A neural progenitor mitotic wave is required for asynchronous axon outgrowth and morphology
Figures
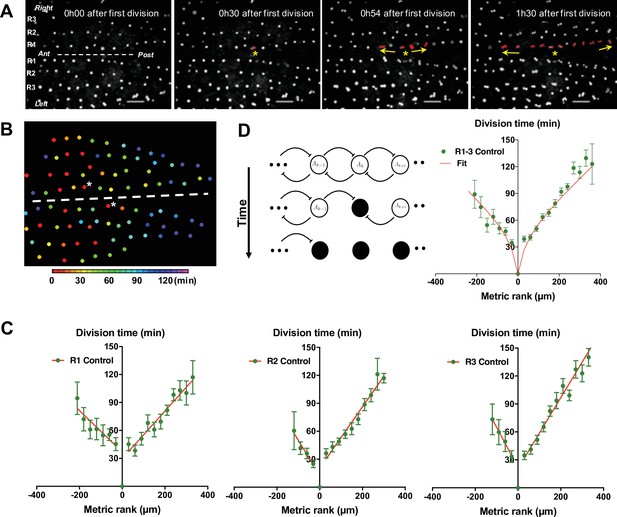
G2-arrested SOP cells of the neuroepithelium resume division in a temporal wave.
(A) Four frames of a time-lapse recording of a control Drosophila melanogaster pupa from Video 1. The first three rows of cells in the most dorsal part of the thorax are indicated as R1, R2, and R3 respectively. The dashed line marks the midline, with anterior (Ant) to the left and posterior (Post) to the right. SOPs that have divided are highlighted in red in R1. The yellow arrows indicate the propagation of the wave from SOP0 marked by a yellow star. Scale bar, 50 µm. (B) Heatmap of a representative control notum depicting time of SOP division. Circles represent SOP cells arranged in rows. The relative time of division is colour coded according to the scale, each colour covering 6 min. Only rows 1–4 are represented. The dashed line shows the midline, with anterior to the left, and SOP0 in row one marked by white stars. (C) The time of SOP cell division (mean time± SEM of n = 16 nota) in row 1 (R1), row 2 (R2), and row 3 (R3) is plotted according to position both relative to the position and time of division of the first cell to divide in each row. Negative and positive metric rank therefore corresponds to SOPs that are anterior and posterior to the SOP0, respectively. Red lines show standard linear regressions. SOPs were identified by GFP expression in a neur> GFP fly line. (D) Modelling the wave of SOP divisions. Left, a schematic view of the model at three consecutive times. Empty circles depict non-dividing cells. The flat-headed arrows indicate the inhibition that one cell exerts on immediate neighbours preventing entry into mitosis. Filled circles represent mitotic or post-mitotic cells which no longer have an inhibitory effect thus allowing neighbours to divide. These interactions allow the progression of mitosis along a row. Right, fit of experimental data with theoretical values obtained with the model using as parameters ρ = 20 and µ = 0.65 (See Model description).
-
Figure 1—source data 1
Related to Figure 1 Raw data.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig1-data1-v2.xlsx
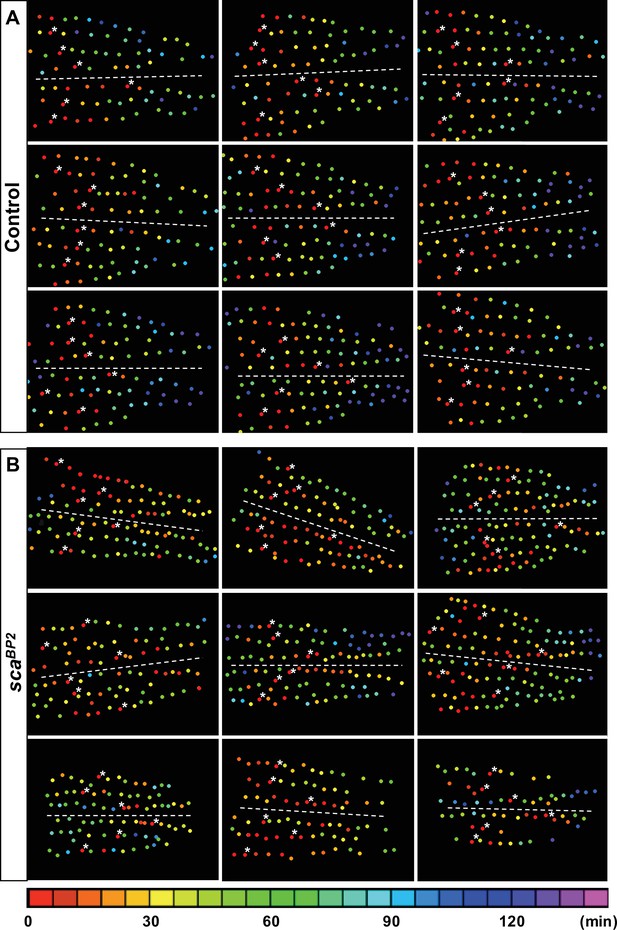
Illustrations of individual mitotic waves in control and scaBP2 mutant.
