YAP1 activation by human papillomavirus E7 promotes basal cell identity in squamous epithelia
Figures
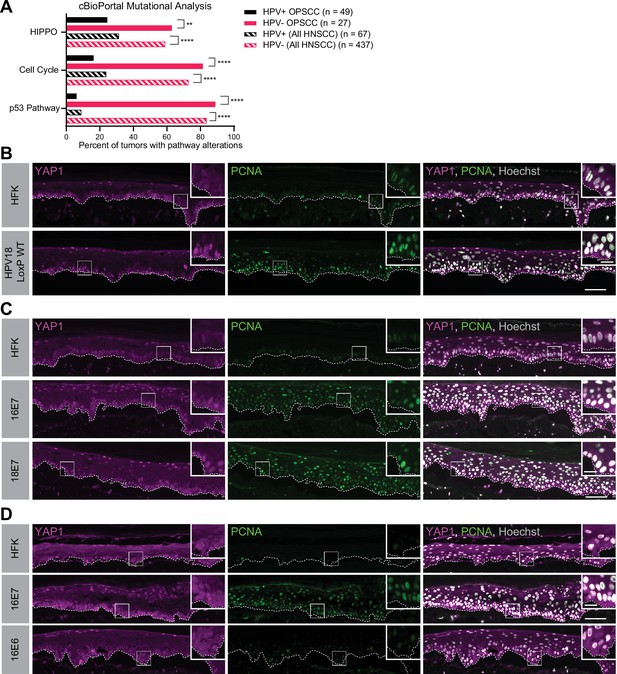
Human papillomavirus (HPV) E7 activates yes-associated protein (YAP1) in basal epithelial keratinocytes.
(A) cBioPortal analysis for total genomic mutations and copy number alterations in HPV-positive and HPV-negative oropharyngeal squamous cell carcinoma (OPSCC) and head and neck squamous cell carcinoma (HNSCC). Graph displays the percent of tumors with alterations in each pathway. Statistical significance was determined by Fisher’s exact test (**p<0.01, ****p<0.0001). (B–D) Organotypic cultures were grown from primary human foreskin keratinocytes (HFK), HFK harboring the HPV18 genome, or HFK transduced with retroviral expression vectors encoding HPV E6 or E7 proteins. Formalin-fixed paraffin-embedded sections of cultures grown from (B) HFK or HFK harboring the HPV18 genome, (C) HFK or HFK expressing HPV16 E7 or HPV18 E7, or (D) HFK or HFK expressing HPV16 E6 or HPV16 E7 were stained for YAP1 (magenta), proliferating cell nuclear antigen (PCNA) (green), and Hoechst (gray). White dashed lines indicate the basement membrane. White boxes indicate the location of insets in main images. Main image scale bars = 100 μm. Inset scale bars = 25 μm.
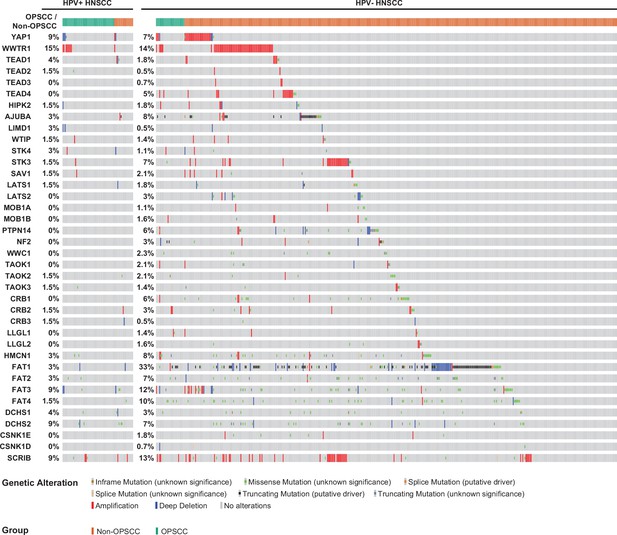
Human papillomavirus (HPV)-positive head and neck squamous cell carcinoma (HNSCC) have fewer Hippo pathway alterations than HPV-negative HNSCC.
cBioPortal analysis for genomic mutations and copy number alterations in HPV-positive and HPV-negative HNSCC and oropharyngeal squamous cell carcinoma (OPSCC). Oncoprint displays specific genomic alterations in individual tumor samples.
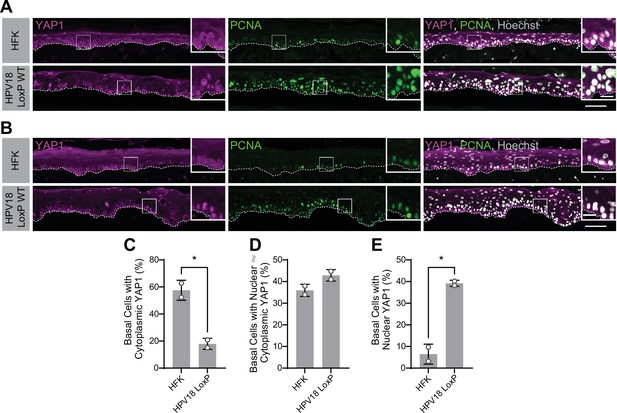
Human papillomavirus type 18 (HPV18) genomes activate yes-associated protein (YAP1) in basal keratinocytes.
