The C. elegans gonadal sheath Sh1 cells extend asymmetrically over a differentiating germ cell population in the proliferative zone
Figures
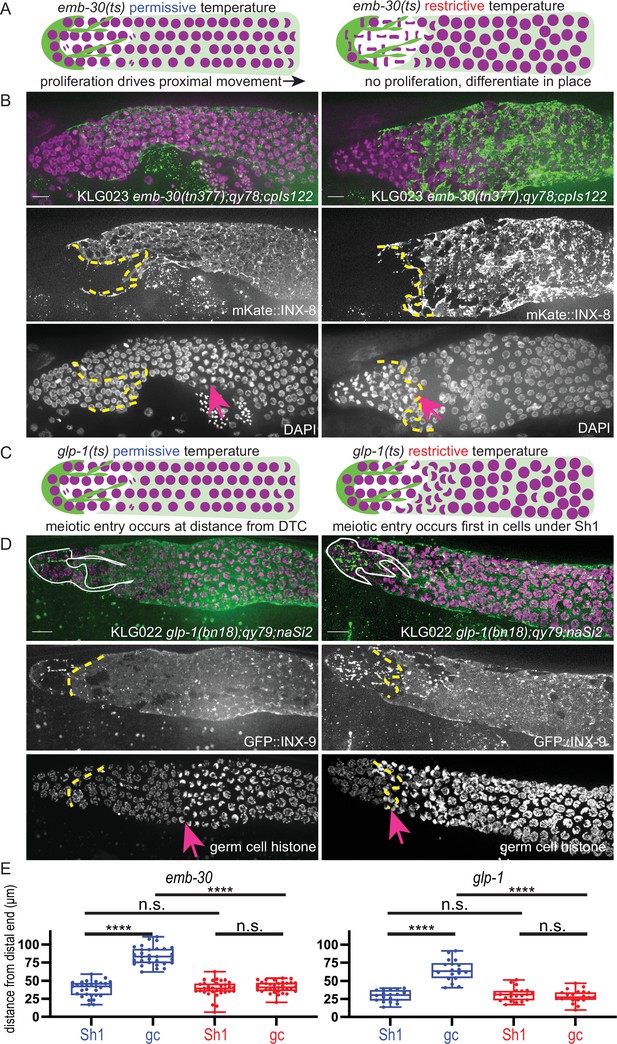
The Sh1 cells associate with proliferative germ cells that are on the path to differentiation.
(A) Schematic of hypothesis for emb-30(tn377) experiment. Germ cell (gc) nuclei shown in magenta, somatic gonad cells shown in green (distal tip cell [DTC]), and transparent green (Sh1). (B) Gonads from KLG023 emb-30(tn377);qy78;cpIs122 worms reared at permissive (left column) and restrictive (right column) temperatures. Top, merged image. Middle, mKate::INX-8 labeling Sh1 (edge outlined with yellow dashed line). Bottom, DAPI staining labeling all nuclei with pink arrow marking gc transition and same yellow dashed line as in middle image showing Sh1 edge. (C) Schematic of hypothesis for glp-1(ts) experiment. (D) Gonads from KLG022 glp-1(bn18);qy79;naSi2 worms reared at permissive (left column) and restrictive (right column) temperatures. Top, merged image. Middle, GFP::INX-9 labeling DTC (outlined in white) and Sh1 (edge outlined with yellow dashed line). Bottom, germ cell histone mCherry (naSi2[mex-5p::H2B::mCherry]) with pink arrow showing gc transition and same yellow dashed line as in middle image showing Sh1 edge. Note that the glp-1(bn18) allele is not fully wild type at permissive temperatures and is known to have a shortened proliferative zone (Fox and Schedl, 2015). (E) Box plots overlaid with all datapoints measuring the distal position of Sh1 and the position of the transition zone in germ cell nuclear morphology. Permissive temperature shown in blue; restrictive temperature shown in red. Permissive emb-30 N=30; restrictive emb-30 N=34. Permissive glp-1 N=18; restrictive glp-1 N=21. A one-way ANOVA to assess the effect of temperature on proximodistal position of gonad features was performed, and was significant for emb-30: F3,124=134.5, p<0.0001. Tukey’s multiple comparison test found that the mean values of the positions of Sh1 and the germ cell transition zone were significantly different at the permissive temperature (mean difference of –45.25 μm, 95% CI –52.38 to –38.12 μm, p<0.0001), but not at the restrictive temperature (mean difference of –2.30 μm, 95% CI –9.00 to 4.40 μm, p=0.808). The position of the germ cell transition zone differed at the permissive vs. restrictive temperatures (mean difference of 42.87 μm, 95% CI 35.95 to 49.79 μm, p<0.0001), but the Sh1 position did not (mean difference of –0.078 μm, 95% CI –7.00 to 6.84 μm, p>0.9999). This pattern is observed across replicates and various controls (Figure 1—figure supplement 1). Similar results were obtained for glp-1: F3,74=52.84, p<0.0001. Tukey’s multiple comparison test found that the mean values of the positions of Sh1 and the germ cell transition zone were significantly different at the permissive temperature (mean difference of –35.51 µm, 95% CI –44.59 to –26.43 µm, p<0.0001) but not at the restrictive temperature (mean difference of 2.514 µm, 95% CI –5.892 to 10.92 µm, p=0.861). The position of the germ cell transition zone differed at permissive vs. restrictive temperatures (mean difference of 36.02 µm, 95% CI 27.27 to 44.77 µm, p<0.0001), but the Sh1 position did not (mean difference of –1.997 µm, 95% CI –10.75 to 6.753 µm, p=0.9318). All scale bars 10 µm.
