Photonic hyperthermia of malignant peripheral nerve sheath tumors at the third near-infrared biowindow
Figures
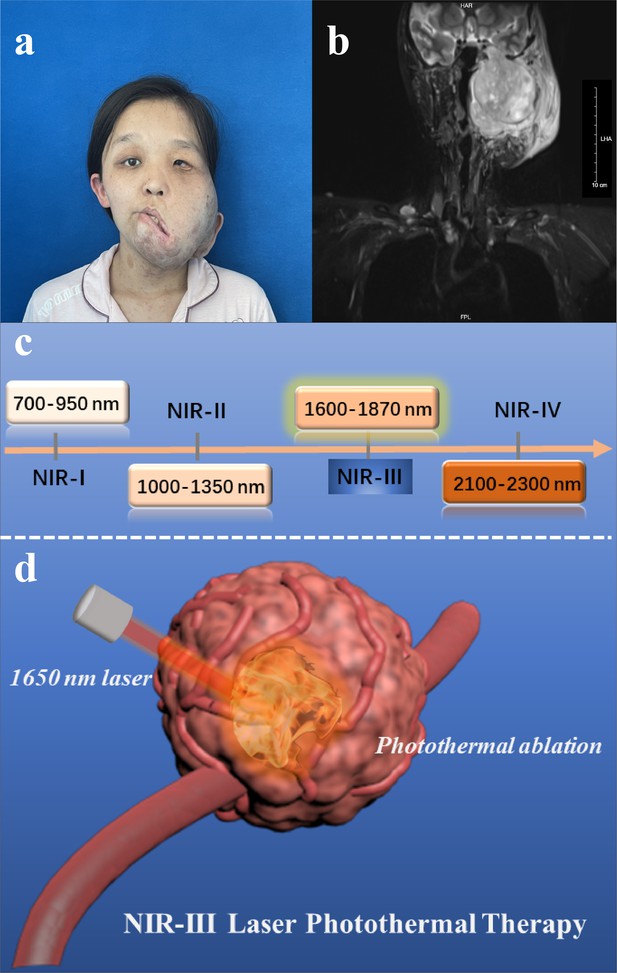
Disease models and therapeutic illustrations.
(a) Photograph and (b) magnetic resonance images of malignant peripheral nerve sheath tumor (MPNST) patient. (c) Classification of four near-infrared (NIR) wavelength regions for biomedical applications. (d) Schematic illustration of NIR-III laser (1650 nm) tumor photothermal therapy (PTT) through photothermal ablation without the addition of photothermal agents (PTAs).
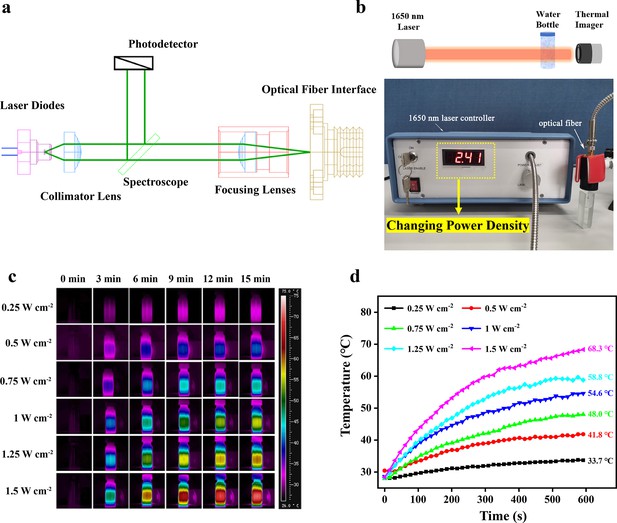
Photothermal properties of the NIR-III laser.
(a) Circuit diagram of the NIR-III laser. (b) Schematic diagram and equipment for detection of the photothermal effectiveness of the NIR-III laser. (c) Photothermal images of aqueous solutions irradiated for various durations with a 1650 nm laser at different power densities. (d) Temperature curves of aqueous solutions irradiated by a 1650 nm laser at different power densities.