Heatmaps of nine nota showing the time of SOP division in control (A) and scaBP2 pupae (B). Circles represent SOP cells arranged in rows, anterior to the left. The relative time of division is colour coded according to the scale along the bottom, each colour covering 6 minutes. In each row, SOP0 are marked by white stars. Dashed lines show the midline. Note that in mutants, many cells are coloured in the reddish part of the spectra showing that several SOP cells divided simultaneously in the same row rather than in control where these cells are located in the anterior region of each row.
-
Figure 1—figure supplement 1—source data 1
Related to Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig1-figsupp1-data1-v2.xlsx
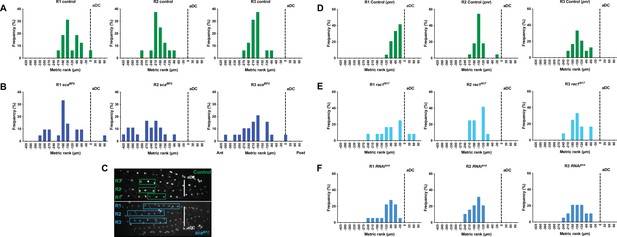
The mitotic wave origin spreads out in several genetic contexts.
Location of the first dividing SOP (SOP0) in rows 1–3 in scaBP2 flies (B, n = 9) and its control, neur> GFP flies, (A, n = 9), after overexpression of rac1N17 (E, n = 6) or RNAi-sca (F, n = 9) and their control, pnr> GAL4 tub> Gal80 ts (D, n = 12). The line relaying the two antero-dorsocentral macrochætes (aDC) was used as a spatial reference (dotted line on graphs A, B, D, E, F and vertical lines in C). Negative and positive metric ranks correspond to SOPs anterior and posterior to the aDC line respectively. Graphs represent the frequency of SOP0 at a given position recorded in row 1 (R1), row 2 (R2) and row 3 (R3). (C) Image of hemi-nota of a control, neur> GFP fly, and a scaBP2 homozygous mutant aligned anterior to the left showing the organisation of the SOPs in the notum. White vertical lines connecting the two aDC were used as spatial reference. Boxes show the locations of 68% of SOP0 in each row, corresponding to one standard deviation from the mean.
-
Figure 1—figure supplement 2—source data 1
Related to Figure 1—figure supplement 2 Raw data.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig1-figsupp2-data1-v2.xlsx
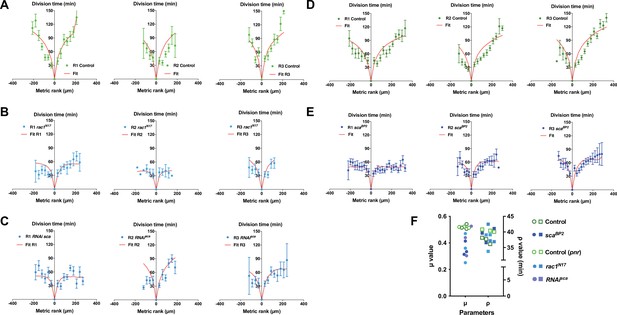
Reduced mitotic waves were better fitted with low values of the inhibitory parameter μ.
(A–E) The time of SOP cell division (mean time ± SEM) in row 1 (R1), row 2 (R2), and row 3 (R3) is plotted relative to the position and time of division of the first cell to divide in each row. The different conditions in which the mitotic wave amplitude was reduced were: (B) over-expression of rac1N17, using the conditional genetic context pnr> gal4 tub GAl80ts UAS-rac1N17 (C) conditional RNAi-sca, using the conditional genetic context pnr> gal4 tub GAl80ts RNAi-sca (E) scaBP2 null mutant. (A) and (D) corresponding controls to (B-C) and (E) respectively. Red lines depict the best fits obtained with our model. (F) Dot plot showing the distribution μ and ρ parameters extracted from the fits displayed in A-E. Note that in all cases the best fit was obtained after the reduction of the μ parameter.
