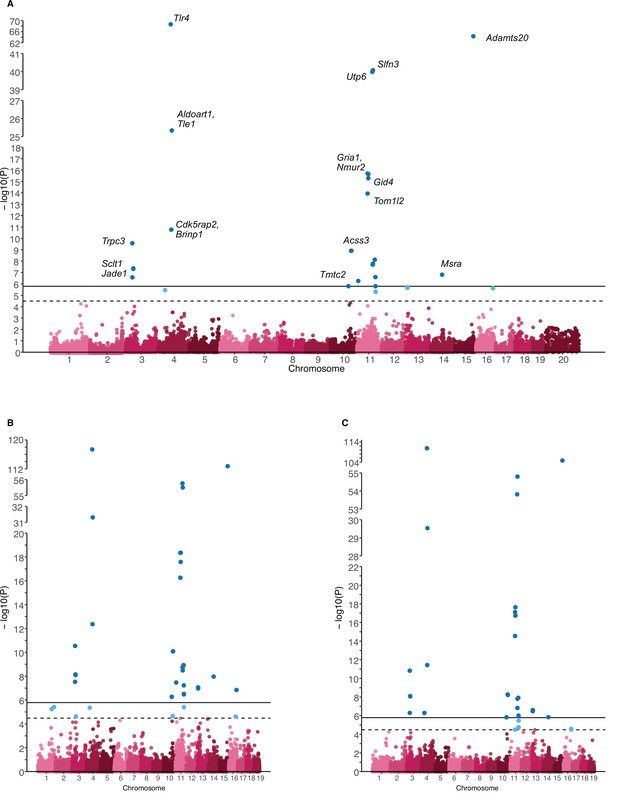
Manhattan plots for ASV184 (Dorea).
The complete model (A), the additive effect (B) or the dominance effect (C) are shown. Single nucleotide polymorphisms (SNPs) passing the study-wide significance threshold (solid line) are shown in dark blue, while genome-wide significant SNPs (dashed line) are shown in light blue. In panel (A), the closest gene to the SNP is shown for a subset of significant SNPs.
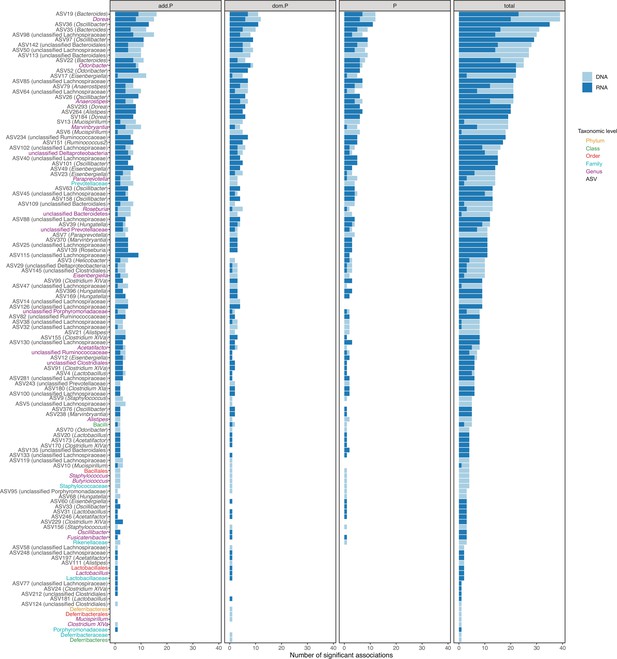
Number of significantly associated loci per bacterial taxon.
Loci with significant additive effects (add.P), dominance effects (dom.P) or effects in full model (P) are indicated.
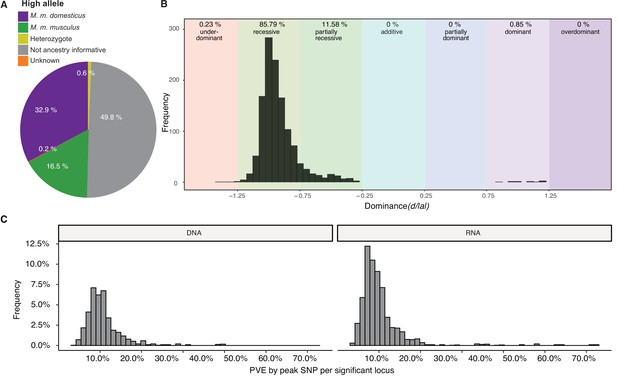
Genetic architecture of significant loci.