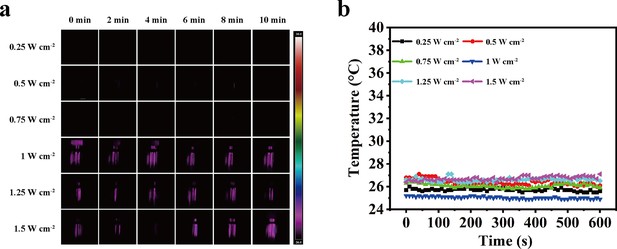
Photothermal behaviors of aqueous solutions by an 808 nm laser.
(a) Photothermal images of aqueous solutions irradiated for various durations by an 808 nm laser (NIR-I) at different power densities. (b) Temperature curves of water irradiated by the NIR-I laser at different power densities.
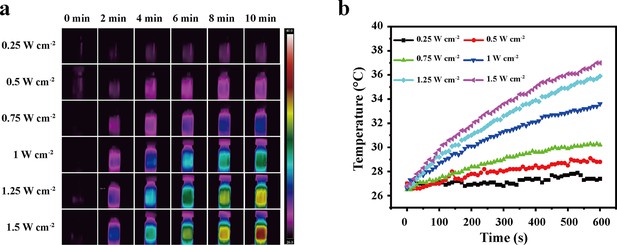
Photothermal behaviors of aqueous solutions by an 980 nm laser.
(a) Photothermal images of aqueous solutions irradiated for various durations by a 980 nm laser (NIR-II) at different power densities. (b) Temperature curves of aqueous solutions irradiated by the NIR-II laser at different power densities.
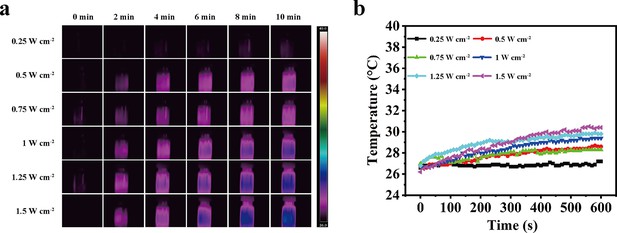
Photothermal behaviors of aqueous solutions by an 1064 nm laser.
(a) Photothermal images of aqueous solutions irradiated for various durations by a 1064 nm laser (NIR-II) at different power densities. (b) Temperature curves of aqueous solutions irradiated by the NIR-II laser at different power densities.
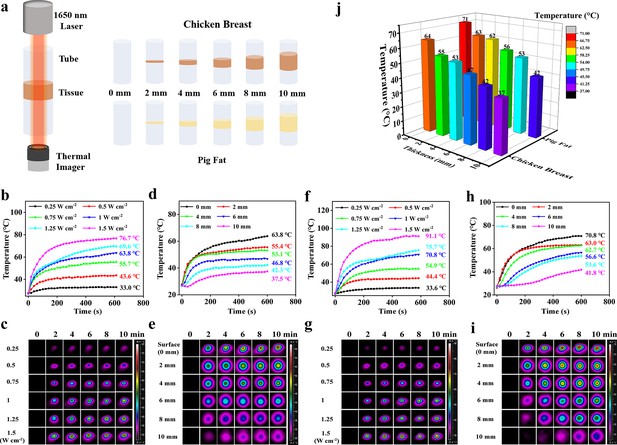
Photothermal performance for skin and subcutaneous tissue penetration.
(a) Schematic diagram for detection of the tissue penetration capability of the NIR-III laser. (b–c) Temperature curves (b) and photothermal images (c) of chicken breast surfaces irradiated by the NIR-III laser at different power densities. (d–e) Temperature curves (d) and photothermal images (e) of chicken breasts with different thicknesses irradiated by the NIR-III laser at 1.0 W cm–2. (f–g) Temperature curves (f) and photothermal images (g) of pig fat surfaces irradiated for various durations with the NIR-III laser at different power densities. (h–i) Temperature curves (h) and photothermal images (i) of pig fat with different thicknesses irradiated for various durations with the NIR-III laser at 1.0 W cm–2. (j) Bar graphs of temperature changes in chicken breast and pig fat of different thicknesses caused by NIR-III laser irradiation for 600 s.