-
Figure 1—figure supplement 3—source data 1
Related to Figure 1—figure supplement 3 Raw data.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig1-figsupp3-data1-v2.xlsx
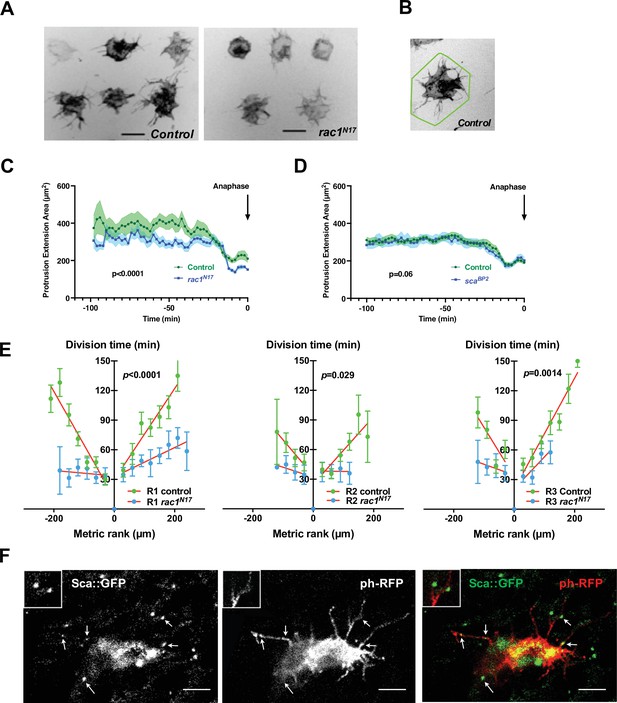
The SOP mitotic wave is affected in conditions where cell protrusions are reduced.
(A) Frames from live recordings showing cell protrusions of SOPs in control (left) and rac1N17 (right) pupae. tubGal80ts was used to conditionally express Gal4 and thus rac1N17 by maintaining flies at 18 °C and shifted them to 30 °C from 12 hr to 19 hr APF. Protrusions were visualised by specific expression of a membrane-tethered RFP form (ph-RFP), invert fluorescence. Scale bar, 20 µm. (B) Illustration of the polygonal line corresponding to the convex hull enclosing the extremities of the longer protrusions (Fiji software) to quantify the cell protrusion length. (C) Protrusion extension area over time in control and after rac1N17 conditional overexpression (mean ± SEM, n = 11 cells for control and n = 14 for rac1N17) measured from live recordings. Anaphase was taken as a temporal reference. (D) Protrusion extension area over time in control and in scaBP2 null mutant (mean ± SEM, n = 20 cells for control and n = 18 for scaBP2) measured from live recordings. Anaphase was taken as a temporal reference. (E) The SOP mitotic wave in row 1 (R1), row 2 (R2) and row 3 (R3) in control and after overexpression of rac1N17. Mean division time± SEM of n = 6 nota. (F) Sca localisation in a SOP cell. Arrows indicate distinct Sca foci along cell protrusions. Scabrous (left panel) in SOP cell marked with a membrane-bound ph-RFP (middle panel), merged in right panel. Images are a maximum projection of three confocal sections. Two Sca foci are magnified in the inserts. Scale bar, 5 µm.
-
Figure 2—source data 1
Related to Figure 2 Raw data.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig2-data1-v2.xlsx

Illustrations of individual mitotic waves after reduction of cell protrusions by overexpression of Rac1N17 and after conditional inactivation of sca.
Heatmaps of six nota showing the time of SOP division in control neur> ph (PLCδ)::RFP/+; pnr> Gal4 tub> Gal80ts/+ pupae (A), when cell protrusions were reduced in neur> ph (PLCδ)::RFP/+; pnr> Gal4 tub> Gal80ts/UAS-rac1N17 pupae (B) and when sca was specifically downregulated between 12 and 19 hr APF, using the conditional genetic context pnr> Gal4 tub Gal80ts RNAi-sca (C). Circles represent SOP cells arranged in rows, anterior to the left. The relative time of division is colour coded according to the scale along the bottom, each colour covering 6 min. In each row, SOP0 are marked by white stars. Dashed lines show the midline. Note that in (B) and (C) many cells are coloured in the reddish part of the spectra showing that several SOP cells divided simultaneously in the same row. Note also that the driver pnr used to overexpress rac1N17 disorganises the pattern of rows.
-
Figure 2—figure supplement 1—source data 1
Related to Figure 2—figure supplement 1 Raw data.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig2-figsupp1-data1-v2.xlsx
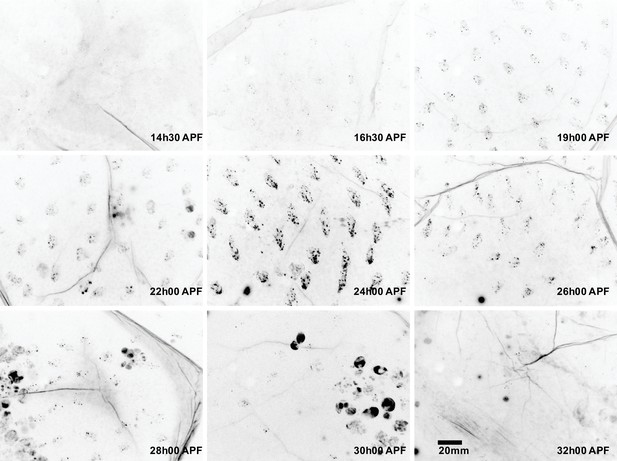
Sca is transiently expressed in the notum.