(A) Source of the allele with the highest phenotypic value. (B) Histogram of dominance values d/a of significant loci reveals a majority of loci acting recessive or partially recessive. (C) Histogram showing the percentage of variance explained (PVE) by the peak single nucleotide polymorphisms (SNP) for DNA (left) and RNA (right).
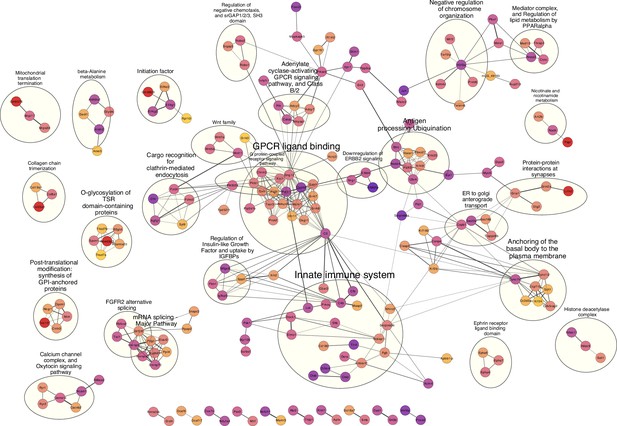
High confidence protein-protein interaction (PPI) network of genes closest to single nucleotide polymorphisms (SNPs) significantly associated with bacterial abundances.
Network clusters are annotated using STRING’s functional enrichment (Doncheva et al., 2019). Nodes represent proteins and edges their respective interactions. Only edges with an interaction score higher than 0.9 are retained. The width of the edge line expresses the interaction score calculated by STRING. The colour of the nodes describes the expression of the protein in the intestine where yellow is not expressed and purple is highly expressed.
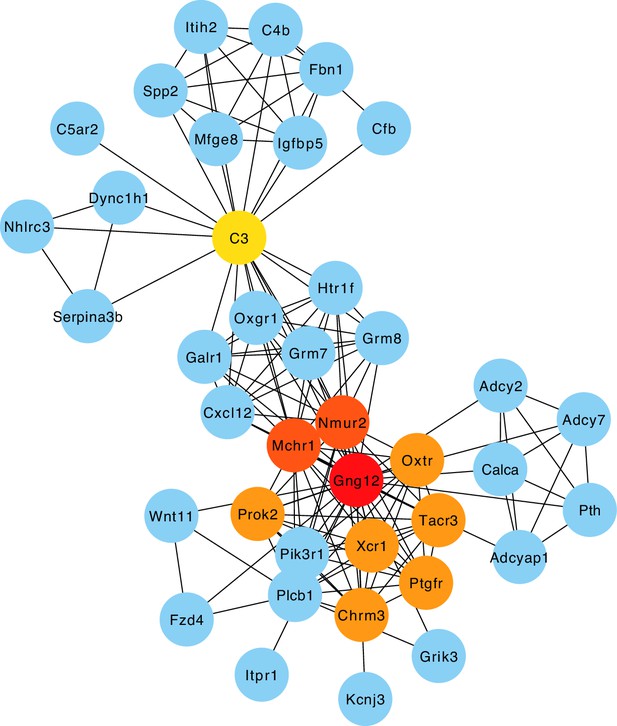
Protein-protein interaction (PPI) network of hub genes of the 'nearest gene' network.
The top ten hub genes of the protein-protein interaction (PPI) network of the closest genes to the host single nucleotide polymorphisms (SNPs) significantly associated with bacterial abundances Figure 4. The nodes are coloured according to hub gene rank from 1 (red) to 10 (yellow). Blue nodes are the first neighbours.
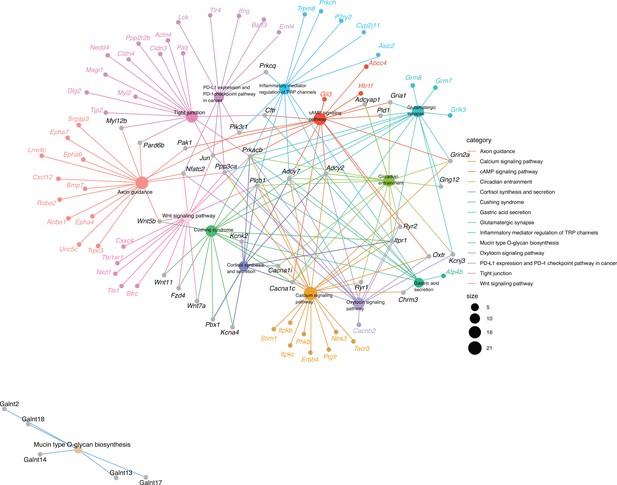
Genes belonging to overrepresented KEGG pathways within the host genes closest to significant single nucleotide polymorphisms (SNPs) from association analysis.