-
Figure 1—source data 1
Source data used to generate plots of distal sheath and germ cell transition zone measurements at permissive and restrictive temperatures for mutant strains shown in Figure 1.
- https://cdn.elifesciences.org/articles/75497/elife-75497-fig1-data1-v1.xlsx
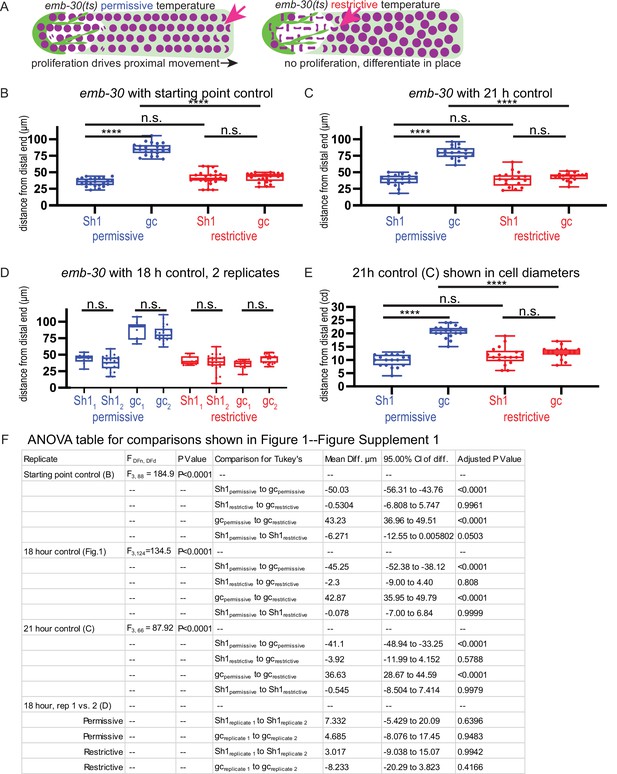
Robustness of emb-30 temperature shift experimental results to timing of control population.
(A) Schematic of hypothesis for emb-30(tn377) experiment. Germ cell (gc) nuclei shown in magenta, somatic gonad cells shown in green (distal tip cell [DTC]), and transparent green (Sh1). (B–F) Results from analysis of strain KLG023 emb-30(tn377);qy78;cpIs122 under different permissive temperature control culture times. Box plots overlaid with all datapoints measuring the distal position of Sh1 and the position of the transition zone in germ cell nuclear morphology. Permissive temperature shown in blue; restrictive temperature shown in red. (B) Controls fixed at starting point, 36 hr after L4 at permissive temperature as in Cinquin et al., 2010. Permissive, N=23; restrictive N=23. (C) Controls fixed after 21 additional hours of culture at the permissive temperature. Permissive N=18; restrictive N=17. (D) Two replicates in which controls were cultured an additional 18 hr at the permissive temperature. Replicate 1, permissive N=9; restrictive N=10. Replicate 2, permissive N=21; restrictive N=24. (E) Same experiment as shown in C, but with distances measured in cell diameters instead of microns. (F) Table of relevant ANOVA values for the significance indicators shown in B, C, and D. For (E), a one-way ANOVA to assess the effect of temperature on proximodistal position of gonad features was performed, and was significant F3,66=58.44, p<0.0001. Tukey’s multiple comparison test found that the mean values of the positions of Sh1 and the germ cell transition zone were significantly different at the permissive temperature (mean difference of –10.78 cell diameters, 95% CI –13.09 to –8.47 cd, p<0.0001), but not at the restrictive temperature (mean difference of –1.24 cd, 95% CI –3.61 to 1.14 cd, p=0.52). The position of the germ cell transition zone differed at the permissive vs. restrictive temperatures (mean difference of 7.73 cd, 95% CI 5.39 to 10.08 cd, p<0.0001), but the Sh1 position did not (mean difference of –1.81 cd, 95% CI –4.15 to 0.53 cd, p>0.19).
-
Figure 1—figure supplement 1—source data 1
Source data used to generate plots of distal sheath and germ cell transition zone measurements at permissive and restrictive temperatures for mutant strains shown in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/75497/elife-75497-fig1-figsupp1-data1-v1.xlsx
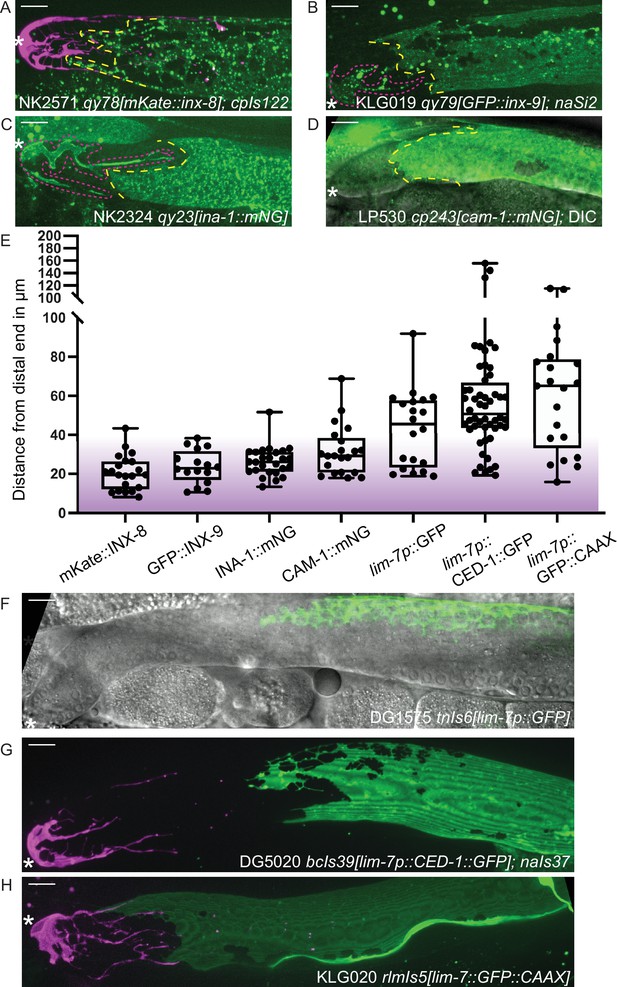
Sheath-expressed fluorescent proteins show consistency among endogenously tagged membrane proteins and greater variability in overexpressed transgenes.