Images of nota in sca::GFSTF protein-trap flies at different developmental times. Sca::GFSTF protein was transiently detected as multiple foci in sensory organ cells. Scale bar, 20 µm.
Time lapse recording showing the dynamic of protrusions in SOP cells in a control pupa.
The membrane was visualised using a membrane tethered red fluorescent protein (ph-RFP). Each frame was obtained by combining a z-stack (composed of 15 optical sections separated by 1 µm) acquired every 2 min. During the in vivo imaging, the pupae was maintained at 30 °C. Anterior is to the left and view is dorsal. Scale bar represents 20 µm.
Time lapse recording showing the dynamics of protrusions of SOPs in pupa overexpressing the negative form of Rac1 (rac1N17).
The membrane was visualised using a membrane tethered red fluorescent protein (ph-RFP). Each frame was obtained by combining a z-stack (composed of 15 optical sections separated by 1 µm) acquired every 2 min. Anterior is to the left and view is dorsal. Scale bar represents 20 µm.
Time lapse showing the dynamics of Sca::GFSTF (in green) in a SOP protrusion visualised using a membrane tethered RFP (in red).
Each frame was obtained by combining a z-stack (composed of three optical sections separated by 1 µm) acquired every 15 s. Time in min:s. Yellow arrowhead indicates a Sca vesicle traveling along the protrusion.
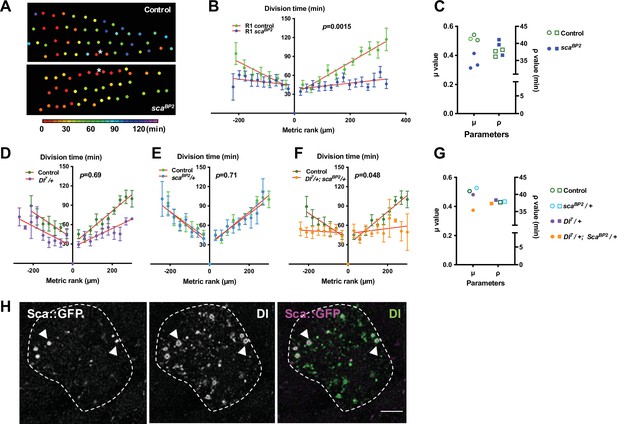
The mitotic wave of SOP cells is controlled by Notch/Delta/Scabrous signals.
(A) Heatmap of a representative heminota in control and scaBP2 homozygous null mutant aligned, anterior to the left. The relative time of SOPs division is colour coded according to the scale. (B) The SOP mitotic wave in row one in control and in scaBP2 homozygous mutant. (C) Dot plot showing the distribution of μ and ρ parameters extracted from the best fits displayed in Figure 1—figure supplement 3D, E. Note that the best fit was obtained in scaBP2 null mutant after reduction of the μ parameter only. (D–F) Genetic interactions between Dl7 and scaBP2. Comparison of the SOP mitotic wave in control and Dl7/+ (D), scaBP2/+ (E) and Dl7/+, scaBP2/+ (F). Mean division time± SEM, n = 9, 8, and 11 nota respectively. (G) Dot plot showing the distribution of μ and ρ parameters extracted from the fits displayed in Figure 1—figure supplement 3D, E. . Note that in simple heterozygous backgrounds, μ and ρ were similar to the control while in Dl7/+, scaBP2/+ double heterozygous background, the best fit was obtained after reduction of the μ parameter only. (H) Confocal image of one SOP in a sca::GFSTF protein-trap fly at the moment of the SOP wave (16 hr APF), immunostained for GFP (left panel) and Dl (middle panel), merged in the right panel (purple and green respectively). Arrowheads show co-localisation of Dl and Sca into vesicle-like structures with Sca::GFSTF as puncta surrounded by Dl staining. SOP soma is delimitated by a dashed line. Scale bar, 5 µm.
-
Figure 3—source data 1
Related to Figure 3 Raw data.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig3-data1-v2.xlsx
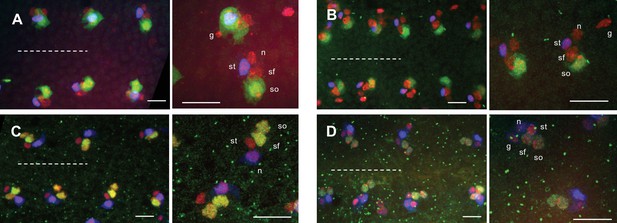
The fate of sensory organ cells is not affected in sca mutant.
