A Notch-dependent transcriptional mechanism controls expression of temporal patterning factors in Drosophila medulla
Figures
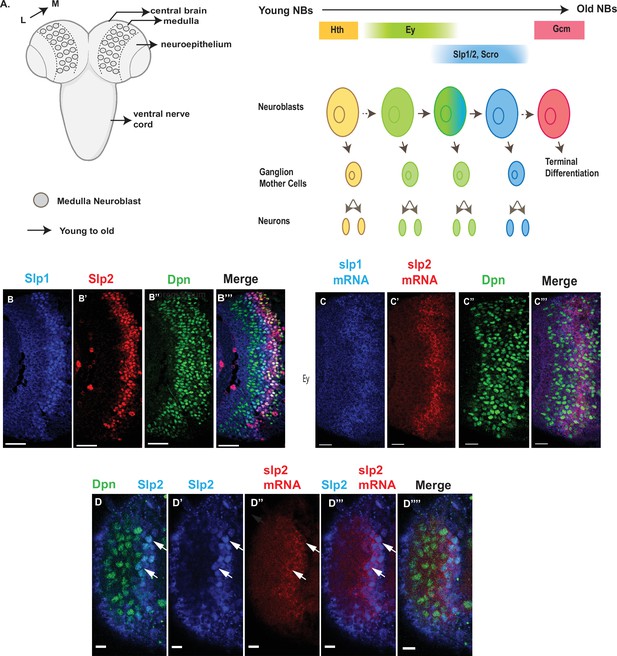
Expression of slp1 and slp2 genes in medulla neuroblasts is controlled at transcription.
(A) Schematic of Drosophila brain at the third instar larval stage highlighting the location of the optic lobe medulla. Medulla NBs (shown as circles), located on the surface of the optic lobe, are transformed from neuroepithelial cells (NE) as a neurogenesis wave spreads in a medial to lateral direction. As a result, medulla NBs of different ages are aligned on the lateral (L) to medial (M) spatial axis. Schematic to the right shows part of the temporal patterning program of medulla NBs with a focus on the Ey to Slp1/2 transition. NBs undergo asymmetric divisions to self-renew and to produce intermediate progenitors called Ganglion Mother Cells that each divide once to produce two neurons. In the Ey stage, NBs undergo a few divisions while gradually activating Slp expression resulting in a significant overlap between Ey and Slp expression in NBs. After Slp level reaches a certain threshold, Ey expression is down-regulated and eventually deactivated completely. This process repeats as subsequent temporal patterning factors are activated and earlier factors down-regulated over several neuroblast division cycles. After several temporal stages that are not shown here indicated by the dashed arrow, neuroblasts express TTF Gcm and exit the cell cycle. The larval brain graphic was created using BioRender. (B-B’’’) Expression patterns of endogenous Slp1 and Slp2 proteins in medulla neuroblasts identified by their expression of Deadpan (Dpn), a neuroblast marker. (C-C’’’) Expression patterns of Slp1 and Slp2 mRNAs in Dpn expressing medulla neuroblasts closely parallels the corresponding protein expression patterns. (D-D’’’’) Detection of slp2 mRNA and Slp2 protein in the same brain shows spatial co-localization of slp2 transcripts and Slp2 protein in the same neuroblasts. Two distinct neuroblasts indicated by arrowheads shown for emphasis. These cells express the neuroblast marker Dpn (D, D’’’’) and Slp2 in the nucleus and the slp2 mRNA is localized to the cytoplasm. Scale bar for panels (B-B’’’) and (C-C’’’): 20 µm. Scale bar for (D-D’’’’) 6 µm.