(A) NK2571 qy78[mKate::inx-8]; cpIs122[lag-2p::mNeonGreen:: PLCδPH] N=21. (B) KLG019 qy79[GFP::inx9];naSi2 (channel not shown) N=16. (C) NK 2324 qy23[ina-1::mNG] N=26. (D) LP530 cp243[cam-1::mNG] N=21. (E) Box plots of Sh1 positions for all strains listed above and below, with fluorescent protein listed on the graph, including transgenes. (F) DG1575 tnIs6[lim-7p::GFP] N=20. (G) Strain DG5020 bcIs39[lim-7p::CED-1::GFP]; naIs37[lag-2p::mCherry-PH] N=52 (note that mean and range agree with those reported in Tolkin et al., 2022). (H) KLG020 rlmIs5[lim-7p::GFP::CAAX];cpIs122 N=21. Purple gradient marks approximate extent of stem cell zone (Lee et al., 2019; Shin et al., 2017). See Figure 2—figure supplement 1 for images of minimum and maximum observed distances for all markers. Figure 2—figure supplement 2 shows comparisons across development of NK2571 and DG5020. All scale bars 10 µm.
-
Figure 2—source data 1
Source data used to generate plots of distal sheath measurements for strains shown in Figure 2.
- https://cdn.elifesciences.org/articles/75497/elife-75497-fig2-data1-v1.csv
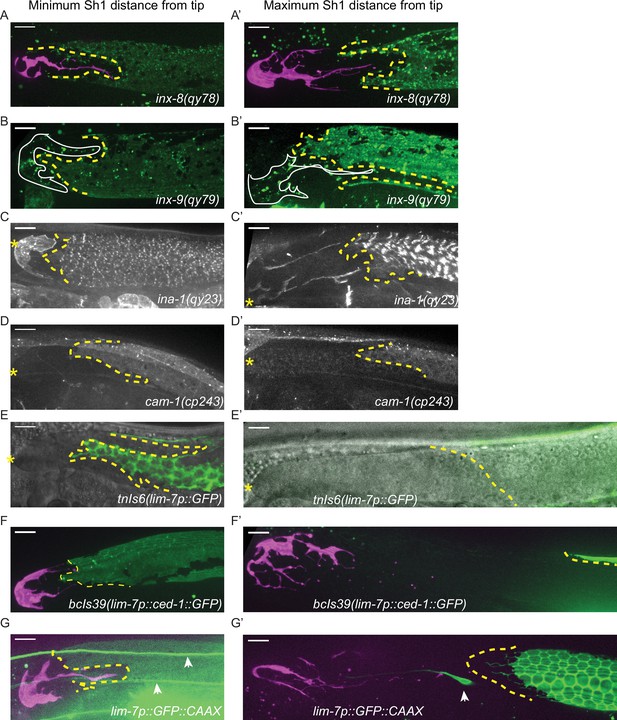
Endogenously tagged fluorescent proteins in the Sh1 membrane are less variable than overexpressed integrated transgenes.
Minimum (left column) and maximum (right column) measurements of the distance between distal Sh1 and the distal end of the gonad for (A, A’) qy78[mKate::inx-8], (B, B’) qy79[GFP::inx9], (C, C’) qy23[ina-1::mNG], (D, D’) cp243[cam-1::mNG] (E, E’) tnIs6[lim-7::GFP], (F, F’) bcIs39[lim-7p::ced-1::GFP], (G, G’) rlmIs5[lim-7p::GFP::CAAX]. Arrowheads in G and G’ mark non-sheath cells positive for lim-7p::GFP::CAAX expression in the plane of the gonad and are unavoidably included in z-projections that capture the gonadal cell surface. Note especially in G how dim the Sh1 expression is at the distal extent, resembling what is sometimes seen for CED-1::GFP expression as reported by Figure 2—figure supplement 2B of Tolkin et al., 2022. In some cases, the selected images are near-minimum or near-maximum due to imaging artifacts like low illumination or sample movement in the true minimum or maximum images. All scale bars 10 µm.
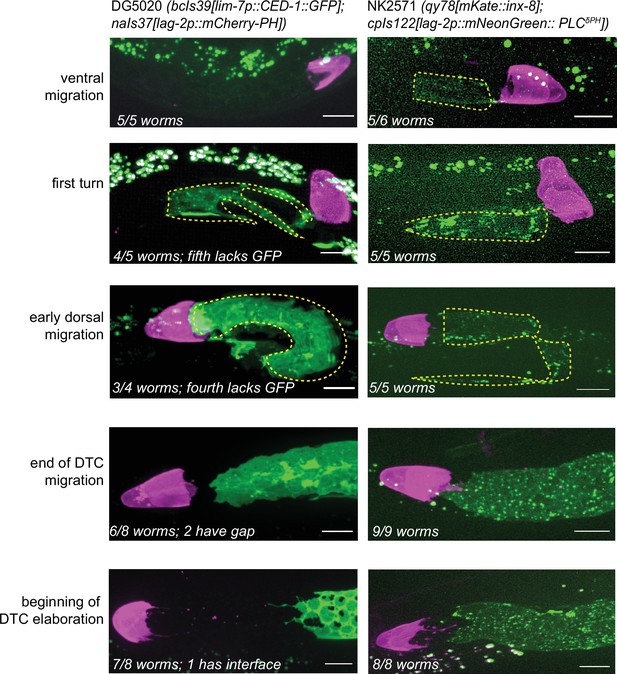
Differences between qy78(mKate::inx-8) and bcIs39(lim-7p::ced-1::GFP) expression in the sheath appear at the L4-young adult transition.