(A–B) Additional replicates of organotypic cultures grown from primary human foreskin keratinocytes (HFK) or HFK harboring the HPV18 genome. Formalin-fixed paraffin-embedded sections were stained for YAP1 (magenta), proliferating cell nuclear antigen (PCNA) (green), and Hoechst (gray). White dashed lines indicate the basement membrane. White boxes indicate the location of insets in main images. Main image scale bars = 100 μm. Inset scale bars = 25 μm. (C–E) Quantification of the percentage of basal cells in which YAP1 is predominantly localized to the cytoplasm (C), comparably distributed between the nucleus and the cytoplasm (D), or predominantly localized to the nucleus (E). Graphs display the mean ± SD and each individual data point (independent organotypic cultures). Statistical significance was determined by t-test (*p<0.05).
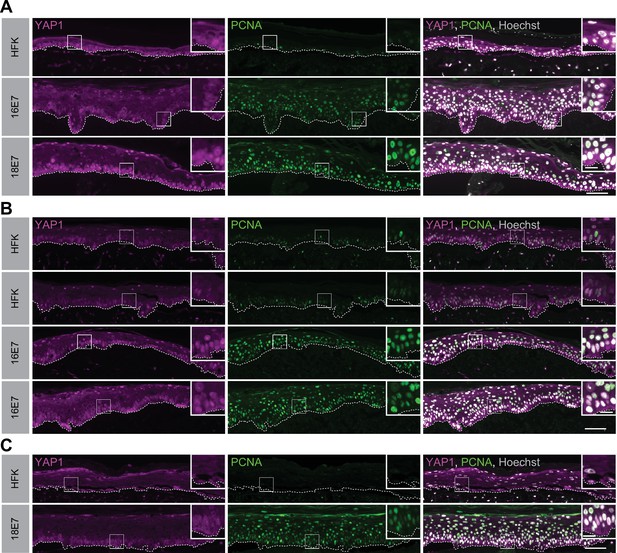
Human papillomavirus (HPV) E7 activates yes-associated protein (YAP1) in basal keratinocytes.
Additional replicates of organotypic cultures grown from primary human foreskin keratinocytes (HFK) or HFK transduced with retroviral expression vectors encoding HPV E7 proteins. Formalin-fixed paraffin-embedded sections of cultures grown from (A) HFK or HFK expressing HPV16 E7 or HPV18 E7, (B) HFK or HFK expressing HPV16 E7, or (E) HFK and HFK expressing HPV18 E7 were stained for YAP1 (magenta), proliferating cell nuclear antigen (PCNA) (green), and Hoechst (gray). White dashed lines indicate the basement membrane. White boxes indicate the location of insets in main images. Main image scale bars = 100 μm. Inset scale bars = 25 μm.
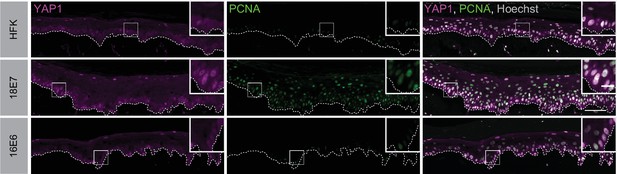
Human papillomavirus (HPV) E6 does not activate yes-associated protein (YAP1) in basal keratinocytes.
Additional replicates of organotypic cultures grown from primary HFK or HFK transduced with retroviral expression vectors encoding HPV E6 or E7 proteins. Formalin-fixed paraffin-embedded sections were stained for YAP1 (magenta), proliferating cell nuclear antigen (PCNA) (green), and Hoechst (gray). White dashed lines indicate the basement membrane. White boxes indicate the location of insets in main images. Main image scale bars = 100 μm. Inset scale bars = 25 μm.
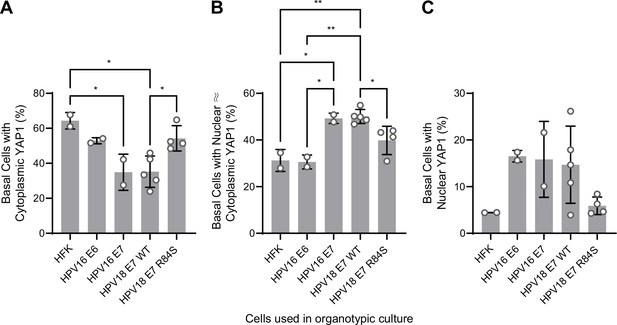
Quantification of yes-associated protein (YAP1) activation by human papillomavirus (HPV) E6 or E7 in basal keratinocytes.
(A–C) Quantification of the percentage of basal cells in which YAP1 is predominantly localized to the cytoplasm (A), comparably distributed between the nucleus and the cytoplasm (B), or predominantly localized to the nucleus (C). Graphs display the mean ± SD and each individual data point (independent organotypic cultures). Statistical significance was determined by ANOVA using the Holm-Sídák correction for multiple comparisons (*p<0.05, **p<0.01).
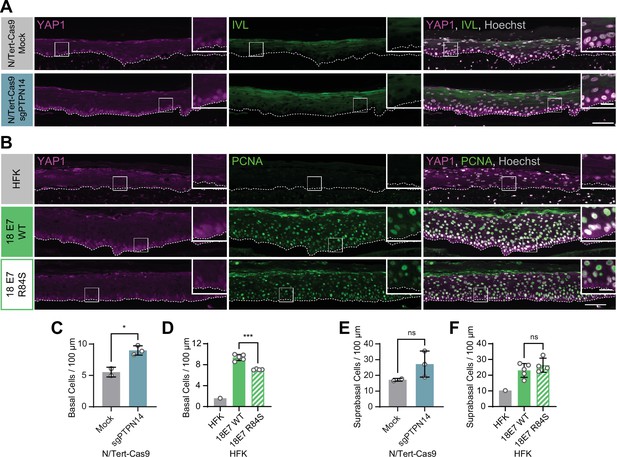
Human papillomavirus (HPV) E7 activates yes-associated protein (YAP1) in basal keratinocytes through protein tyrosine phosphatase non-receptor type 14 (PTPN14) degradation.