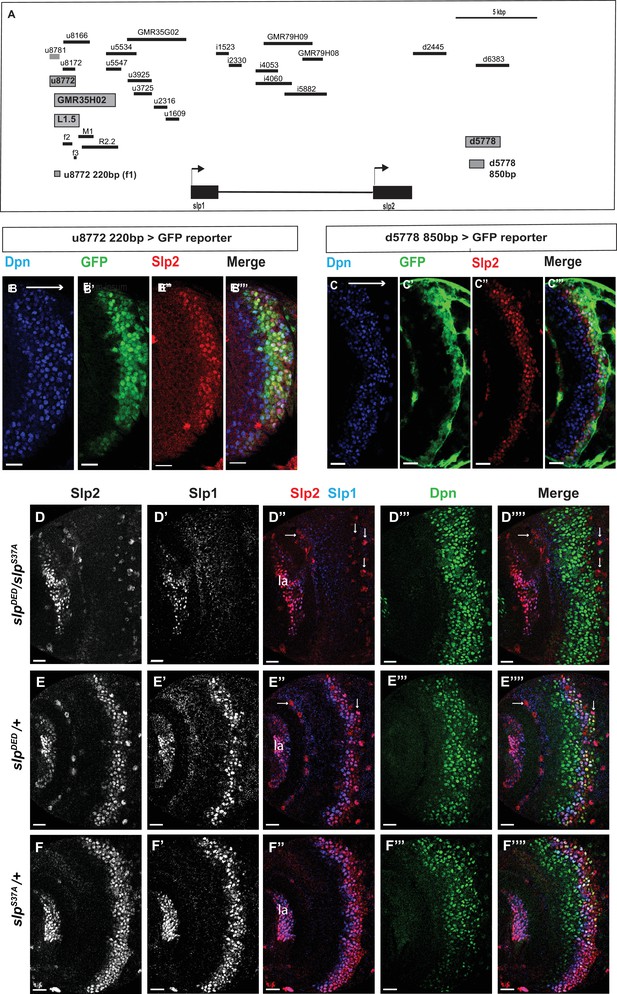
Transcription of slp1 and slp2 in medulla neuroblasts is regulated by two enhancers of lengths 220 bp and 850 bp, respectively.
(A) A schematic of the enhancer screening. slp1 and slp2 genes and transcriptional enhancers identified as regulating their expression are shown as black and grey boxes, respectively. Positions of slp1 coding locus (2 L:3,825,675.3,827,099 [+]), slp2 coding locus (2 L:3,836,840.3,839,185 [+]), GMR35H02 (2 L:3,817,265.3,821,014) and the REDfly enhancers u8772 (2 L:3,816,967.3,818,532) and d5778 (2 L:3,842,530.3,844,660) are shown relative to one another. A 220 bp enhancer located within the genomic segment covered by GMR35H02 and an 850 base pair enhancer element within the REDFly enhancer d5778 were identified as potential cis-regulatory elements of slp1 and slp2 genes. The distance between the start of GMR35H02 and the end of d5778 is around 27.394 kbp. Other fragments that were screened /cloned are shown as black bars with names on top. (B-B’’’) A 220 bp enhancer element located within the REDfly enhancer u8772 activates GFP expression in medulla neuroblasts in the same pattern as endogenous Slp1 and Slp2 in reporter assays. (C-C’’’) Reporter GFP expression driven by an 850 bp enhancer segment located within the REDfly enhancer d5778 also closely coincides with the expressions of endogenous Slp1 and Slp2, although it is initiated at a slightly later temporal stage than the 220 bp enhancer. (D-D’’’’) A CRISPR-Cas9 edited chromosome 2 with both enhancers deleted (indicated as SlpDED) when placed over the SlpS37A chromosome results in loss of Slp1 and Slp2 expression in medulla neuroblasts in affected flies (n=11). The effect is confined to neuroblasts, as Slp2 expression is retained in both lamina (la) and in surface glial cells (arrows), and Slp1 is also seen in laminar cells. This also confirms that coding sequences of slp1 and slp2 genes are unaffected by the CRISPR-Cas9 editing procedure. Control experiments where SlpDED was placed against a wild-type chromosome 2 (indicated by ‘+’) (E-E’’’’) (n=9) and where the SlpS37A chromosome was placed against a wild-type chromosome 2 (F-F’’’’) (n=11) show normal expression of Slp1 and Slp2 in medulla neuroblasts. Scale bars 20 µm.