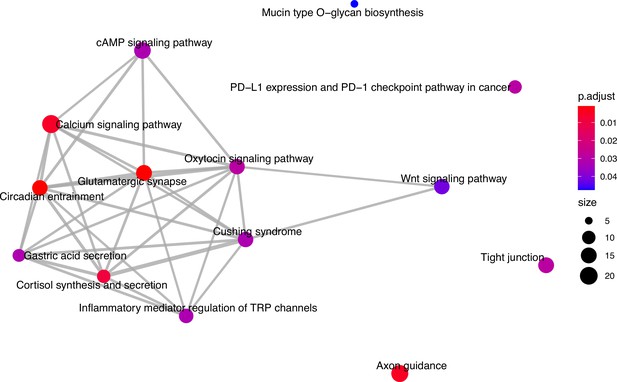
Enriched KEGG pathways.
KEGG pathways enriched among closest genes to significant single nucleotide polymorphisms (SNPs) from association analysis. Node colour indicates false discovery rate (FDR)-adjusted p-value of enrichment and node size indicates number of candidate genes in pathway.
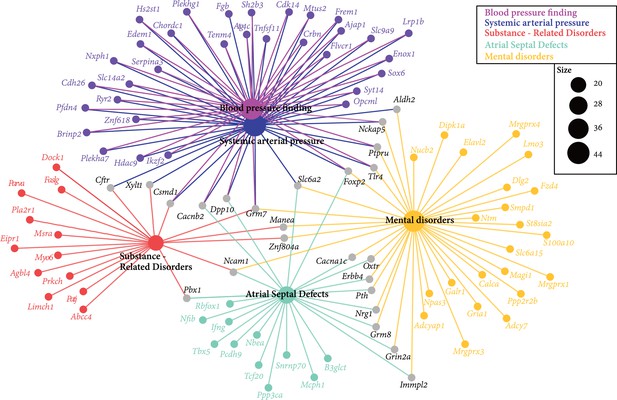
Enriched human diseases among genes closest to significant single nucleotide polymorphisms (SNPs) from association analysis.
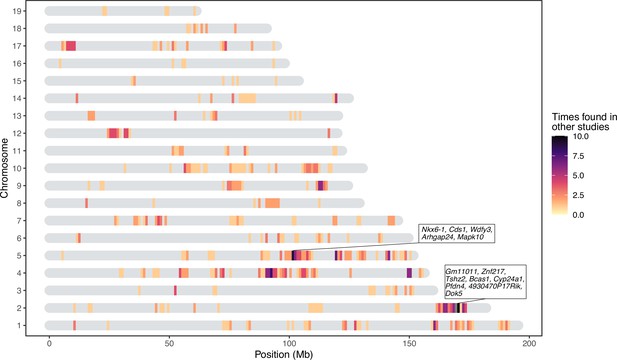
Significant loci in this study previously found in quantitative trait loci (QTL) studies of the mouse gut microbiome.
The genes present in two repeatedly identified regions are depicted in boxes.
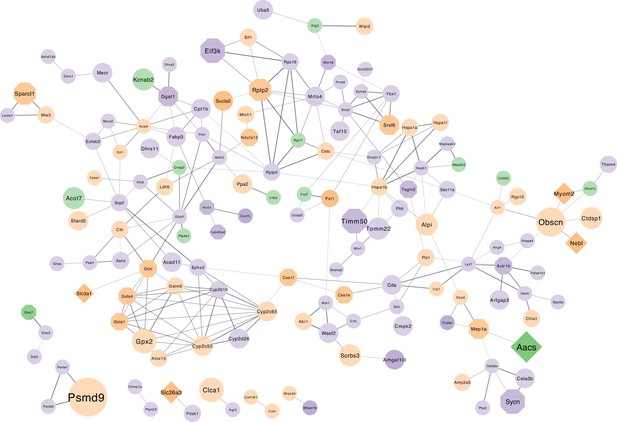
Protein-protein interaction (PPI) network of overlapping differentially expressed proteins according to colonization status.