Mechanosensory organ cells at two magnifications in control (A and C) and scaBP2 null mutant (B and D) pupae. Cells from sensory organs are identified by the neur> RFP construction (red). Fate of socket (so), shaft (sf), sheath (st), neuron (n) and glial (g) cells were characterised by nuclear size, relative position and specific markers: Su(H) (green in A and B) to identify socket cells. Prospero (blue in A and B) specifics of sheath cells. Pdm (green in C and D) to identify the two external cells, the socket and shaft cells. ELAV (blue in C and D) to identify neurons. Glial cells, when present, show a condensed nucleus. Nota from pupae at 24 hr APF (n = 8). Anterior to the left. The midline is represented by the dashed line. Scale bar = 10 μm.

The amplitude of the mitotic wave is reduced in sca loss of function.
(A) The relative time of SOP cell division in control and scaBP2 homozygous mutant in row 1 (R1), row 2 (R2) and row 3 (R3) is plotted according to their relative position (mean time± SEM, of 16 control nota and nine scaBP2 nota). The position and the absolute time of division of SOP0 in each row was taken as spatial and temporal references. The red lines correspond to standard linear regressions. (B) Efficiency of RNAi-sca. Immunostaining of Sca in control and after overexpression of RNAi-sca using the conditional genetic context pnr> gal4 tub GAl80ts RNAi-sca in the protein trap line sca::GFSTF. Pupae at 16 hr APF. Note that Sca spots (green, arrowheads) in SOP cells, identified by Cut staining (red), are not detected in RNAi-sca conditions. (C) The SOP mitotic wave in control and in flies where sca was specifically downregulated between 12 and 19 hr APF, starting just before and throughout the mitotic wave, using the conditional genetic context pnr> gal4 tub Gal80ts UAS-RNAi-sca in row 1 (R1), row 2, (R2) and row 3 (R3). Mean time± SEM of 10 control nota and 10 RNAi-sca nota. The position and the absolute time of division of the first cell which divided in each row was taken as spatial and temporal reference. Red lines correspond to standard linear regressions.
-
Figure 3—figure supplement 2—source data 1
Related to Figure 3—figure supplement 2 Raw data.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig3-figsupp2-data1-v2.xlsx
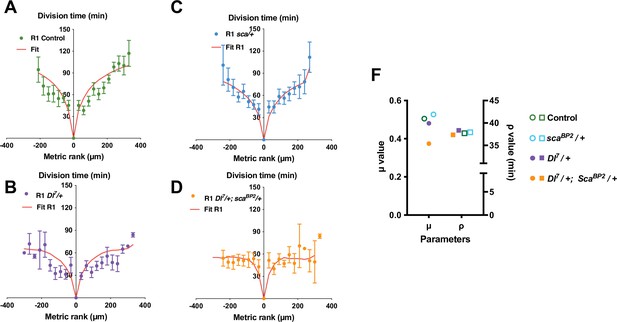
The best fit of the mitotic wave with a reduced amplitude is obtained after preferential lowering the inhibitory parameter μ.
The time of SOP cell division (mean time± SEM) in row 1 (R1), row 2 (R2) and row 3 (R3) is plotted relative to the position and time of division of the first cell to divide in each row. The different conditions were: (A) control, (B) Dl7/+ heterozygous background, (C) scaBP2/+ heterozygous background, (D) Dl7/+, scaBP2/+ double heterozygous background. Red lines depict the best fits obtained with our model. (E) Dot plot showing the distribution μ and ρ parameters extracted from the fits displayed in A-D. Note that in simple heterozygous backgrounds, μ and ρ were similar to the control while in Dl7/+, scaBP2/+ double heterozygous background, the best fit was obtained after reduction of the μ parameter only.
-
Figure 3—figure supplement 3—source data 1
Related to Figure 3—figure supplement 3 Raw data.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig3-figsupp3-data1-v2.xlsx
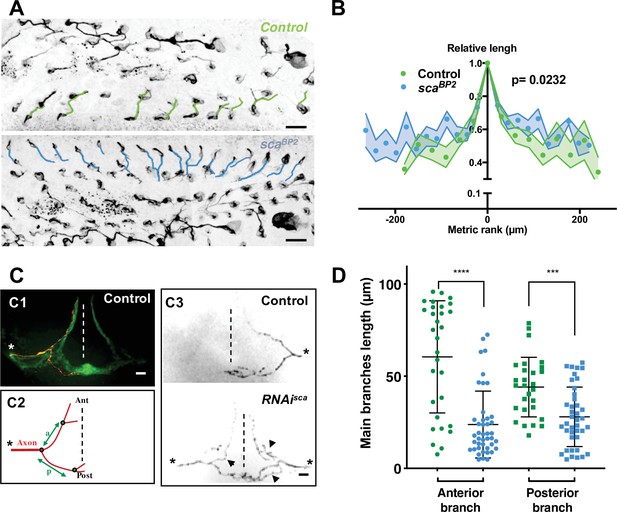
Microchæte axonal projections are affected after SOP mitotic wave disruption.