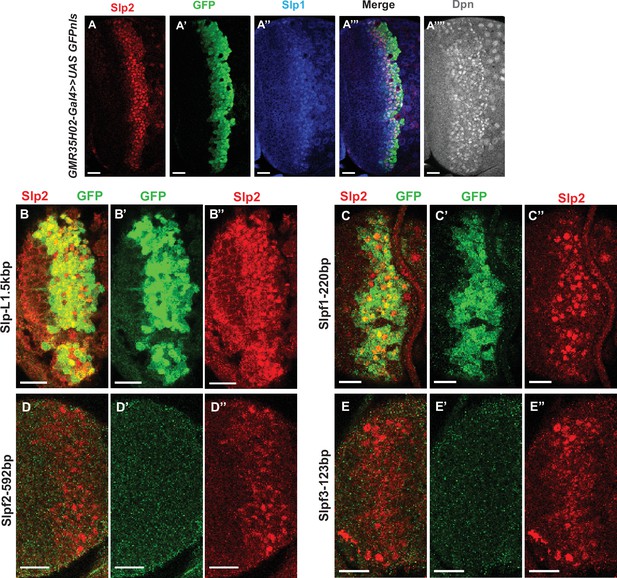
The identification of the 220 bp enhancer by promoter bashing.
(A-A”’’) UAS-GFP driven by GMR35H02-Gal4 is initiated at the same time as endogenous Slp1 and Slp2 in neuroblasts marked by Dpn. (B-D’’) Expression of GFP reporters driven by SlpL1.5 (B-B’’) and its sub-segments Slpf1-220bp(C-C’’), Slpf2-592bp (D-D’’), and Slpf3-123bp (E-E’’). Of the three sub-segments of SlpL1.5, only the Slpf1 fragment (220 bp in size) shows enhancer activity. Scale bars 20 µm.
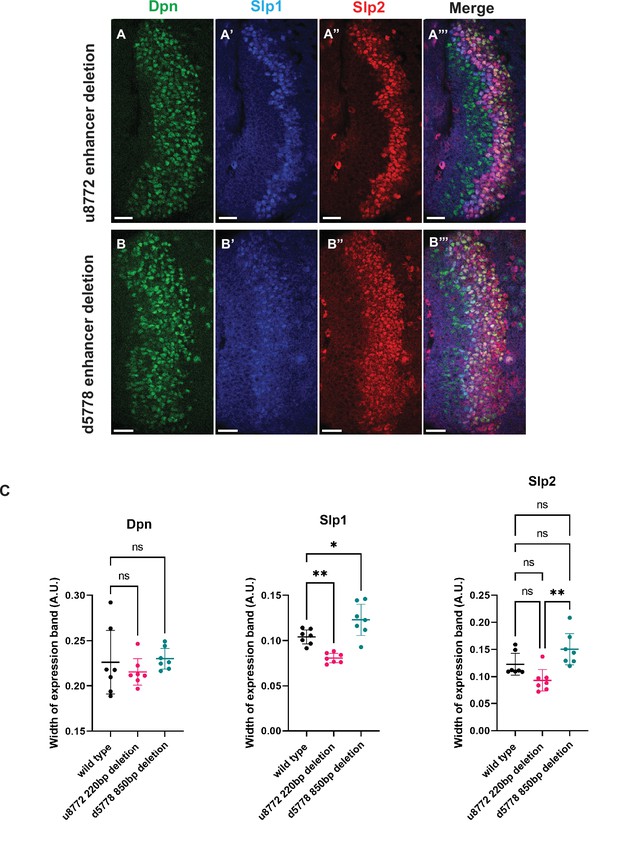
The two enhancers act partially redundantly.