STRING (Szklarczyk et al., 2019) protein-protein interaction (PPI) network of proteins that are differentially expressed in the intestine (small intestine and colon) of germ-free (GF) mice compared to conventionally raised mice, found in the present study. The colour of the network nodes indicates whether the quantitative trait loci (QTL) hit was found using the DNA abundances (green), RNA abundances (purple) or was found in both (orange). The shape represents if the gene of the protein was the closest gene to the significant single nucleotide polymorphisms (SNP) (rectangle), if the gene was also found in QTLs of other studies (octagon), a combination of both (diamond), or only differentially expressed in GF mice vs conventionally raised mice. The node size expresses the number of taxa where the gene was found in a QTL. The edges represent PPI, where the line thickness indicates the strength of the data support from text mining, experiments, databases, coexpression, gene-fusion, and co-occurrence.
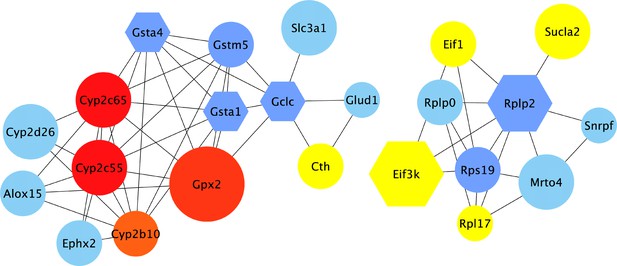
Protein-protein interaction network of hub genes of 'differentially expressed according to colonization status'-network.
Visualisation of the top hub genes calculated with the MCC algorithm and their first neighbours from the protein-protein interaction (PPI) network of genes found in intervals in present study that are also differentially expressed in germ-free versus conventionally raised mice (Figure 5—figure supplement 1). Edges represent the protein-protein associations. The red nodes represent genes with a high degree ( = hub genes), and the yellow nodes with a low degree, while the blue nodes represent their first neighbours. All nodes shown are differentially expressed in GF mice. Hexagon shaped nodes are genes/proteins also found associated with gut microbiome abundances in other mouse quantitative trait loci (QTL) studies, and round nodes are ‘only’ differentially expressed in GF mice. The size of the node is an indication of the number of taxa associated with the gene.
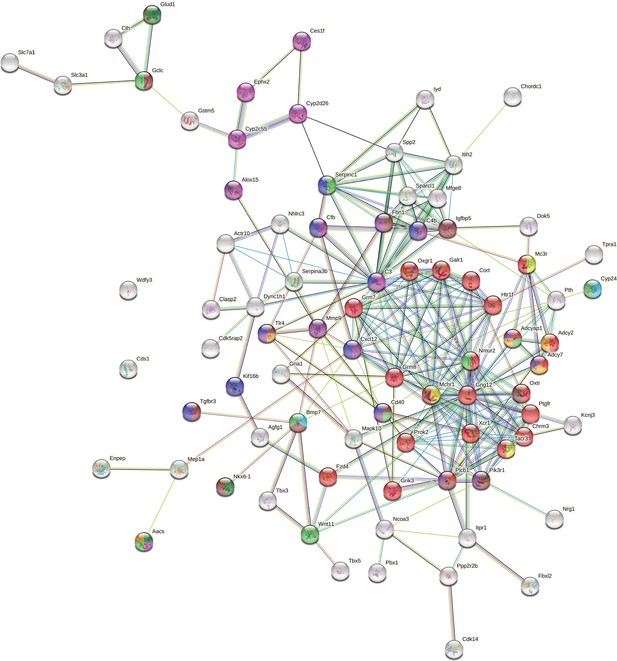
Network of host candidate genes influencing bacterial traits using STRING (https://string-db.org).
The nodes represent proteins and are coloured according to a selection of enriched GO terms and pathways: G-protein coupled receptor (GPCR) signalling (red), regulation of the immune system process (blue), response to nutrient levels (light green), fatty acid metabolic process (pink), glucose homeostasis (purple), response to antibiotic (orange), regulation of feeding behaviour (yellow), positive regulation of insulin secretion (dark green), circadian entrainment (brown), and response to vitamin D (turquoise). The colour of the edges represents the interaction type: known interactions from curated databases (turquoise) or experimentally determined (pink); predicted interactions from gene neighbourhood (green), gene fusions (red), gene co-occurrence (blue); other interactions from text-mining (light green), coexpression (black), and protein homology (purple).