(A) Axon immunostaining using anti-Futsch/22C10 antibodies in control (w1118) and scaBP2 heminota at 24 hr APF aligned, anterior to the left. Axons in row one were artificially coloured in green for w1118 and blue for scaBP2 homozygous flies. (B) Relative axon length of neurons belonging to sensory organs on row one plotted against relative position (mean length± SEM of n = 18 rows). The neuron with the longest axon on the row was taken as temporal and spatial reference in control (w1118) and scaBP2 homozygous pupae at 24 hr APF. (C) Axon projections of single sensory organ neurons in an adult thoracic ganglion identified with MCFO strategy. The midline is indicated by vertical dotted lines, anterior (Ant) up and posterior (Post) down. The asterisks indicate the point of entry of the sensory nerve into the thoracic ganglion. (C1) A single axon projection (red) counterstained with the neuropile formed by all axon terminals from sensory organs on the central notum (green). (C2) Schematic drawing of an axon projection. The length of the anterior (a) and posterior (p) primary branches were used to quantify the degree of branching of axon terminals. (C3) Representative examples of axon projections into the thoracic ganglion in control fly (top panel) and when the SOP mitotic wave was impaired (UAS-RNAi-sca, tubGal80ts was used to conditionally express Gal4 by maintaining flies at 18 °C and shifted them to 30 °C from 12 hr to 19 hr APF, bottom panel). Arrowheads point to additional branches in RNAi-sca. (D) Lengths of anterior and posterior primary branches in single axon terminals of control (green, n = 30) and in flies where the SOP mitotic wave was impaired (conditional RNAi-sca) (blue, n = 43). Mean ± SEM ****p < 0.0001, ***p = 0.0001, two-tailed unpaired t-test.
-
Figure 4—source data 1
Related to Figure 4 Raw data.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig4-data1-v2.xlsx
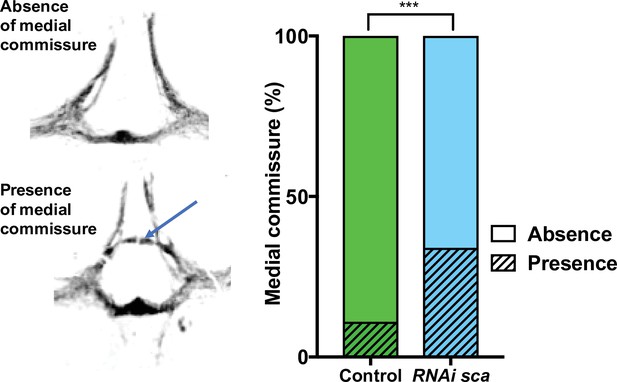
sca downregulation modify the sensory neuropile structure.
On the left, the neuropile formed by all terminal axons from sensory organs on the central notum illustrating the absence (top) and presence (bottom) of the medial commissure (arrow) in thoracic ganglia of adult flies. On the right, quantification of the medial commissure occurrence in the thoracic ganglion of control and after overexpression of RNAi-sca using the conditional genetic context pnr> gal4 tub GAl80ts UAS-RNAi-sca; ***p = 0.0002, Fisher’s exact test, control n = 64, RNAi-sca n = 63.
-
Figure 4—figure supplement 1—source data 1
Related to Figure 4—figure supplement 1 Raw data.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig4-figsupp1-data1-v2.xlsx
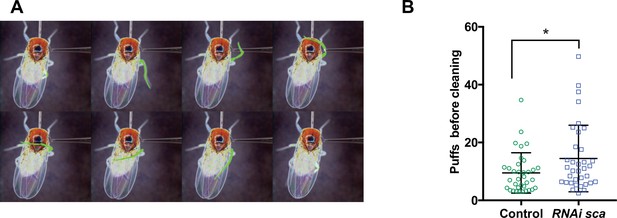
Disruption of the SOP mitotic wave leads to changes in fly behaviour.
(A) Eight frames of a time-lapse recording of a decapitated control fly showing the sequence of movements of the metathoracic leg (artificially coloured in green) upon stimulation of the dorsal bristles. (B) Number of air puffs required to elicit a cleaning reflex in control flies (n = 35) and in flies where the SOP wave was impaired (UAS-RNAi-sca, tubGal80ts was used to conditionally express Gal4 by maintaining flies at 18 °C and shifted them to 30 °C from 12 hr to 19 hr APF) (n = 35). Mean ± SEM. *p = 0.0238; ANOVA.