Organotypic cultures were grown from N/Tert-Cas9 keratinocytes transfected with sgRNA or from primary human foreskin keratinocytes (HFK) transduced with retroviral expression vectors encoding HPV18 E7 WT or R84S. (A) Formalin-fixed paraffin-embedded (FFPE) sections of cultures grown from mock or sgPTPN14 transfected N/Tert-Cas9 keratinocytes were stained for YAP1 (magenta), involucrin (IVL) (green), and Hoechst (gray). (B) FFPE sections of cultures grown from parental HFK, HPV18 E7 WT, or HPV18 E7 R84S expressing HFK were stained for YAP1 (magenta), proliferating cell nuclear antigen (PCNA) (green), and Hoechst (gray). White dashed lines indicate the basement membrane. White boxes indicate the location of insets in main images. Main image scale bars = 100 μm. Inset scale bars = 25 μm. (C–F) Quantification of the number of (C and D) basal cells and (E and F) suprabasal cells per 100 μm of epidermis. Graphs display the mean ± SD and each individual data point (independent organotypic cultures). (C and E) Statistical significance was determined by t-test. (D and F) Statistical significance was determined by ANOVA using the Holm-Sídák correction for multiple comparisons (*p<0.05, ***p<0.001).
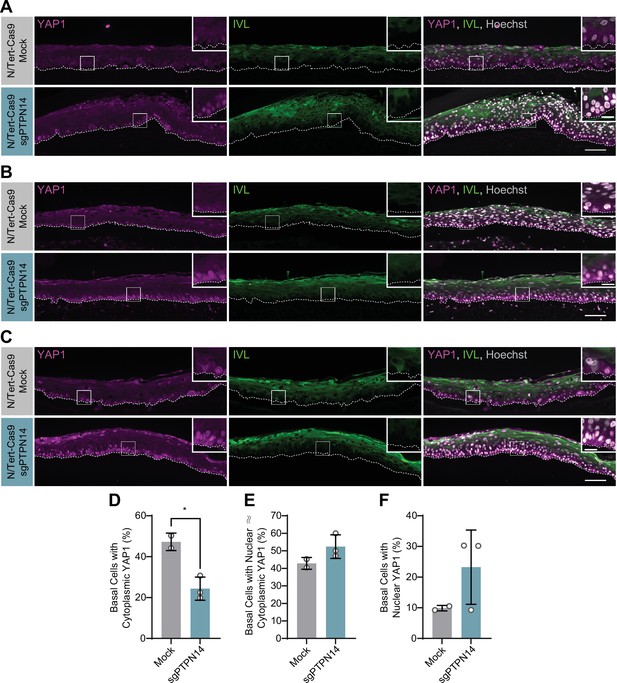
Protein tyrosine phosphatase non-receptor type 14 (PTPN14) knockout activates yes-associated protein (YAP1) in basal keratinocytes.
Additional replicates of organotypic cultures grown from N/Tert-Cas9 keratinocytes. (A–C) Formalin-fixed paraffin-embedded sections from mock or sgPTPN14 transfected N/Tert-Cas9 keratinocytes were stained for YAP1 (magenta), IVL (green), and Hoechst (gray). White dashed lines indicate the basement membrane. White boxes indicate the location of insets in main images. Main image scale bars = 100 μm. Inset scale bars = 25 μm. (D–F) Quantification of the percentage of basal cells in which YAP1 is predominantly localized to the cytoplasm (D), comparably distributed between the nucleus and cytoplasm (E), or predominantly localized to the nucleus (F). Graphs display the mean ± SD and each individual data point (independent organotypic cultures). Statistical significance was determined by t-test (*p<0.05).
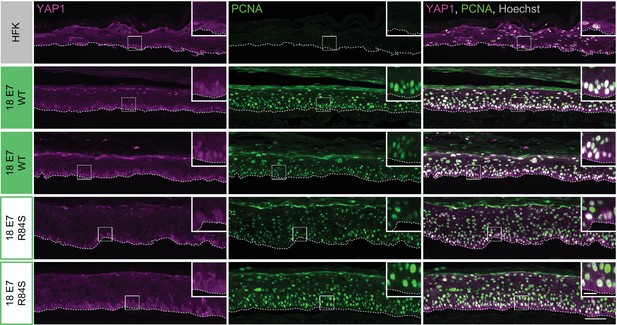
Human papillomavirus (HPV) E7 activates yes-associated protein (YAP1) in basal keratinocytes through protein tyrosine phosphatase non-receptor type 14 degradation.
Additional replicates of organotypic cultures grown from primary human foreskin keratinocytes (HFK) transduced with retroviral expression vectors encoding HPV18 E7 WT or R84S. Formalin-fixed paraffin-embedded sections from parental HFK, HPV18 E7 WT, or HPV18 E7 R84S expressing HFK were stained for YAP1 (magenta), proliferating cell nuclear antigen (PCNA) (green), and Hoechst (gray). White dashed lines indicate the basement membrane. White boxes indicate the location of insets in main images. Main image scale bars = 100 μm. Inset scale bars = 25 μm.