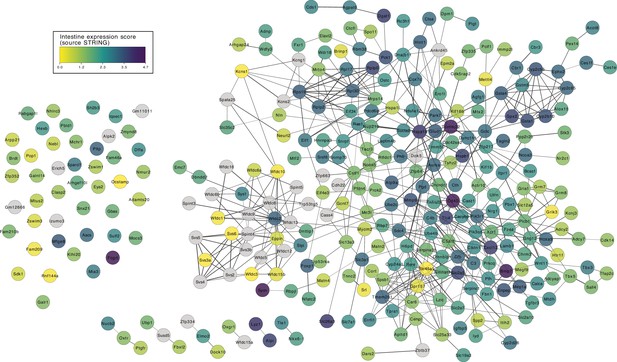
Protein protein interaction (PPI) network of 304 filtered candidate genes.
This network is generated in STRING (Szklarczyk et al., 2019) and Cytoscape. Nodes are coloured according to the intestine expression score (source: STRING).
Visualization of the random effects: mating pair identifier (A) and mating pair identifier nested within the sub-cross ID (B).
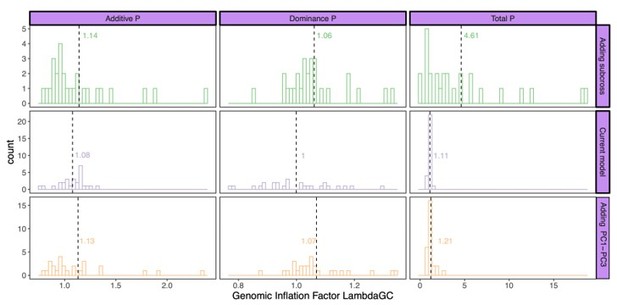
Genomic inflation factor for different P values (additive effect, left; dominance effect, middle; total model, right) calculated by the different models: current model including the mating pair id nested in the sub-cross ID (top), current model (middle), and the current model including PC1-PC3 as fixed effects (bottom).
Dashed line and values indicate the average genomic inflation factor.
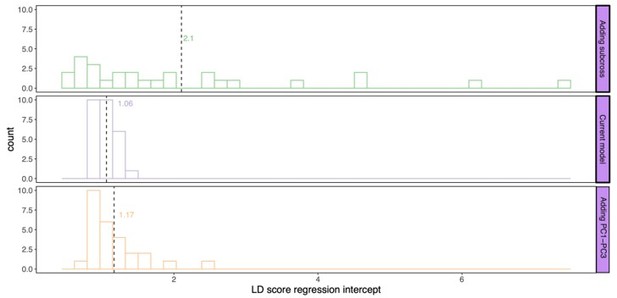
LD score regression intercept for P values calculated by the different models: current model including the mating pair id nested in the sub-cross ID (top), current model (middle), and the current model including PC1-PC3 as fixed effects (bottom).
Dashed line and values indicate the average genomic inflation factor.
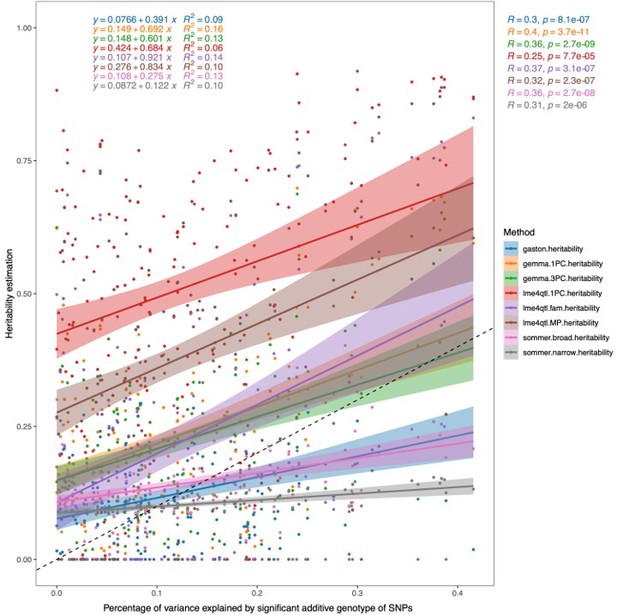
Correlation of heritability estimates calculated using different methods/programs and models to the percentage of phenotypic variance explained by the additive genotypes of all significant SNPs within the trait.