-
Figure 5—source data 1
Related to Figure 5 Raw data.
- https://cdn.elifesciences.org/articles/75746/elife-75746-fig5-data1-v2.xlsx
Recording of cleaning reflex assay in a decapitated control adult fly.
The leg movements observed were elicited after stimulation by air puffs of sensory organs in the more posterior central part of the notum. Air puffs were materialised by a red dot.
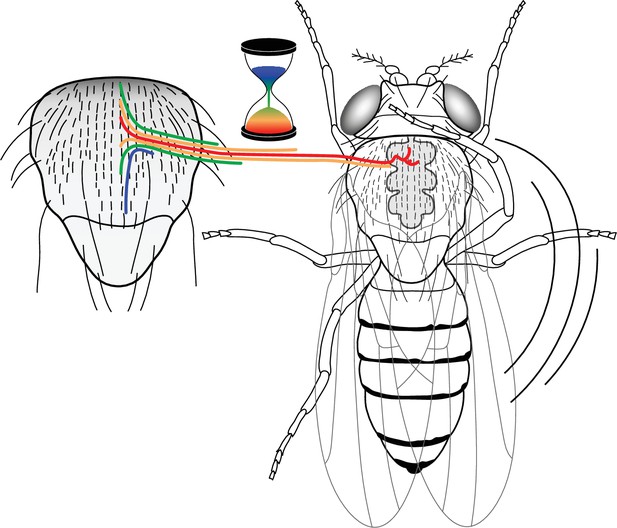
A neural progenitor mitotic wave is required for asynchronous axon outgrowth.
The mitotic wave of precursor cells induces an asynchronous arrival of sensory axons into the thoracic ganglion. This is depicted here by axons coloured according to the developmental time of sensory organs that in turn is controlled by the wave of precursor cell divisions. Red, early-formed; blue, late-formed sensory organs. This asynchrony would be associated with a particular axon connectivity in the thoracic central ganglion required for a normal cleaning behaviour in the fly.
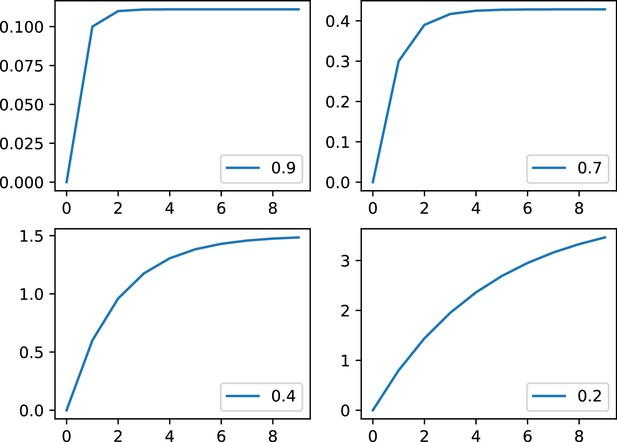
Example of the phenomenological model with and for .
For for low to strong inhibition. Please note that the y scales are different.
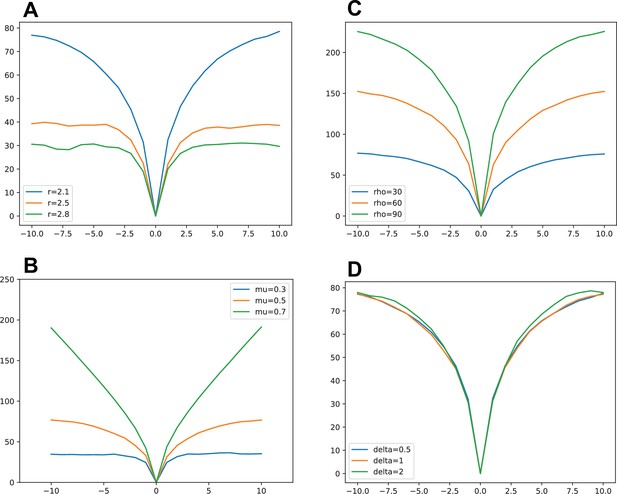
TOP LEFT: Simulation different production rate (other parameters are , , , ).
TOP RIGHT: Simulation with various temporal constant (other parameters are, , ,, ). This adjusts the temporal scale but not the shape of the wave BOTTOM LEFT simulation with various inhibition strength with (other parameters are, , ,, ). Strong inhibition here (0.7) imposes a more linear division time shape whereas low inhibition allows for quicker division with neighboring cells dividing almost simultaneously. BOTTOM RIGHT: simulation with various inhibition time constant with (other parameters are, , ,, ).
-
Appendix 1—figure 2—source code 1
Source code for model description.