CRISPR-Cas9 deletions of the u8772 enhancer (A-A’’’) and the d5778 enhancer (B-B’’’) individually do not abolish Slp1/2 expression suggesting possible redundance. Scale bars: 20 µm. (C) Middle panel: comparison of widths of expression domains normalized to brain size for Slp1 in wild type and enhancer deleted brains. p Value of ordinary one way ANOVA is <0.0001 and is statistically significant because p<0.05. Adjusted p values from Dunnett’s multiple comparison test between wild type and the u8772 220 bp enhancer deletion is 0.0041, between wild type and d5778 850 bp enhancer deletion is 0.0187. Both are statistically significant since p<0.05. Data from seven brains of each genotype are quantified (n=7). Right panel: expression domain width normalized to brain size for Slp2 compared between wild type and enhancer deleted brains shows no statistically significant differences relative to wild type (Adjusted p values from Dunnett’s multiple comparison test relative to wild type are as follows: for u8772 220 bp enhancer deletion p=0.0730, for d5778 850 bp enhancer deletion p=0.0965. Neither are statistically significant since p>0.05). The difference between u8772 deletion and d5778 deletion is significant (p value from ordinary one way ANOVA = 0.0019, statistically significant because p<0.05). Data from seven brains of each genotype are quantified (n=7). Left panel: expression domain width normalized to brain size for Dpn also shows no significant differences between wild type and enhancer deletion mutants (p value from ordinary one-way ANOVA is 0.5277 and is >0.05, hence not statistically significant. Adjusted p values from Dunnett’s multiple comparisons test between enhancer deletion mutants and the wild type are as follows: for the u8772 220 bp enhancer deletion p=0.6310, for the d5778 850 bp enhancer deletion p=0.9414. Neither are statistically significant being >0.05). Data from seven brains of each genotype are quantified (n=7). This shows that deletion of the enhancers doesn’t affect Dpn expression.
-
Figure 2—figure supplement 2—source data 1
Quantification and comparison of Dpn expression in wild type control, u8772 enhancer deletion, and d5778 enhancer deletion brains.
- https://cdn.elifesciences.org/articles/75879/elife-75879-fig2-figsupp2-data1-v1.xlsx
-
Figure 2—figure supplement 2—source data 2
Quantification and comparison of Slp1 expression in wild type control, u8772 enhancer deletion, and d5778 enhancer deletion brains.
- https://cdn.elifesciences.org/articles/75879/elife-75879-fig2-figsupp2-data2-v1.xlsx
-
Figure 2—figure supplement 2—source data 3
Quantification and comparison of Slp2 expression in wild type control, u8772 enhancer deletion, and d5778 enhancer deletion brains.
- https://cdn.elifesciences.org/articles/75879/elife-75879-fig2-figsupp2-data3-v1.xlsx
-
Figure 2—figure supplement 2—source data 4
Expression level quantification of Dpn, Slp1 and Slp2 in wild type control, u8772 enhancer deletion, and d5778 enhancer deletion brains.
- https://cdn.elifesciences.org/articles/75879/elife-75879-fig2-figsupp2-data4-v1.xlsx
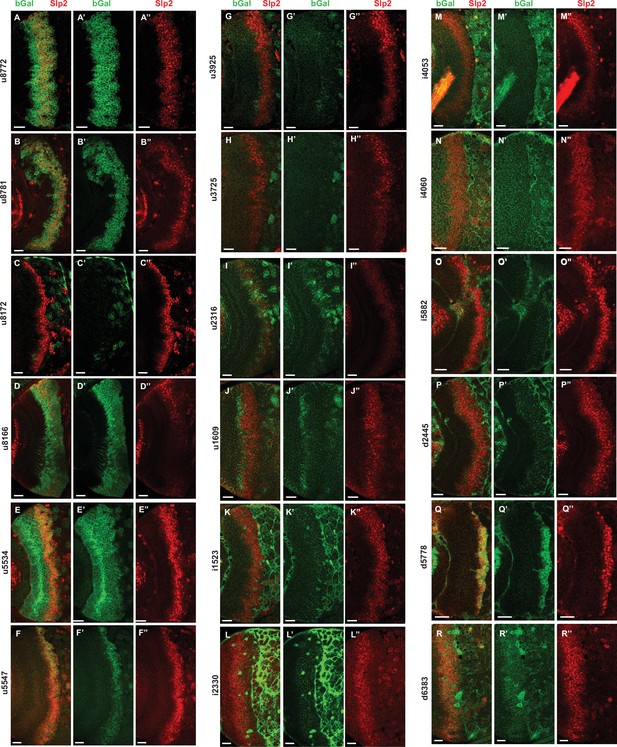
Screen of REDfly enhancers identifies d5778 as an enhancer of slp1/2 transcription in medulla neuroblasts.