For Gaston, Sommer, Gemma 1PC, and lme4QTL.1PC, a linear mixed model was used with the first PC of the genotypes as a fixed effect and a genomic relatedness matrix (GRM) as a random effect. For Gemma.3PC, the first three PCs of the genotype data were used as fixed effects and a GRM as a random effect. For lme4QTL.MP, the mating pair ID was used as a random effect together with the GRM, while in lme4QTL.fam the mating pair ID was nested in the sub-cross ID as random effects together with a GRM to estimate the additive heritability.
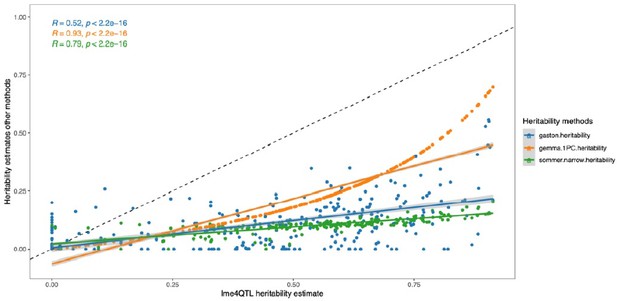
Heritability estimates calculated with lme4QTL compared to other methods.
The dashed line represents the identity line with a slope of 1. The colored lines are the corresponding linear regression lines.
Correlation of heritability estimates, calculated with GEMMA using the first principal component as a fixed effect, with co-speciation rates reported in Groussin et al., 2017.
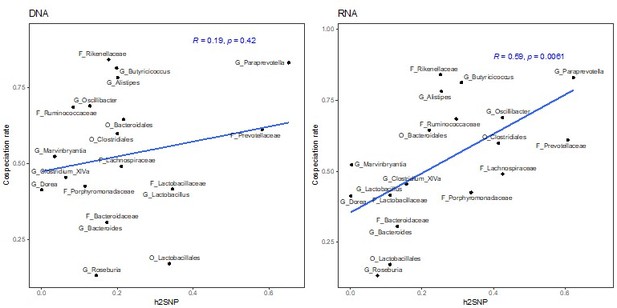
Correlation of heritability estimates, calculated with GEMMA using the first three principal component as a fixed effect, with co-speciation rates reported in Groussin et al., 2017.
Tables
Overview of mapping statistics.
Loci with a P-value below the study-wide threshold (P<1.29 × 10–8) are considered significant. ‘Significant loci total P’ are the loci having a significant P-value from the total model (additive and dominance effect), ‘Significant loci additive P’ have a significant additive effect, and ‘Significant loci dominance P’ have a significant dominance effect.
| DNA | RNA | Total | |
|---|---|---|---|
| Mapped taxa | 101 | 142 | 153 |
| Taxa with significant loci | 65 | 93 | 120 |
| Median interval size (Mb) | 1.32 | 2.5 | 1.91 |
| Total significant SNPs | 316 | 596 | 782 |
| Total significant loci | 443 | 746 | 1184 |
| Unique significant loci | 172 | 305 | 428 |
| Significant loci total P | 83 | 144 | 204 |
| Significant loci additive P | 144 | 244 | 351 |
| Significant loci dominance P | 88 | 171 | 230 |
| Median significant loci per trait | 5 | 5 | 8 |
| Median unique significant loci per trait | 3 | 3 | 4 |
| Median unique significant SNPs per locus | 2 | 2 | 2 |
| Median number of genes per locus | 32 | 54 | 43 |
| Median protein coding genes per locus | 11 | 17 | 14 |
-
SNPs: single nucleotide polymorphisms.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (Mus musculus musculus-Mus musculus-domesticus, males) | (HZ)A-(HZ)H | This paper | Second generation wild-derived inter-crossed hybrid mouse line, Mus musculus musculus-Mus musculus domesticus originating from four breeding stocks captured in the wild hybrid zone around Freising, Germany. | |
| biological sample (Mus musculus) | Cecum tissue (mucosa) | This paper | Collected and stored in RNAlater overnight. After RNAlater removal tissue was stored in –20°C | |
| biological sample (Mus musculus) | Ear clips | This paper | For genotyping | |
| commercial assay or kit | Allprep DNA/RNA 96-well | Qiagen (Hilden, Germany) | Cat. No.: 80,284 | DNA/RNA extraction |
| commercial assay or kit | Lysing matrix E | MP Biomedical (Eschwege, Germany) | SKU:116914050-CF | Lysing |
| commercial assay or kit | High-capacity cDNA Reverse Transcription Kit | Applied Biosystems (Darmstadt, Germany) | Cat. No.: 368,814 | cDNA transcription |