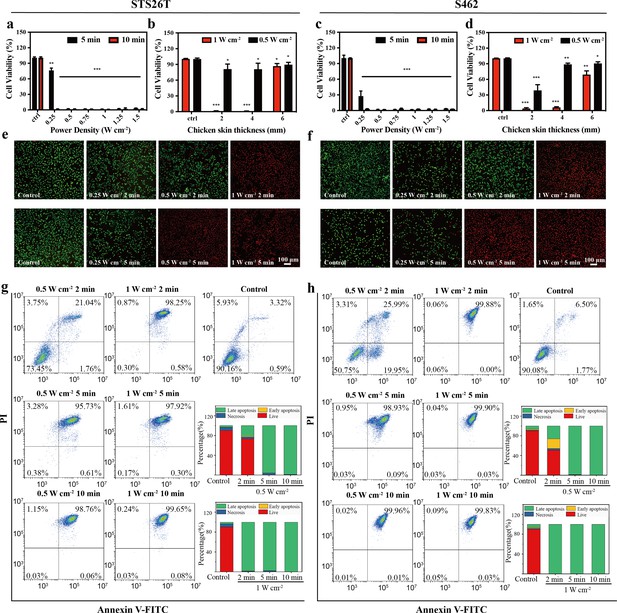
In vitro photothermal therapy (PTT) in the NIR-III biowindow.
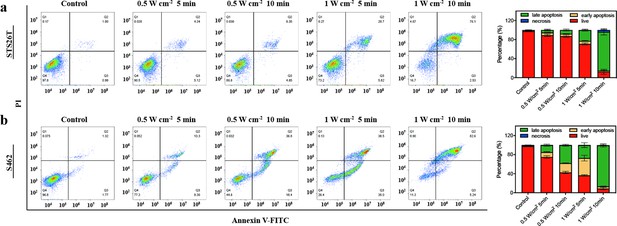
(a, c) Relative viabilities of STS26T cells (a) and S462 cells (c) seeded in 96-well culture plates and subjected to various durations of irradiation with the NIR-III laser at different power densities. (b, d) Relative viabilities of STS26T cells (b) and S462 cells (d) treated with the NIR-III laser (0.5 and 1 W cm–2) under different chicken skin thickness. All data are presented as mean ± SD and were analyzed with the unpaired t-test (n=3). (e–f) Confocal laser scanning microscopy (CLSM) images of STS26T cells (e) and S462 cells (f) costained with calcein-AM (green) and propidium iodide (PI) (red) after different treatments. (g–h) Flow cytometric analysis using Annexin V-FITC/PI kits in STS26T cells (g) and S462 cells (h) after different treatments. *p<0.05,**p<0.01, and ***p<0.001.
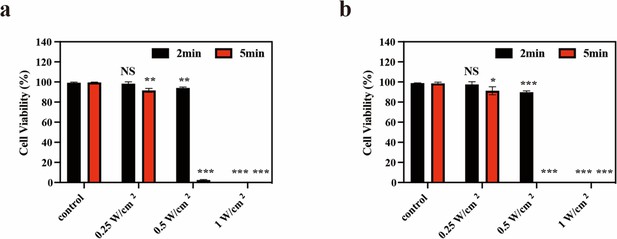
Cell viability analysis of STS26T cells (a) and S462 cells (b) using calcein-AM (green) and propidium iodide (PI) (red) staining after different treatments.
All data are presented as mean ± SD and were analyzed with the unpaired t-test (n=3). ‘NS’ indicates ‘not significant’. *p<0.05, **p<0.01, and ***p<0.001.