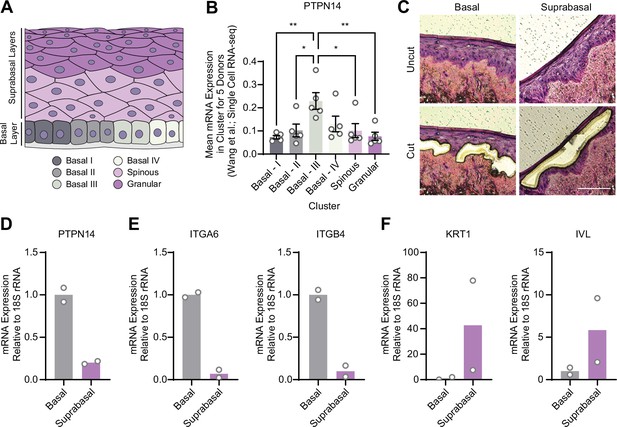
Protein tyrosine phosphatase non-receptor type 14 (PTPN14) expression is enriched in basal keratinocytes.
(A–B) Single-cell RNA sequencing data and clustering analysis from Wang et al. was reanalyzed to assess PTPN14 expression in different subsets of epidermal cells. (A) Diagram of epidermis; shading depicts tissue localization of cell clusters. (B) For each donor, the mean of PTPN14 mRNA expression was calculated for each cell cluster. Graphs display the mean of PTPN14 mRNA expression for each donor (circles) as well as the mean of all five donors ± SEM (bars and error bars). Statistical significance was determined by ANOVA using the Holm-Sídák correction for multiple comparisons (*p<0.05, **p<0.01). (C–F) Basal and suprabasal layers from organotypic cultures were dissected using laser capture microdissection. (C) Representative images of HFK cultures before and after individual laser dissections. Hundreds of such cuts were performed per sample. (D–F) RNA was purified from isolated layers and qRT-PCR was used to assess the expression of PTPN14 (D), basal cell markers ITGA6 and ITGB4 (E), and differentiation markers KRT1 and IVL (F). Graphs display the mean and each individual data point.
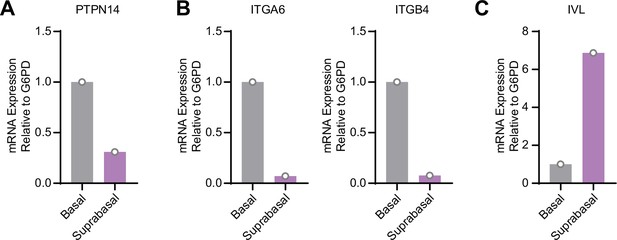
Protein tyrosine phosphatase non-receptor type 14 (PTPN14) expression is enriched in basal keratinocytes in human papillomavirus type 18 (HPV18) E7 expressing organotypic cultures.
Basal and suprabasal layers from a 3D organotypic culture grown from human foreskin keratinocytes transduced with a retroviral expression vector encoding HPV18 E7 were dissected using laser capture microdissection. RNA was purified from isolated layers and qRT-PCR was used to assess the expression of PTPN14 (A), the basal cell markers ITGA6 and ITGB4 (B), and the differentiation marker IVL (C). Graphs display individual data points.
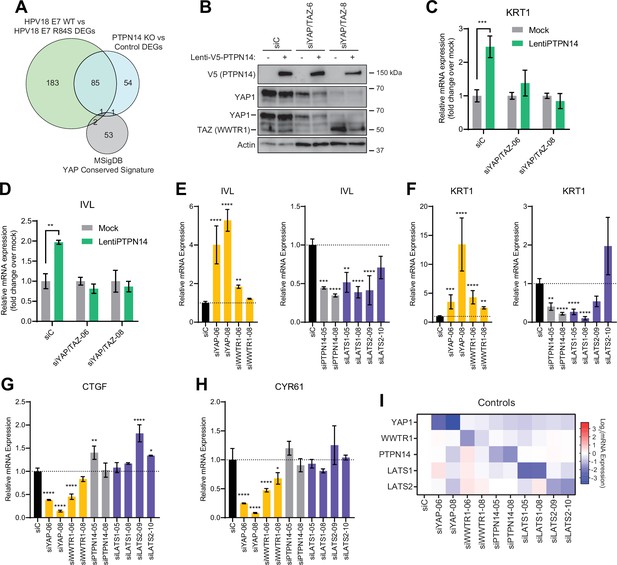
Yes-associated protein (YAP1)/TAZ regulate differentiation downstream of protein tyrosine phosphatase non-receptor type 14 (PTPN14).
(A) Venn diagram comparing the MSigDB YAP conserved signature to the differentially expressed genes (DEG) from our two published experiments that reflect PTPN14 loss in keratinocytes. (B–D) YAP1 and WWTR1 were simultaneously knocked down by siRNA transfection in human foreskin keratinocytes (HFK). Transfected HFK were then transduced with PTPN14 lentivirus at 24 hr post-transfection. Cells were lysed for protein and total cellular RNA at 72 hr post-transfection. (B) Cell lysates were subjected to SDS-PAGE/Western blot analysis and probed with antibodies to PTPN14, YAP1, TAZ, and actin (Figure 4—source data 1). TAZ blot was generated by reprobing the membrane that was originally probed for YAP1. (C and D) qRT-PCR was used to measure the expression of the differentiation markers KRT1 and IVL relative to G6PD. Graphs display fold change in gene expression relative to the mock transduced cells. (E–I) Primary HFK were transfected with siRNAs targeting YAP1, WWTR1 (TAZ), PTPN14, LATS1, and LATS2. Two siRNAs were used per target. qRT-PCR was used to measure gene expression for: the differentiation markers IVL (E) and KRT1 (F), and the canonical YAP1/TAZ targets CTGF (G) and CYR61 (H). Data confirming that individual siRNA transfections depleted intended transcripts are summarized in a heatmap of log2(fold-change) levels (I). Bar graphs display the mean ± SD of three independent replicates. Statistical significance of each treatment compared to siC was determined by ANOVA using the Holm-Sídák correction for multiple comparisons (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
-
Figure 4—source data 1
Original images for Western blots in Figure 4B.