Side-by-side developmental comparisons between the two favored marker strains. Left column, DG5020 (bcIs39[lim-7p::CED-1::GFP]; naIs37[lag-2p::mCherry-PH]); right column, NK2571 (qy78[mKate::inx-8]; cpIs122[lag-2p::mNeonGreen:: PLCδPH]). Number of worms examined for each strain at each stage given in the figure annotations. Note that both strains show a close association of the distal tip cell (DTC) and sheath cells during larval development; fluorescence signal from lim-7p::ced-1::GFP appears later in development than mKate::inx-8 signal (top row). During dorsal DTC migration, highly variable gaps often appear between the DTC and Sh1 in both marker strains (not pictured). Rapid Sh1 growth during this stage has been observed (Gordon et al., 2020), so we attribute these variable gaps to our taking snapshots of a dynamic process. By the end of gonad elongation, most Sh1 cells come to rest within a germ cell diameter of the DTC for both strains. The large gap between the DTC and Sh1 only appears in the lim-7p::ced-1::GFP strain after larval gonad migration is complete. This stage is also when the lim-7p::ced-1::GFP expressing sheath develops two other unique characteristics—holes over the germ cell bodies and a frilly distal edge with small, thin distal projections.
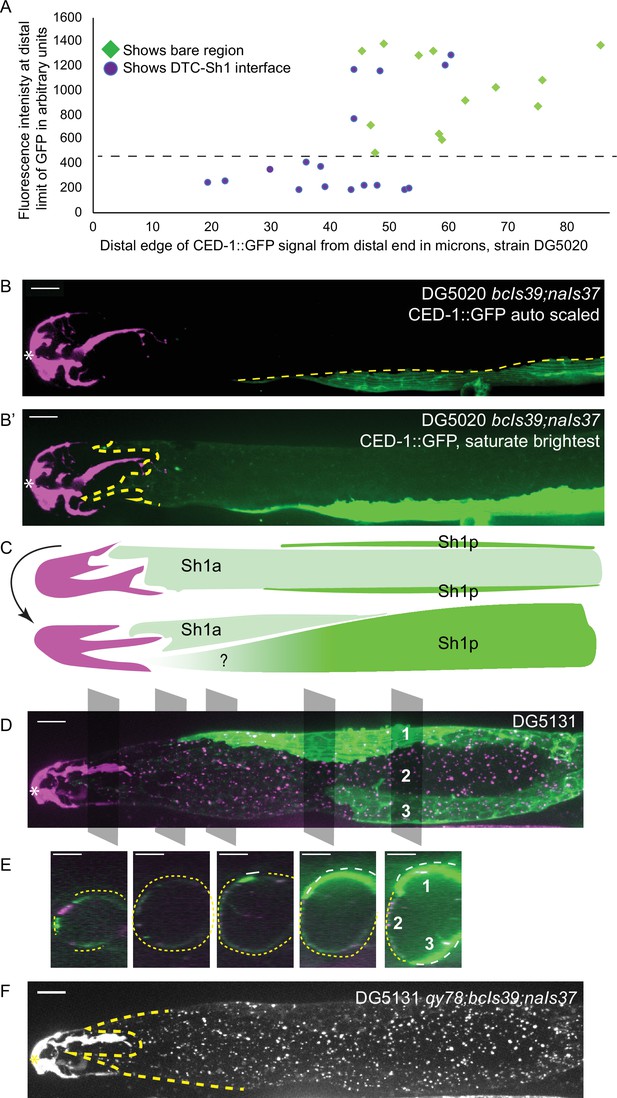
lim-7p::CED-1::GFP has variable expression intensity that conceals distal position of Sh1.
(A) Plot of distal position vs. fluorescence intensity in arbitrary units of CED-1::GFP at the distal limit of its domain in N=30 DG5020 bcIs39[lim-7p::CED-1::GFP]; naIs37[lag-2p::mCherry-PH] animals. Dashed black line: all of the lowly expressing gonads (under ~400 AU, or <30% maximum brightness of brightest sample) have a distal tip cell (DTC)-Sh1 interface detected. (B) DG5020 sample in which disparate expression levels in the two Sh1 cells of a single gonad arm obscure detection of the DTC-Sh1 interface. The GFP channel is scaled automatically in B; B’ is scaled to saturate the brightest pixels and reveal the dim second Sh1 cell. Dashed yellow link marks the edge of the bright Sh1 cell. (C) Schematic showing Sh1 pair configuration over distal germline, with the distal extent of Sh1p uncertain in superficial projection. The two Sh1 cells of a pair descend from the anterior and posterior daughters of Z1 and Z4, so the two Sh1 cells are here labeled Sh1a and Sh1p (arbitrarily). Top, superficial view. Bottom, side view. (D) DG5131 qy78[mKate::inx-8]; bcIs39[lim-7p::CED-1::GFP]; naIs37[lag-2p::mCherry-PH] sample in which one Sh1 cell contacts the DTC around the circumference of the germline and the other Sh1 cell lies at some distance from the distal end. Gray boxes and numbers mark planes and landmarks shown in (E). (E) Five cross sections through gonad in (E) made by projecting through two 1 µm re-slices at the positions shown by gray boxes in (D). Same analysis for DG5020 shown in Figure 3—figure supplement 1. (F) Same worm as in (D,E); signal from endogenously tagged allele qy78[mKate::inx-8] more uniformly labels the Sh1 cells, obscuring their individual shapes. All scale bars 10 µm.