- https://cdn.elifesciences.org/articles/75746/elife-75746-app1-fig2-code1-v2.zip
-
Appendix 1—figure 2—source code 2
Source code for minimal model description.
- https://cdn.elifesciences.org/articles/75746/elife-75746-app1-fig2-code2-v2.zip
Videos
Time lapse recording of a control pupa during a 4 hr period beginning at 15 hr after pupal formation.
The wave of SOP division is highlight in red in the row 1. Each frame was obtained by combining a z-stack (composed of 10 optical sections separated by 2 µm) acquired every 3 min. During the in vivo imaging, the pupae was maintained at 30 °C. Anterior is to the left and view is dorsal. Scale bar represents 50 µm.
Time lapse recording of the SOP mitotic wave in control pupa (top panel) and in scaBP2 mutant pupa (bottom panel).
For comparison, two hemi-nota are aligned. The observed difference in SOP nuclei size in the two movies is due to the lineage markers used (nuclear GFP and histone-RFP). The waves of SOP divisions are highlighted in red in the rows 1. Anterior is to the left and view is dorsal. Scale bar represents 50 µm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-Futsch (clone 22C10, mouse monoclonal) | DSHB | AB_528403 | IF (1:200) |
| Antibody | Anti-HA (clone 3F10, rabbit polyclonal) | Roche | AB_231 4,622 | IF (1:250) |
| Antibody | Anti-Flag (M2, mouse monoclonal) | Sigma | AB_259529 | IF (1:250) |
| Antibody | Anti-V5 (E10/V4RR), DyLight650 (mouse monoclonal) | ThermoFisher | AB_2537642 | IF (1:100) |
| Antibody | Anti-cut (mouse monoclonal) | DSHB | AB_528186 | IF (1:500) |
| Antibody | Anti-ELAV (rat monoclonal) | DSHB | AB_7E8A10 | IF (1:500) |
| Antibody | Anti-Su(H) (rat polyclonal) | Gift F. Schweisguth | IF (1:500) | |
| Antibody | Anti-Prospero (mouse monoclonal) | DSHB | AB_528440 | IF (1:500) |
| Antibody | Anti-pdm (rabbit polyclonal) | Gift T. Preat | IF (1:2000) | |
| Antibody | Anti-Dl (mouse monoclonal) | DSHB | AB_528194 | IF (1: 500) |
| Antibody | Anti-GFP (B-2, rabbit polyclonal) | Santa Cruz | sc-9996 | IF (1: 500) |
| Antibody | Goat anti-rabbit AlexaFluor 488 | Molecular Probes | AB_2576217 | IF (1: 1000) |
| Antibody | Goat anti-rat AlexaFluor 488 | Molecular Probes | AB_2534074 | IF (1: 1000) |
| Antibody | Goat anti-mouse AlexaFluor 488 | Molecular Probes | AB_2534088 | IF (1: 1000) |
| Antibody | Goat anti-rat AlexaFluor 568 | Molecular Probes | AB_2534121 | IF (1: 1000) |
| Antibody | Goat anti-mouse AlexaFluor 568 | Molecular Probes | AB_144696 | IF (1: 1000) |
| genetic reagent (D. melanogaster) | w1118 | Bloomington Stock Center | BDSC_3605 | |
| genetic reagent (D. melanogaster) | neurD> GFP | F.Schweisguth | ||
| genetic reagent (D. melanogaster) | scaBP2 | Bloomington Stock Center | BDSC_7320 | |
| genetic reagent (D. melanogaster) | UAS sca-RNAi | Vienna Drosophila Resource Center | VDRC#44,527 | |
| genetic reagent (D. melanogaster) | sca::GFSTF | Bloomington Stock Center | BDSC_64443 | |
| genetic reagent (D. melanogaster) | neur> ph (PLCd)::RFP | F.Schweisguth | ||
| genetic reagent (D. melanogaster) | UAS-MCFO-1 | Bloomington Stock Center | BDSC_64085 | |
| genetic reagent (D. melanogaster) | UAS-rac1N17 | Bloomington Stock Center | BDSC_6292 | |
| genetic reagent (D. melanogaster) | Dl7 | Bloomington Stock Center | BDSC_485 | |
| genetic reagent (D. melanogaster) | UAS-ph::GFP | Y.Bellaiche | ||
| genetic reagent (D. melanogaster) | neurp72> Gal4 | Y.Bellaiche |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/75746/elife-75746-transrepform1-v2.docx
-
Appendix 1—figure 2—source code 1
Source code for model description.
- https://cdn.elifesciences.org/articles/75746/elife-75746-app1-fig2-code1-v2.zip
-
Appendix 1—figure 2—source code 2
Source code for minimal model description.
- https://cdn.elifesciences.org/articles/75746/elife-75746-app1-fig2-code2-v2.zip