Original uncropped images from YAP1, TAZ, V5, and actin Western blots presented in Figure 4B are included as individual image files and as a compiled summary document.
- https://cdn.elifesciences.org/articles/75466/elife-75466-fig4-data1-v2.zip
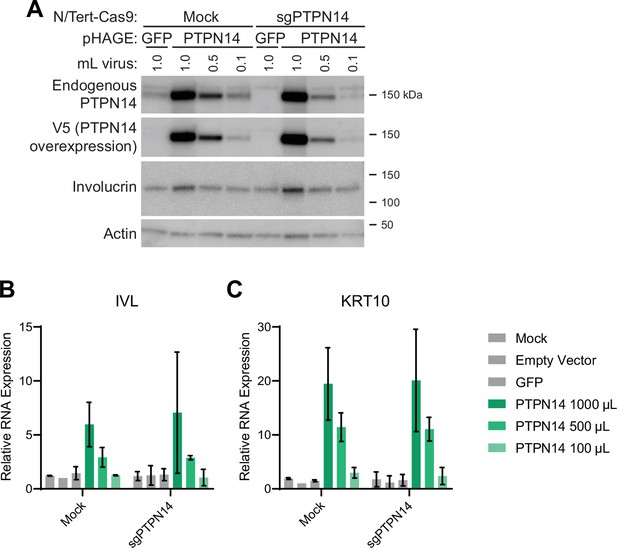
Protein tyrosine phosphatase non-receptor type 14 (PTPN14) overexpression promotes differentiation in keratinocytes.
NTert-Cas9 Mock and sgPTPN14-1 keratinocytes were transduced with lentiviruses encoding green fluorescent protein (GFP) or PTPN14 or the empty vector control. (A) Cell lysates were subjected to SDS-PAGE/Western blot analysis and probed with antibodies to PTPN14, V5-tag, involucrin, and actin (Figure 4—figure supplement 1—source data 1). (B) qRT-PCR was used to measure the expression of the differentiation markers IVL and KRT10 relative to G6PD. Graphs display the mean ± SD of two independent replicates.
-
Figure 4—figure supplement 1—source data 1
Original images for Western blots in Figure 4—figure supplement 1A.
Original uncropped images from PTPN14, involucrin, V5, and actin Western blots presented in Figure 4—figure supplement 1A are included as individual image files (cropped to individual gels and uncropped versions) and as a compiled summary document.
- https://cdn.elifesciences.org/articles/75466/elife-75466-fig4-figsupp1-data1-v2.zip
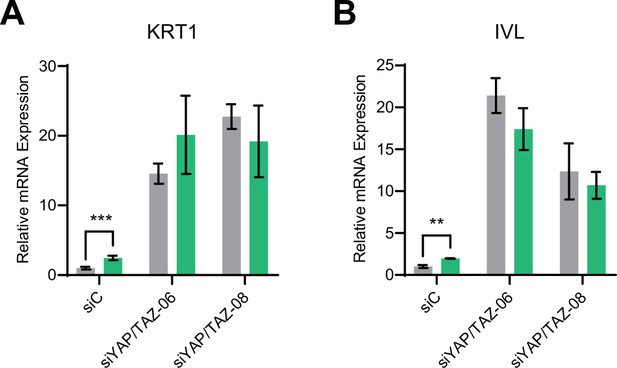
Yes-associated protein (YAP1) and TAZ are required for protein tyrosine phosphatase non-receptor type 14 (PTPN14) to promote keratinocyte differentiation.
Primary human foreskin keratinocytes (HFK) were transfected with control or YAP1 and WWTR1 targeting siRNAs then transduced with PTPN14 encoding lentivirus. qRT-PCR was used to measure the expression of the differentiation markers (A) KRT1 and (B) IVL relative to G6PD. Graphs portray the change in gene expression relative to siC. Graphs display the mean ± SD of three independent replicates. Statistical significance was determined by ANOVA using the Holm-Sídák correction for multiple comparisons (**p<0.01, ***p<0.001).
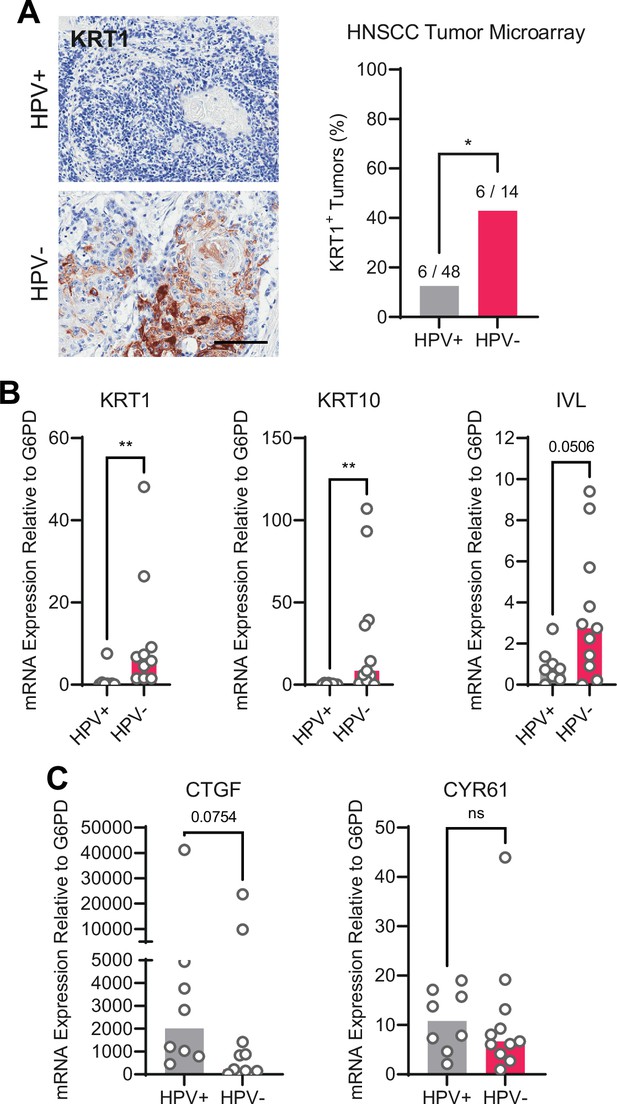
Human papillomavirus (HPV)-positive head and neck squamous cell carcinoma (HNSCC) are less differentiated than HPV-negative HNSCC.