-
Figure 3—source data 1
Source data used to generate plots of distal sheath position and fluorescence intensity measurements for samples shown in Figure 3A and Figure 4D.
- https://cdn.elifesciences.org/articles/75497/elife-75497-fig3-data1-v1.xlsx
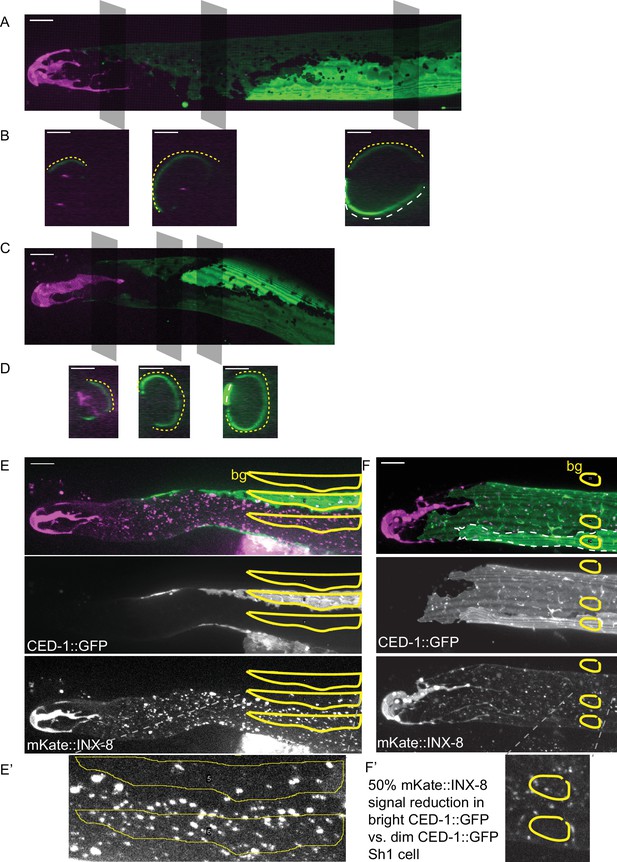
The Sh1 cells of a pair can take two distinct configurations over the distal germline.
(A) Example of a gonad from DG5020 lim-7p::ced-1::GFP animal with dramatically different CED-1::GFP signals revealing the shapes of the two Sh1 cells of the pair. Gray boxes show planes depicted in (B). (B) Three cross sections through gonad in (A) made by maximum projection through two 1 µm re-slices (FIJI) at the positions shown by gray boxes in (A). Dashed yellow and white lines mark the two Sh1 cells. Depending on the proximodistal position of the gonad, one or the other Sh1 cell may surround more of the germline. (C) Example of another gonad from DG5020. (D) Three cross sections through gonad in (C) made by projecting through two 1 µm re-slices at the positions shown by gray boxes in (C). (E,F) Gonads from strain DG5131 with merged images on top, CED-1::GFP channel in the middle, and mKate::INX-8 and distal tip marker channel on the bottom. Yellow outlines show regions of interest in which fluorescence intensity was measured. bg = background, subtracted from fluorescence intensity measured in each of the two Sh1 cells, which express CED-1::GFP at disparate levels. (E’ and F’) Insets from E and F. In both cases, mKate::INX-8 signal is half as strong in the Sh1 cells with more CED-1::GFP. Note also in E and F that mKate::INX-8 and bright CED-1::GFP mark a different distal extent of Sh1. All scale bars 10 µm.
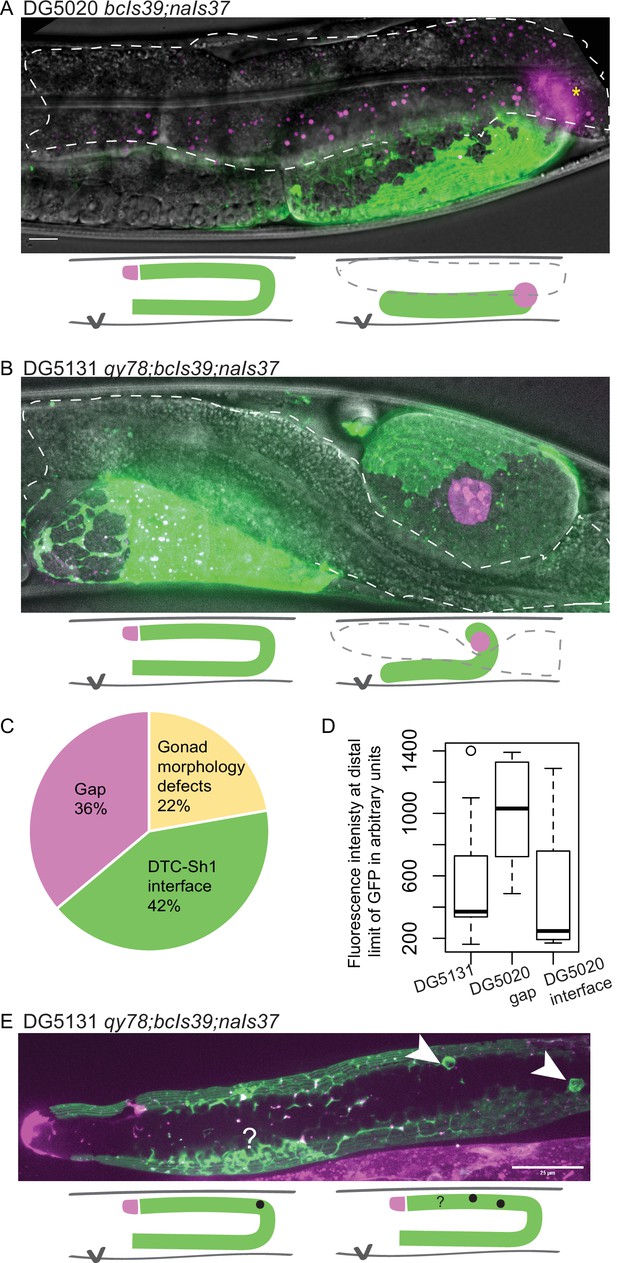
lim-7p::ced-1::GFP is correlated with gonad defects.