(A-R’’) Results of the screen showing lacZ reporter expressions driven by REDfly enhancers. Of the reporters tested, u8772 (A-A’’) and u8781 (B-B’’) recapitulated the expression patterns driven by the previously identified 220 bp slp enhancer, thereby supporting our findings. Both u8772 and u8781 contain within them the 220 bp enhancer element. Additionally, patterns of lacZ expression driven by u8166 (D-D’’) and u5547 (F-F’’) are also interesting in that they somewhat overlap with the expression of endogenous Slp2. However, the u5547 enhancer was contained within the GMR35H02 sequence, and enhancer bashing experiments encoding this part of GMR35H02 did not show reporter expression. The u8166 line drove reporter expression primarily in neurons than in neuroblasts. For these reasons, the u5547 and u8166 enhancers were not considered for further analysis. However, the screen of REDfly enhancers identified d5778 as a potential enhancer of slp1 and slp2 transcription since it drove reporter expression in neuroblasts in a pattern overlapping endogenous Slp2 (Q-Q’’). Scale bars 20 µm.
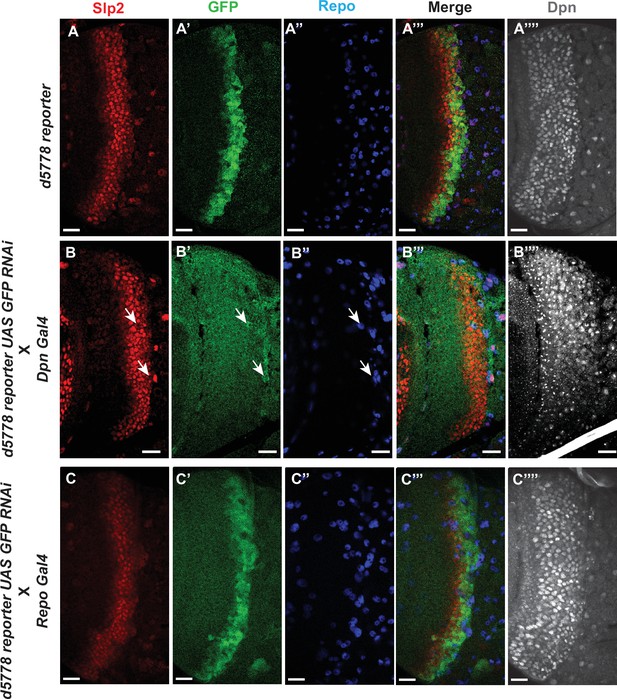
The d5778 850 bp enhancer drives reporter expression predominantly in neuroblasts.
(A-A’’’) The d5778 850 bp enhancer is activated slightly after the initiation of Slp2 expression. The wild type d5778 850 bp reporter drives GFP expression (A’) predominantly in neuroblasts identifiable by their expression of the neuroblast marker Dpn (A’’’’). Slp2 is expressed in some surface glial cells expressing the marker Repo (A, A’’, A’’’). (B-B’’’’) Neuroblast-specific suppression of 850 bp reporter GFP expression by GFP-RNAi results in no visible GFP signals from neuroblasts. (C-C’’’’) Glial specific suppression of 850 bp reporter GFP by GFP-RNAi preserves GFP expression in neuroblasts and establishes that the d5778 enhancer is indeed prominently active in medulla neuroblasts. Scale bars: 20 µm.