(A) Human HNSCC tumor samples were stained for KRT1 (left). Scale bar = 100 μm. Graph displays the percentage of tumors that were KRT1+ (right). Statistical significance was determined by Fisher’s exact test. (B–C) Total RNA was purified from patient-derived xenograft samples and qRT-PCR was used to assess gene expression of (B) the differentiation markers KRT1, KRT10, and IVL and (C) the canonical yes-associated protein 1/TAZ targets CTGF and CYR61. Statistical significance was determined by Mann-Whitney nonparametric test (*p<0.05, **p<0.01).
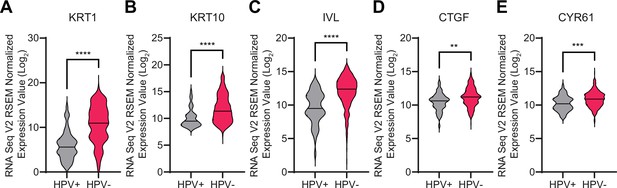
Human papillomavirus (HPV)-positive head and neck squamous cell carcinomas (HNSCC) expresse lower levels of differentiation genes than HPV-negative HSNCC.
RNA-seq data from The Cancer Genome Atlas were accessed through cBioPortal. Violin plots display the distribution in log2 mRNA expression of differentiation markers (A) KRT1, (B) KRT10, and (C) IVL, and the canonical YAP1/TAZ targets (D) CTGF and (E) CYR61. Statistical significance was determined by Mann-Whitney nonparametric test (**p<0.01, ***p<0.001, ****p<0.0001).
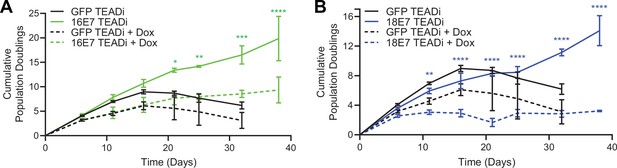
High-risk human papillomavirus (HPV) E7 requires yes-associated protein 1/TAZ-TEAD transcriptional activity to extend the lifespan of primary keratinocytes.
Primary human foreskin keratinocytes (HFK) were transduced with retroviruses encoding HPV16 E7, HPV18 E7, or GFP, plus pInducer20 TEADi lentivirus. Each cell population was cultured with or without 1 μg/mL doxycycline (dox) in the media for 38 days and population doublings were tracked with each passage. Graph displays the mean ± SD of two independently transduced cell populations per condition. Statistical significance when comparing cell growth with and without doxycycline was determined by two-way ANOVA using the Sídák correction for multiple comparisons (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
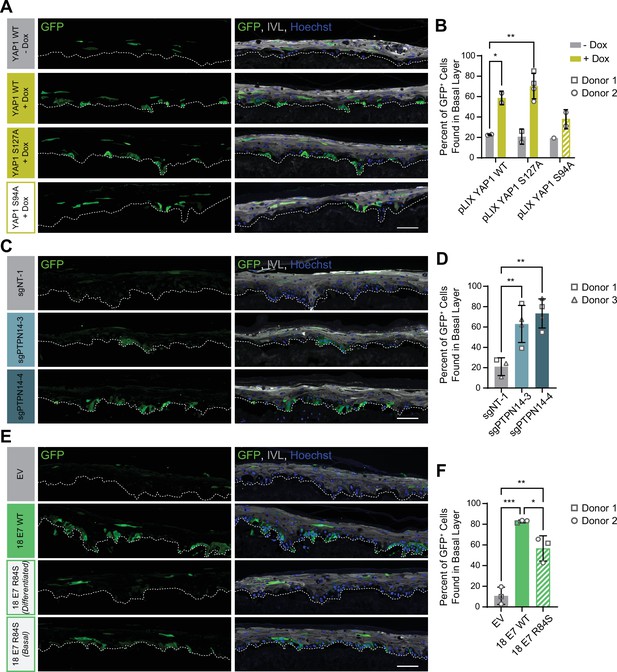
Protein tyrosine phosphatase non-receptor type 14 (PTPN14) loss and yes-associated protein (YAP1) activation by human papillomavirus (HPV) E7 promote basal cell retention in organotypic cultures.