(A) Example of gonad morphology defect in DG5020 bcIs39[lim-7p::CED-1::GFP]; naIs37[lag-2p::mCherry-PH] strain, in which the gonad failed to turn. Gut outlined in dashed shape; magenta puncta in that domain are autofluorescent gut granules. (B) Example of gonad morphology defect in DG5131 qy78[mKate::inx-8]; bcIs39[lim-7p::CED-1::GFP]; naIs37[lag-2p::mCherry-PH] strain, in which gonad turned once and arrested without elongating along the dorsal body wall. Schematics in A and B show wild-type gonad morphology with two turns and a distal tip cell (DTC) that arrives at the dorsal midbody, left, beside schematics of defective gonad migration shown in micrographs. (C) Relative proportions of phenotypes observed in DG5020 animals (N=72). (D) Boxplot comparing fluorescence intensity for coexpressing strain DG5131 in addition to data shown in Figure 3 for DG5020. Fluorescence intensity of the lim-7p::ced-1::GFP transgene in this background is statistically indistinguishable from expression levels of this transgene in an otherwise wild-type background in the subset of samples that display a DTC-Sh1 interface, shown here segregated from samples from this strain that show a gap between the DTC and Sh1 cells. DG5131 N=17. DG5020 gap N=13. DG5020 interface N=17. A one-way ANOVA to determine the effect of category (genotype or presence of an interface) and fluorescence intensity was performed and was significant, F2,44=7.70, p=0.001. Tukey’s multiple comparison test finds that the mean fluorescence intensity of DG5020 gonads with a DTC-Sh1 interface differs from DG5020 gonads with a gap between Sh1 and the distal end (p=0.002) and does not differ from DG5131 worms (p=0.908). (E) Gonad from DG5131 strain with white arrowheads indicating aberrant engulfment of germ cells in the distal gonad. Closer to the distal end, a large mass of germ cells showing substantial localization of the CED-1::GFP protein may also reflect ectopic engulfment. Schematics show location of germ cell engulfment in wild-type gonads on the left and locations of the features marked in the micrograph in E on the right. Scale bars in A and B, 10 µm; scale bar in E, 25 µm.
-
Figure 4—source data 1
Classifications of 72 gonads from strain DG5020 that display a defect, a gap, or an interface used to generate pie chart in Figure 4C.
- https://cdn.elifesciences.org/articles/75497/elife-75497-fig4-data1-v1.xlsx
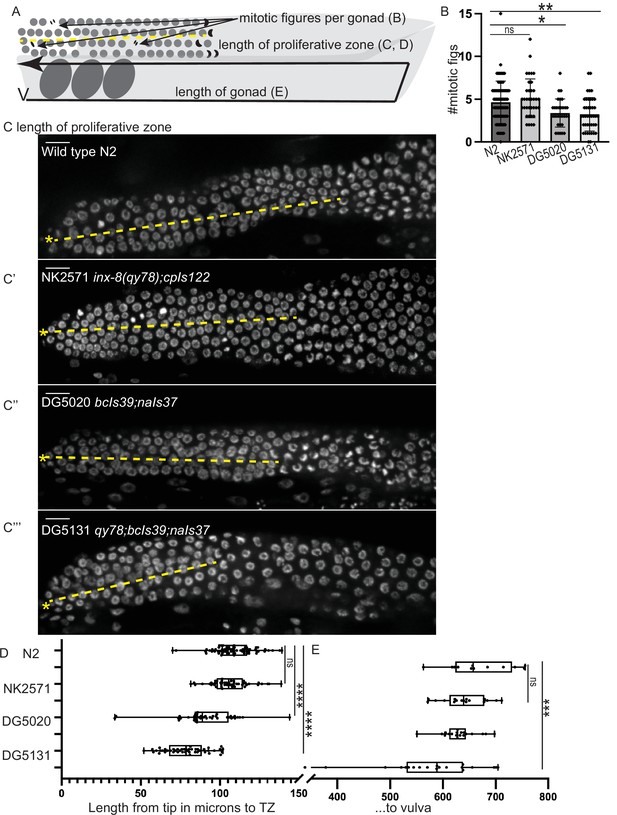
A synthetic interaction between lim-7p::ced-1::GFP and the tagged innexin qy78 shortens the proliferative zone.