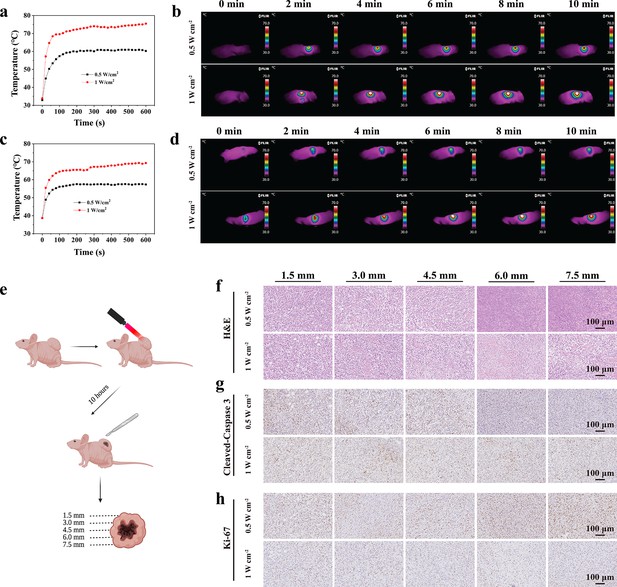
In vivo photothermal ablation of malignant peripheral nerve sheath tumors (MPNSTs) in the NIR-III biowindow.
(a–b) Temperature curves (a) and photothermal images (b) of STS26T tumor-bearing mice irradiated with the NIR-III laser at different power densities. Temperature curves (c) and photothermal images (d) of S462 tumor-bearing mice irradiated with the NIR-III laser at different power densities. (e) Schematic diagram for detection of the STS26T tumor tissue penetration capability of the NIR-III laser. (f–h) Hematoxylin and eosin (H&E) staining for pathological changes (f) anti-Cleaved-Caspase 3 immunohistochemical staining for apoptosis (g) and anti-Ki-67 immunohistochemical staining for cellular proliferation (h) at different depths (1.5, 3, 4.5, 6, and 7.5 mm) of dissected STS26T tumor tissues from the NIR-III (0.5 W cm–2, 5 min) group and the NIR-III (1 W cm–2, 5 min) group.
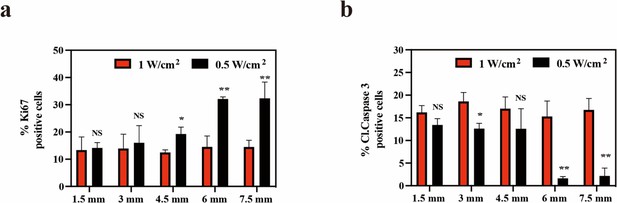
The statistical analysis of (a) Ki67-positive cells and (b) Cleaved-Caspase 3-positive cells after immunohistochemical staining.
All data are presented as mean ± SD and were analyzed with the unpaired t-test (n=3). ‘NS’ indicates ‘not significant’. *p<0.05 and **p<0.01.
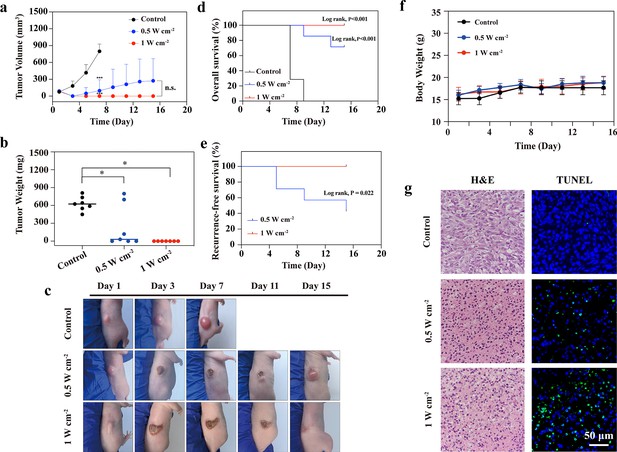
In vivo photothermal therapy (PTT) in the NIR-III biowindow.
(a) Time-dependent tumor growth curves for the control, NIR-III (0.5 W cm–2, 5 min), and NIR-III (1 W cm–2, 5 min) groups. (b) Tumor weights for tumor-bearing nude mice after different treatments. All data are presented as mean ± SD and were analyzed with the unpaired t-test (n=7). ‘NS’ indicates ‘not significant’. (c) Photographs of STS26T tumor-bearing mice and their tumor regions at 1, 3, 7, 11, and 15 days after different treatments. (d–e) Overall survival (OS) curves (d) and recurrence-free survival (RFS) curves (e) of mice after various treatments (n=7, p-value, log-rank test). (f) Time-dependent body weight curves of nude mice after different treatments (n=7). (g) Hematoxylin and eosin (H&E) staining and TdT-mediated dUTP-biotin nick and labeling (TUNEL) staining for pathological changes in tumor tissues from each group. *p<0.05.