Organotypic cultures were grown from GFP-labeled human foreskin keratinocytes (HFK) mixed with unmodified HFK. (A–B) GFP-labeled HFK were transduced with lentiviral vectors encoding YAP1 WT, YAP1 S127A, or YAP1 S94A under the control of a doxycycline (dox) inducible promoter. GFP-labeled YAP1 cells were mixed 1:25 into unmodified HFK and organotypic cultures were grown from the mixture. Cultures were grown ±1 μg/mL doxycycline. (C–D) GFP-labeled HFK were transduced with LentiCRISPR v2 vectors encoding control or PTPN14 targeting sgRNAs. GFP-labeled cells were mixed 1:25 into unmodified HFK and organotypic cultures were grown from the mixture. (E–F) GFP-labeled HFK were transduced with HPV18 E7 WT, HPV18 E7 R84S, or the empty vector (EV). GFP-labeled HPV18 E7 cells were mixed 1:50 into unmodified HFK and organotypic cultures were grown from the mixture. Two different images for 18E7 R84S reflect cases in which all tracer cells migrated to the suprabasal layers (differentiated) or in which some tracer cells remained in the basal layer (basal). (A, C, E) Formalin-fixed paraffin-embedded sections of cultures were stained for GFP (green), IVL (gray), and Hoechst (blue). Scale bar = 100 μm. (B, D, F) Quantification of the percentage of GFP+ cells found in the basal layer. Graphs display the mean ± SD and each individual data point (independent organotypic cultures). Shapes indicate cultures grown from different HFK donors. Statistical significance was determined by ANOVA using the Holm-Sídák correction for multiple comparisons (*p<0.05, **p<0.01).
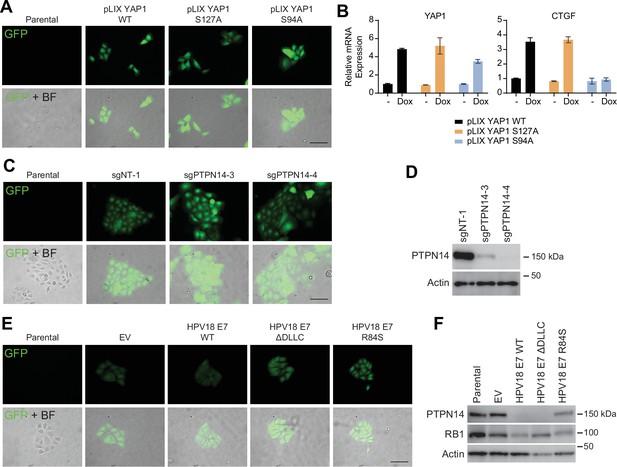
Protein tyrosine phosphatase non-receptor type 14 (PTPN14) degradation by human papillomavirus (HPV) E7 promotes basal cell retention.
(A–B) GFP-labeled human foreskin keratinocytes (HFK) were transduced with yes-associated protein (YAP1) WT, YAP1 S127A, or YAP1 S94A under the control of a doxycycline inducible promoter. (A) GFP expression was confirmed by fluorescence microscopy. Scale bar = 100 μm. (B) Total RNA was purified from monolayer cells ± treatment with 1 μg/mL doxycycline for 72 hr. qRT-PCR was used to assess gene expression of YAP1 and CTGF. (C–D) GFP-labeled HFK were transduced with LentiCRISPR v2 sgNT-1, sgPTPN14-3, or sgPTPN14-4 vectors. (C) GFP expression was confirmed by fluorescence microscopy. Scale bar = 100 μm (D) Cell lysates were subjected to SDS-PAGE/Western blot analysis and probed with antibodies to PTPN14 and actin (Figure 7—figure supplement 1—source data 1). (E–F) GFP-labeled HFK were transduced with retroviral vectors encoding HPV18 WT, HPV18 ΔDLLC, HPV18 E7 R84S, or the empty vector control (EV). (E) GFP expression was confirmed by fluorescence microscopy. Scale bar = 100 μm. (F) Cell lysates were subjected to SDS-PAGE/Western blot analysis and probed with antibodies to PTPN14, RB1, and actin (Figure 7—figure supplement 1—source data 2).
-
Figure 7—figure supplement 1—source data 1
Original images for Western blots in Figure 7—figure supplement 1D.
Original uncropped images from PTPN14 and actin Western blots presented in Figure 7—figure supplement 1D are included as individual image files and as a compiled summary document.
- https://cdn.elifesciences.org/articles/75466/elife-75466-fig7-figsupp1-data1-v2.zip
-
Figure 7—figure supplement 1—source data 2
Original images for Western blots in Figure 7—figure supplement 1F.
Original uncropped images from PTPN14, RB1, and actin Western blots presented in Figure 7—figure supplement 1F are included as individual image files and as a compiled summary document.
- https://cdn.elifesciences.org/articles/75466/elife-75466-fig7-figsupp1-data2-v2.zip
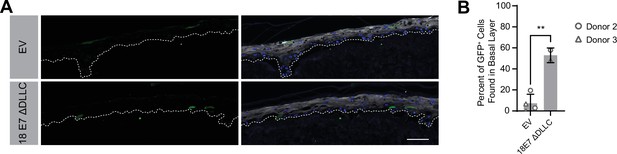
Human papillomavirus type 18 (HPV18) E7 can promote basal cell retention in the absence of retinoblastoma 1 protein binding.