(A) Illustration of measurements made for Figure 5. (B) Number of mitotic figures observed in DAPI stained animals of the four strains. Numbers of dividing cells and gonads examined are as follows: wild-type N2 (N=311 dividing cells/67 gonads), the NK2571 strain with the tagged innexin qy78 (N=184 dividing cells/36 gonads), the DG5020 strain with lim-7p::ced-1::gfp (N=105 dividing cells/31 gonads), the DG5131 strain combining these sheath markers (N=136 dividing cells/42 gonads). A one-way ANOVA to determine the effect of genotype on number of mitotic figures was significant F3, 172=7.081, p=0.0002. Tukey’s multiple comparison test revealed that NK2571 did not differ from wild type (mean difference –0.47 cells per gonad, 95% CI –1.65 to 0.71, p=0.7291), DG5020 differed from wild type by 1.26 germ cells per gonad, 95% CI 0.02–2.50, p=0.0453, and DG5131 differed from wild type (mean difference of 1.40 cells per gonad, 95% 0.28–2.52, p=0.0075). (C-C””) DAPI stained distal gonads for measurement of proliferative zone for the four strains. Asterisk marks tip of gonad, dashed line marks example of lengths measured. (C) Wild-type N2 (N=68), (C’) the NK2571 strain with the tagged innexin qy78 (N=49), (C”) the DG5020 strain with lim-7p::ced-1::gfp (N=40), (C”’) and the DG5131 strain combining these sheath markers (N=45). Asterisk marks tip of gonad. (D) Plots of proliferative zone length (left) and whole gonad length (right) for the four strains. A one-way ANOVA to determine the effect of genotype on length of proliferative zone was significant F3,198=49.15, p<0.0001. Tukey’s multiple comparison test revealed that NK2571 did not differ from wild type (mean difference 2.69 μm, 95% CI –4.283 to 9.663 μm, p=0.750), DG5020 differed from wild type by ~2–5 germ cell diameters (mean difference of 17.94 μm, 95% CI 10.53 to 25.36 μm, p<0.0001), and DG5131 dramatically differed from wild type (mean difference of 30.36 μm, 95% CI 23.21 to 37.51 μm, p<0.0001). The proliferative zone length of DG5131 was also significantly different from both of its parent strains (NK2571 vs. DG5131 mean difference of 27.67 μm, 95% CI 19.99 to 35.35 μm, p<0.0001; DG5020 vs. DG5131 mean difference of 12.42 μm, 95% CI 4.331 to 20.50 μm, p=0.0006). (E) Plots of length of entire gonad from tip to vulva. N2 (N=12), NK2571 (N=17), DG5020 (N=19), DG5131 (N=20). A one-way ANOVA to determine the effect of genotype on gonad length was significant F3,64=6.27, p=0.0009. Tukey’s multiple comparison test revealed that NK2571 did not differ from wild type (mean difference 31.16 μm, 95% CI –32.99 to 95.32 μm, p=0.578), DG5020 also did not differ from wild type (mean difference of 39.42 μm, 95% CI –23.32 to 102.2 μm, p=0.3546), and DG5131 did differ from wild type (mean difference of 95.15 μm, 95% CI 33.02 to 157.3 μm, p=0.0008). All scale bars 10 μm.
-
Figure 5—source data 1
Measurements used to generate plots of proliferative zone length, gonad length, and number of mitotic figures for Figure 5.
- https://cdn.elifesciences.org/articles/75497/elife-75497-fig5-data1-v1.xlsx
Tables
Brood size assays.
| Strain name | Full genotype | Live brood* | Reduction vs. wt % | Unhatched eggs | Embryonic lethality % |
|---|---|---|---|---|---|
| N2 | Wild type | 295±39 (n=57) | NA | NA | NA† |
| KLG019 | qy79[GFP::inx-9];nasi2‡, § | 226±22 (n=13) | 23% | 165±58 | 41 ± 8% |
| NK2571¶ | qy78[mKate::inx-8];cpIs122§ | 220±41 (n=15) | 25% | 20±14 | 9 ± 5% |
| DG5020¶ | bcIs39[lim-7p::ced-1::GFP];naIs37§ | 202±29 (n=12) | 32% | 62±47 | 20 ± 14% |
| DG5131 | qy78[mKate::inx-8];bcIs39[lim-7p::ced-1::GFP];naIs37§ | 187±45 (n=14) | 37% | 40±25 | 18 ± 11% |
| LP530 | cp243[cam-1::mNG] | 260±31 (n=10) | 12% | NA | NA |
| NK2324 | qy23[ina-1::mNG] | 237±37 (n=8) | 20% | NA | NA |
-
*
Viable offspring that hatch from a single parent.
-
†
N2 numbers come from multiple trials, not all of which were scored for embryonic lethality, including the trial in which ina-1(qy23) and cam-1(cp243) were counted.
-
‡
qy79[GFP::inx-9] allele in strains NK2572 and NK2573 from Gordon et al., 2020, with germ cell nuclear marker naSi2; this combination of alleles was used in the cross to glp-1(bn18) in Figure 1D.
-
§
Full transgene descriptions in Methods for germ cell (naSi2) and DTC (cpIs122, naIs37) markers.