| sequence-based reagent | 27 F | https://doi.org/10.1016/j.ijmm.2016.03.004 | PCR primers | Forward primer V1-V2 hypervariable region |
| sequence-based reagent | 338 R | https://doi.org/10.1016/j.ijmm.2016.03.004 | PCR primers | Reverse primer V1-V2 hypervariable region |
| software, algorithm | DADA2 | https://doi.org/10.1038/nmeth.3869 | 16S rRNA gene processing | |
| software, algorithm | phyloseq | https://doi.org/10.1371/journal.pone.0061217 | 16S rRNA gene analysis | |
| commercial assay or kit | DNAeasy Blood and Tissue 96-well | Qiagen (Hilden, Germany) | Cat. No.: 69,504 | DNA extraction ear clips for genotyping |
| Other | GigaMUGA | Neogen, Lincoln, NE | Illumina Infinium II array containing 141,090 SNP probes | |
| software, algorithm | plink 1.9 | https://doi.org/10.1186/s13742-015-0047-8 | Quality control genotypes | |
| software, algorithm | GEMMA (v 0.98.1) | https://doi.org/10.1038/ng.2310 | Genetic relatedness matrix | |
| software, algorithm | lme4QTL | https://doi.org/10.1186/s12859-018-2057-x | SNP-based heritability, GWAS | |
| software, algorithm | exactLRT (RLRsim, v 3.1–6) | Scheipl et al., 2008 | Significance heritability estimates | |
| software, algorithm | inv.logit (Gtools, v 3.9.2) | Grieneisen et al., 2021 | Inverse logistic transformation | |
| software, algorithm | matSpDlite | Nyholt, 2019; https://doi.org/10.1038/sj.hdy.6800717 | Study-wide significance threshold | |
| software, algorithm | biomaRt (mm10) | Durinck et al., 2009 | Gene annotation | |
| software, algorithm | r.squaredGLMM (MuMIn, v 1.37.17) | Kamil, 2020 | Percentage of variance explained | |
| software, algorithm | locateVariants (VariansAnnotation, v 1.34.0) | https://doi.org/10.1093/bioinformatics/btu168 | Nearest gene | |
| software, algorithm | STRING (v 11) | https://doi.org/10.1093/nar/gky1131 | Protein-protein interaction networks | |
| software, algorithm | Cytoscape (v 3.8.2) | https://doi.org/10.1101/gr.1239303 | Network analysis and visualisation | |
| software, algorithm | MCODE | https://doi.org/10.1186/1471-2105-4-2 | Cytoscape plugin for identifying network clusters | |
| software, algorithm | stringApp | https://doi.org/10.1021/acs.jproteome.8b00702 | Cytoscape plugin for functional annotation | |
| software, algorithm | clusterprofiler (v 3.16.1) | https://doi.org/10.1089/omi.2011.0118 | Enrichment analysis | |
| software, algorithm | poverlap | Pedersen and Brown, 2013 | To determine significant overlap between studies | |
| software, algorithm | R (v 3.5.3) | https://www.R-project.org/ |
BIC scores models for RNA-based genera.
| Taxa | Current | +PC1 | +PC2 | +PC3 | +PC4 | +PC5 | Current +subcross |
|---|---|---|---|---|---|---|---|
| G_Acetatifactor | -2651.9 | -2637.7 | -2628.1 | -2613.8 | -2599.6 | -2584.5 | -2646.2 |
| G_Alistipes | -2714 | -2711.2 | -2701.5 | -2687.3 | -2673.9 | -2658.9 | -2712.8 |
| G_Anaerostipes | -3416.6 | -3400.9 | -3385.2 | -3369.2 | -3352.7 | -3336.1 | -3410.8 |
| G_Bacteroides | -2076.6 | -2065.9 | -2053.9 | -2045.2 | -2033.5 | -2021.5 | -2070.9 |
| G_Butyricicoccus | -3676.1 | -3661.1 | -3645.3 | -3628.4 | -3612.7 | -3596.6 | -3671.4 |
| G_Clostridium_XlVa | -2356.8 | -2354.2 | -2340.6 | -2327.2 | -2314.8 | -2301 | -2351.1 |
| G_Dorea | -2979.7 | -2965.4 | -2952 | -2936.6 | -2926.1 | -2916.2 | -2974.4 |