Organotypic cultures were grown from GFP-labeled cells mixed with unmodified human foreskin keratinocytes (HFK). GFP-labeled HFK were transduced with HPV18 E7 ΔDLLC or the empty vector (EV). GFP-labeled cells were mixed 1:50 into unmodified HFK. (A) Formalin-fixed paraffin-embedded sections were stained for GFP (green), IVL (gray), and Hoechst (blue). Scale bar = 100 μm (B) Quantification of the percentage of GFP+ cells found in the basal layer. Graphs display the mean ± SD and each individual data point (independent cultures). Statistical significance was determined by t-test. (**p<0.01).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-Actin (Mouse monoclonal) | Sigma-Aldrich | Cat#: MAB1501; RRID:AB_2223041 | WB (1:20,000) |
| Antibody | anti-GFP (Rabbit polyclonal) | Invitrogen | Cat#: A6455; RRID:AB_221570 | WB (1:1,000); IHC-P (1:2000) |
| Antibody | anti-Mouse IgG Alexa Fluor 488 (Goat polyclonal) | Invitrogen | Cat#: A11001; RRID:AB_2534069 | IHC-P (1:250) |
| Antibody | anti-Mouse IgG HRP (Horse monoclonal) | Cell Signaling Technologies | Cat#: 7076; RRID:AB_330924 | WB (1:2000) |
| Antibody | anti-Rabbit IgG Alexa Fluor 594 (Goat polyclonal) | Invitrogen | Cat#: A11012; RRID:AB_2534079 | IHC-P (1:250) |
| Antibody | anti-Rabbit IgG HRP (Goat monoclonal) | Cell Signaling Technologies | Cat#: 7074; RRID:AB_2099233 | WB (1:2000) |
| Antibody | anti-HA-Peroxidase (Rat monoclonal) | Roche | Cat#: 12013819001; RRID:AB_390917 | WB (1:500) |
| Antibody | anti-ITGB4 (Rabbit polyclonal) | Sigma-Aldrich | Cat#:HPA036348; RRID:AB_2675077 | IHC-P (1:100) |
| Antibody | anti-IVL (Mouse monoclonal) | Santa Cruz Biotechnology | Cat#: sc-398952 | IHC-P (1:100) |
| Antibody | anti-KRT1 (Mouse monoclonal) | Enzo Life Sciences | Cat#:C34904; RRID:AB_2265594 | |
| Antibody | anti-PCNA | Santa Cruz Biotechnology | Cat#: sc-56; RRID:AB_628110 | IHC-P (1:100) |
| Antibody | Anti-PTPN14 (Rabbit monoclonal) | Cell Signaling Technology | D5T6Y; Cat#:13808; RRID:AB_2798318 | WB (1:500) |
| Antibody | anti-TAZ (Rabbit monoclonal) | Cell Signaling Technology | D3I6D; Cat#: 70148; RRID:AB_2799776 | WB (1:1000) |
| Antibody | anti-V5 (Mouse monoclonal) | Invitrogen | Cat#: 46–0705 | WB (1:1000) |
| Antibody | anti-YAP1 (Rabbit monoclonal) | Cell Signaling Technology | D8H1X; Cat#: 14074; RRID:AB_2650491 | WB (1:1000); IHC-P (1:50) |
| Transfected construct (human) | nontargeting siRNA | Dharmacon | Cat#: D-001810–01 | |
| Transfected construct (human) | siRNA to YAP1 (OnTarget Plus) | Dharmacon | Cat#: J-012200–06 | |
| Transfected construct (human) | siRNA to YAP1 (OnTarget Plus) | Dharmacon | Cat#: J-012200–08 | |
| Transfected construct (human) | siRNA to WWTR1 (OnTarget Plus) | Dharmacon | Cat#: J-016083–06 | |
| Transfected construct (human) | siRNA to WWTR1 (OnTarget Plus) | Dharmacon | Cat#: J-016083–08 | |
| Transfected construct (human) | siRNA to PTPN14 (OnTarget Plus) | Dharmacon | Cat#: J-008509–05 | |
| Transfected construct (human) | siRNA to PTPN14 (OnTarget Plus) | Dharmacon | Cat#: J-008509–08 | |
| Transfected construct (human) | siRNA to LATS1 (OnTarget Plus) | Dharmacon | Cat#: J-004632–05 | |
| Transfected construct (human) | siRNA to LATS1 (OnTarget Plus) | Dharmacon | Cat#: J-004632–08 | |
| Transfected construct (human) | siRNA to LATS2 (OnTarget Plus) | Dharmacon | Cat#: J-003865–09 | |
| Transfected construct (human) | siRNA to LATS2 (OnTarget Plus) | Dharmacon | Cat#: J-003865–10 |
Additional files
-
Supplementary file 1
Plasmids and antibodies used in this study.
Sheet one includes a list of all plasmids used in this study including details on the encoded genes, sgRNAs, promoters, antibiotic resistance markers, epitope tags, tag locations, selectable markers as well as Addgene plasmid numbers and citations of the original sources. Sheet two includes a list of all antibodies used in this study including details on the company, product number, and experimental conditions in which they were used.
- https://cdn.elifesciences.org/articles/75466/elife-75466-supp1-v2.xlsx
-
Supplementary file 2
Organotypic cultures used in this study.
Lists all organotypic cultures analyzed in this study and includes details on the expresion vectors employed, the cell backgrounds used, and all figures and panels that portray each culture.
- https://cdn.elifesciences.org/articles/75466/elife-75466-supp2-v2.xlsx
-
Supplementary file 3
Tumor microarray specimen information.
Lists the characteristics of the tumors included in the tumor microarray including the primary tumor site, HPV-positive/negative status, primary tumor T-stage, presence of nodal metastasis, and overall pathological stage.
- https://cdn.elifesciences.org/articles/75466/elife-75466-supp3-v2.xlsx
-
Supplementary file 4
Gene lists used for pathway mutational analyses.
Lists the genes included in each pathway used in the pathway mutational analyses.
- https://cdn.elifesciences.org/articles/75466/elife-75466-supp4-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/75466/elife-75466-transrepform1-v2.pdf