-
¶
See Appendix 1—table 1 for replicates and statistical analysis of NK2571 and DG5020.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Caenorhabditis elegans) | inx-8 | https://wormbase.org/ | Sequence CELE_ZK792.2 | Encodes gap junction hemichannel subunit |
| Gene (Caenorhabditis elegans) | inx-9 | https://wormbase.org/ | Sequence CELE_ZK792.3 | Encodes gap junction hemichannel subunit |
| Gene (Caenorhabditis elegans) | glp-1 | https://wormbase.org/ | Sequence CELE_F02A9.6 | Encodes Notch receptor |
| Gene (Caenorhabditis elegans) | emb-30 | https://wormbase.org/ | Sequence CELE_F54C8.3 | Encodes putative member of APC |
| Gene (Caenorhabditis elegans) | ina-1 | https://wormbase.org/ | Sequence CELE_F54G8.3 | Encodes worm alpha integrin ortholog |
| Gene (Caenorhabditis elegans) | cam-1 | https://wormbase.org/ | Sequence CELE_C01G6.8 | Encodes worm Wnt receptor |
| Gene (Caenorhabditis elegans) | ced-1 | https://wormbase.org/ | Sequence CELE_Y47H9C.4 | Encodes worm cell death receptor |
| Genetic reagent (Caenorhabditis elegans) | inx-8(qy78(mKate::inx-8)) IV; cpIs122(lag-2p::mNeonGreen:: PLCdPH) | Gordon et al., 2020 | NK2571 | Can be obtained from K Gordon lab |
| Genetic reagent (Caenorhabditis elegans) | inx-9(qy79(GFP::inx-9)) IV; naSi2(mex-5p::H2B::mCherry::nos-2 3′UTR) II | nasi2 transgene from Roy et al., 2018; qy79 from Gordon et al., 2020 | KLG019 | Can be obtained from K Gordon lab |
| Genetic reagent (Caenorhabditis elegans) | rlmIs5[lim-7p::GFP::CAAX] | This study | KLG020 | Can be obtained from K Gordon lab |
| Genetic reagent (Caenorhabditis elegans) | qy78(mKate::inx-8) IV | This study | KLG021 | ×2 outcross of NK2571 to N2 |
| Genetic reagent (Caenorhabditis elegans) | inx-9(qy79(GFP::inx-9)) IV; naSi2(mex-5p::H2B::mCherry::nos-2 3′UTR) II; glp-1(bn18) III | glp-1(bn18) from Kodoyianni et al., 1992 doi: 10.1091/mbc.3.11.1199 | KLG022 | Mutant obtained from CGC, crossed to KLG019 |
| Genetic reagent (Caenorhabditis elegans) | inx-8(qy78(mKate::inx-8)) IV; cpIs122(lag-2p::mNeonGreen:: PLCdPH); emb-30(tn377) III | emb-30(tn377) from Cinquin et al., 2010 doi: 10.1073/pnas.0912704107 | KLG023 | Mutant obtained from CGC, crossed to NK2571 |
| Genetic reagent (Caenorhabditis elegans) | cp243[cam-1::mNG] | Heppert et al., 2018 doi:10.1534/GENETICS.117.300487 | LP530 | Can be obtained from B Goldstein lab |
| Genetic reagent (Caenorhabditis elegans) | qy23[ina-1::mNG] | Jayadev et al., 2019 doi: 10.1083/jcb.201903124 | NK2324 | Can be obtained from D Sherwood lab |
| Genetic reagent (Caenorhabditis elegans) | tnIs6[lim-7p::GFP] | Hall et al., 1999 | DG1575 | Obtained from CGC |
| Genetic reagent (Caenorhabditis elegans) | bcIs39[lim-7p::ced-1::GFP];naIs37 | Tolkin et al., 2022; naIs37 originally from Pekar et al., 2017 | DG5020 | See Tolkin et al., 2022 |
| Genetic reagent (Caenorhabditis elegans) | qy78[mKate::inx-8];bcIs39[lim-7p::ced-1::GFP];naIs37 | Tolkin et al., 2022 | DG5131 | See Tolkin et al., 2022 |
| Software, algorithm | μManager software v1.4.18 | (Edelstein et al., 2010) doi: 10.1002/0471142727.mb1420s92 | RRID:SCR_016865 | https://micro-manager.org/ |
| Software, algorithm | FIJI 2.0 | Schindelin et al., 2012 doi: 10.1038/nmeth.2019 | RRID:SCR_002285 | https://fiji.sc/ |
| Software, algorithm | GraphPad Prism version 9.20 (283) for macOS | GraphPad Software, San Diego, CA | RRID:SCR_002798 | https://www.graphpad.com/ |
| Software, algorithm | Adobe Illustrator CC | Adobe Systems Inc | RRID:SCR_010279 |
Replicated, anonymized brood size assay (Dowen Lab).
| Strain | Tolkin (v1) | Tolkin (revised) | Li (submitted) | Dowen (anon.) | Li vs. Dowen |
|---|---|---|---|---|---|
| NK2571 (qy78; cpIs122) | 155±24.4 (n=19) | 212.8±27.5 (n=23) | 220±41 (n=15) | 233±32 (n=11) | t=0.899, p=0.378, n.s. |
| DG5020 (lim-7p::ced-1::GFP; naIs37) | 235.8±43.2 (n=56) | 237.5±46.5 (n=24) | 202±29 (n=12) | 213±52 (n=12) | t=0.643, p=0.527, n.s. |
| Strain, Genotype | Tolkin Figure 4 label | Tolkin CYE-1(+) domain | Mohammed Figure 2 CYE-1(+) domain | Mohammed Figure 2 DAPI, est. from 1 sample | Li Figure 5 label | Li DAPI mean measurement of PZ |
|---|---|---|---|---|---|---|
| N2 wild-type | NA | NA | ~95 um ~21 gcd | ~113 um ~28 gcd | Wild type N2 | 109 +/- 13 um 27 +/- 3 gcd |
| NK2571 (qy78;cpIs122) | NA | NA | NA | NA | inx-8(qy78);cpIs122 | 106 +/- 12 um 26 +/- 2 gcd |
| DG5020 (bcIs39; naIs37) | “markers only” (4A) “Wild-type” (4C) | ~70 um ~17 gcd | NA | NA | DG5020(bcIs39, naIs37) | 91 +/- 19 um 23 +/- 3 gcd |
| DG5131 (qy78;bcIs39; naIs37) | inx-8(qy78) | ~60 um ~15 gcd | NA | NA | DG5131(qy78;bcIs39; naIs37) | 79 +/- 13 um 20 +/- 2 gcd |