| G_Eisenbergiella | -1500.5 | -1492 | -1489.3 | -1479.8 | -1469.3 | -1458.2 | -1495.3 |
| G_Fusicatenibacter | -2932.8 | -2918.3 | -2902.8 | -2888.3 | -2873.5 | -2857.8 | -2927.1 |
| G_Helicobacter | -1145.4 | -1138.4 | -1131 | -1122.1 | -1114.8 | -1104.8 | -1140.3 |
| G_Hungatella | -2415.2 | -2402.3 | -2390.2 | -2376.7 | -2364.5 | -2351.1 | -2409.5 |
| G_Intestinimonas | -3286.5 | -3272.5 | -3256.3 | -3240.1 | -3228.2 | -3213.9 | -3282.4 |
| G_Lactobacillus | -2407 | -2395.6 | -2384.4 | -2380.5 | -2366.8 | -2352.6 | -2403.5 |
| G_Marvinbryantia | -2961.6 | -2950.7 | -2934.7 | -2918.8 | -2903.2 | -2887.3 | -2956.4 |
| G_Mucispirillum | -1938.8 | -1930.8 | -1918.9 | -1907.3 | -1896.7 | -1884.8 | -1933.1 |
| G_Odoribacter | -3183.2 | -3170.8 | -3156 | -3143.5 | -3134.7 | -3120 | -3186.1 |
| G_Oscillibacter | -2117.2 | -2105.8 | -2095.2 | -2084.3 | -2072.1 | -2060 | -2111.9 |
| G_Paraprevotella | -2230.2 | -2222.8 | -2219.2 | -2207.7 | -2195.4 | -2182.8 | -2228.4 |
| G_Roseburia | -2719 | -2704 | -2689.4 | -2674.8 | -2661.3 | -2647.6 | -2713.5 |
| G_Staphylococcus | -7666.6 | -7636.9 | -7607.6 | -7576.8 | -7546.5 | -7515.8 | -7660.9 |
| unclassified_C_Deltaproteobacteria | -3208.6 | -3197.8 | -3183.4 | -3167.3 | -3158.7 | -3142.1 | -3205.5 |
| unclassified_F_Lachnospiraceae | -1435.4 | -1433.5 | -1423 | -1413.9 | -1407.2 | -1396.8 | -1433.1 |
| unclassified_F_Porphyromonadaceae | -2997.7 | -2984.6 | -2970.3 | -2956.3 | -2943.4 | -2928.9 | -2991.9 |
| unclassified_F_Prevotellaceae | -4216.4 | -4198.5 | -4182 | -4175.1 | -4156.4 | -4136.9 | -4212.7 |
| unclassified_F_Ruminococcaceae | -2590.6 | -2578.8 | -2565.6 | -2554.4 | -2540.7 | -2525.9 | -2586.1 |
| unclassified_O_Bacteroidales | -2348 | -2339.5 | -2328 | -2317 | -2305.3 | -2291.7 | -2344.4 |
| unclassified_O_Clostridiales | -2721.2 | -2708 | -2694.8 | -2681 | -2667.4 | -2654.7 | -2715.4 |
| unclassified_P_Bacteroidetes | -3260.7 | -3252.8 | -3237.9 | -3222.6 | -3207 | -3194.2 | -3256.9 |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/75419/elife-75419-transrepform1-v1.docx
-
Supplementary file 1
Narrow-sense and chip heritability estimates from the present study and previous mQTL studies combined with PVE by additive and additive and dominance effects of significant SNPs, and cospeciation rates reported by Groussin et al., (2017).
- https://cdn.elifesciences.org/articles/75419/elife-75419-supp1-v1.xlsx
-
Supplementary file 2
Genome-wide significant associations.
- https://cdn.elifesciences.org/articles/75419/elife-75419-supp2-v1.xlsx
-
Supplementary file 3
Study-wide significant associations.
- https://cdn.elifesciences.org/articles/75419/elife-75419-supp3-v1.xlsx
-
Supplementary file 4
Study-wide significant association with an interval size smaller than 1Mb.
- https://cdn.elifesciences.org/articles/75419/elife-75419-supp4-v1.xlsx
-
Supplementary file 5
Over-represented KEGG pathways within the genes neighbouring the peak SNP.
- https://cdn.elifesciences.org/articles/75419/elife-75419-supp5-v1.xlsx
-
Supplementary file 6
IBD genes found within the study-wide significant regions.
- https://cdn.elifesciences.org/articles/75419/elife-75419-supp6-v1.xlsx
-
Supplementary file 7
List of promising candidate genes.
- https://cdn.elifesciences.org/articles/75419/elife-75419-supp7-v1.xlsx