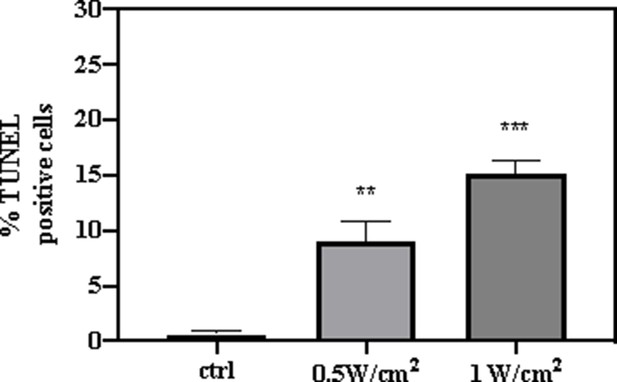
The statistical analysis of TdT-mediated dUTP-biotin nick and labeling (TUNEL)-positive cells according to TUNEL staining.
All data are presented as mean ± SD and were analyzed with the unpaired t-test (n=3). **p<0.01 and ***p<0.001.
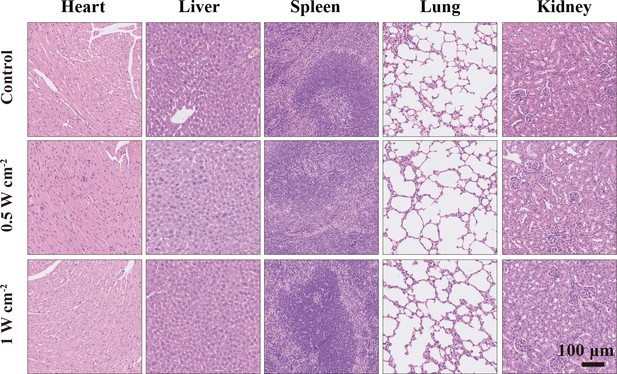
Hematoxylin and eosin (H&E) staining of major organs (heart, liver, spleen, lung, and kidney) in the control, NIR-III (0.5 W cm–2, 5 min), and NIR-III (1 W cm–2, 5 min) groups.
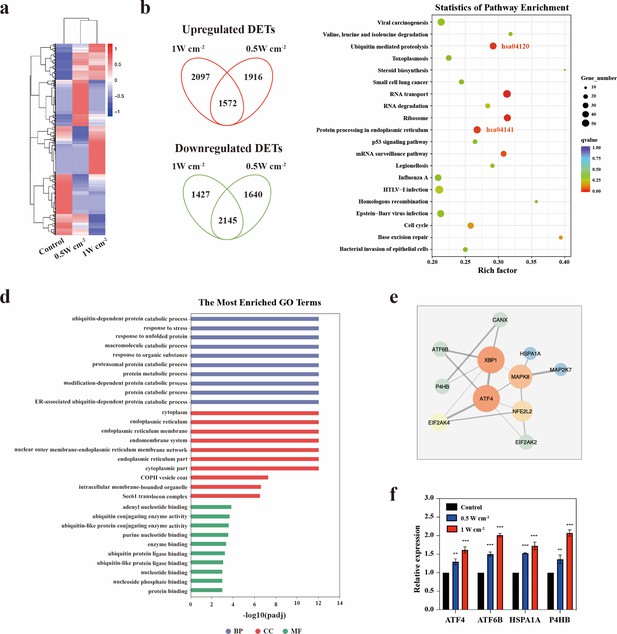
Endoplasmic reticulum stress triggered by NIR-III laser treatment.
(a) Heatmap of the mRNA screening data for STS26T tumors with or without NIR-III laser treatment at the indicated power densities. Blue indicates a low abundance of mRNA, and red indicates a high abundance. (b) Venn diagram showing the intersection of differentially expressed transcripts (DETs) in 0.5 and 1 W cm–2 NIR-III laser treatment group. (c) Top 20 pathways revealed by Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment among DETs simultaneously upregulated or downregulated in the NIR-III (0.5 W cm–2, 5 min) and NIR-III (1 W cm–2, 5 min) groups. The color indicates the q value; red represents a lower q value, while green represents a higher q value. A lower q value indicates more significant enrichment. The point size indicates the number of DET-related genes. (d-e) Gene Ontology (GO) functional enrichment (d) and protein-protein interaction network (e) of the genes enriched in the protein processing in endoplasmic reticulum pathway. (f) mRNA expression levels of ATF4, ATF6B, HSPA1A, and P4HB were measured by RT-qPCR in the indicated groups. All data are presented as mean ± SD and were analyzed with the unpaired t-test (n=3). **p<0.01 and ***p<0.001.
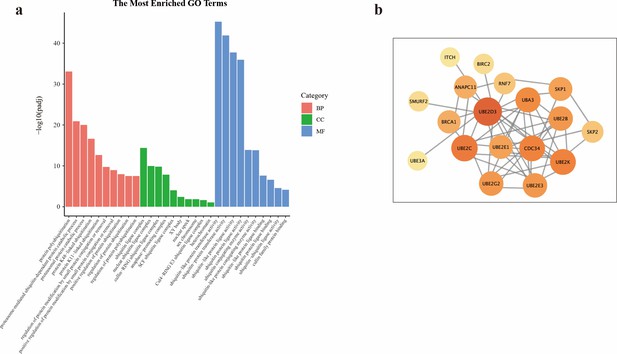
Alterations in ubiquitin-mediated proteolysis pathway triggered by NIR-III laser treatment.
(a) Gene Ontology (GO) functional enrichment and (b) protein-protein interaction network of the genes enriched in the protein processing in ubiquitin-mediated proteolysis pathway.
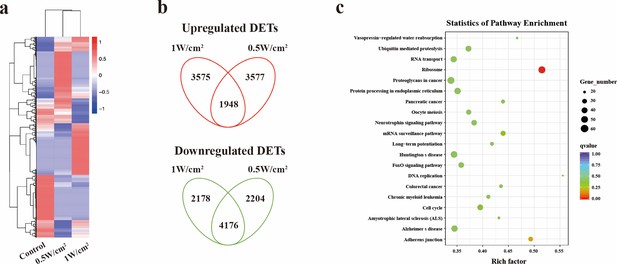
Therapeutic mechanisms exploration on S462 cells by transcriptome high-throughput sequencing.
(a) Heatmap of the mRNA screening data for S462 tumors with or without NIR-III laser treatment at the indicated power density. (b) Venn diagram showing the intersections of genes induced and repressed by 0.5 and 1 W cm–2 NIR-III laser treatment. (c) Top 20 pathways revealed by Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment among the differentially expressed transcripts (DETs) simultaneously upregulated or downregulated in the NIR-III (0.5 W cm–2, 5 min) and NIR-III (1 W cm–2, 5 min) groups.
Tables
STS26T NIR-III 1 W cm-2.
| Pathway term | qvalue | Gene number |
|---|---|---|
| Protein processing in endoplasmic reticulum | 0.03501825 | 68 |
| Ubiquitin mediated proteolysis | 0.01292571 | 62 |
STS26T NIR-III 0.
5 W cm-2
| Pathway term | qvalue | Gene number |
|---|---|---|
| Protein processing in endoplasmic reticulum | 0.02231676 | 55 |
| Ubiquitin mediated proteolysis | 0.00044313 | 56 |
Additional files
-
Supplementary file 1
Laser parameters (test after preheating for 5 min).
- https://cdn.elifesciences.org/articles/75473/elife-75473-supp1-v2.doc
-
Supplementary file 2
Top 10 upregulated genes related to hsa04141.
- https://cdn.elifesciences.org/articles/75473/elife-75473-supp2-v2.doc
-
Supplementary file 3
Top 10 upregulated genes related to hsa04120.
- https://cdn.elifesciences.org/articles/75473/elife-75473-supp3-v2.doc
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/75473/elife-75473-transrepform1-v2.docx