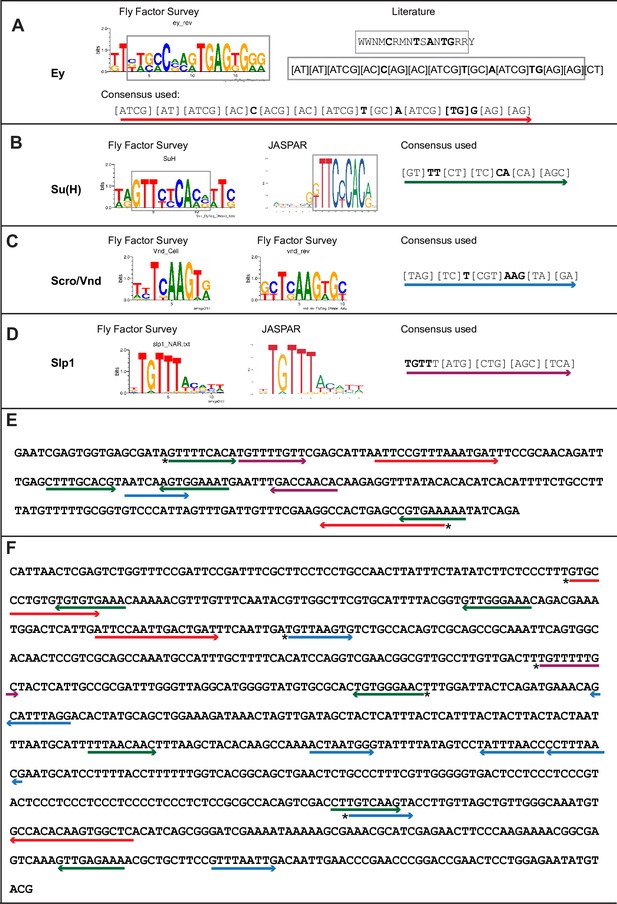
Predicted binding sites for Ey, Su(H), Scro, and Slp1 in the two enhancers.
(A–D) Position weight matrix of for Ey (A), Su(H) (B), Scro /Vnd (C) and Slp1 (D) from Fly Factor Survey, JASPAR and literature are considered together to make a consensus sequence. Arrows of different colors are underlying these consensus binding sites. Critical invariant bases are in bold. Grey boxes indicate the corresponding positions of the Position weight matrix from two sources. (E,F) Predicted binding sites for Ey, Su(H), Scro, and Slp1 in the 220 bp enhancer (E) and 850 bp enhancer (F). Arrows are color coded as in (A–D). Right-pointing arrow indicates the binding site is on direct strand, while left-pointing arrow indicated the binding site is on reverse strand. Star* indicates that the binding site is also predicted by the FIMO pipeline.
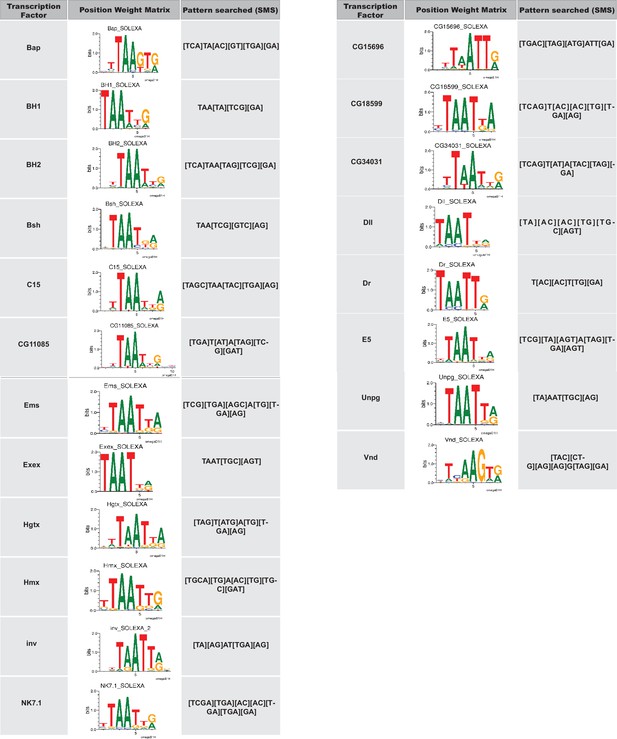
NK-2 family transcription factors with known binding consensus motifs were used to identify potential Scro binding sites in the d5778 850 bp enhancer.
The transcription factor is listed in the column to the left. Its position weight matrix, either from JASPAR or Fly Factor Survey, is shown in the column in the middle. The consensus binding motif of the transcription factor inferred from the position weight matrix and used as input for the Sequence Manipulation Suite DNA Pattern Find program is listed in the column on the right.
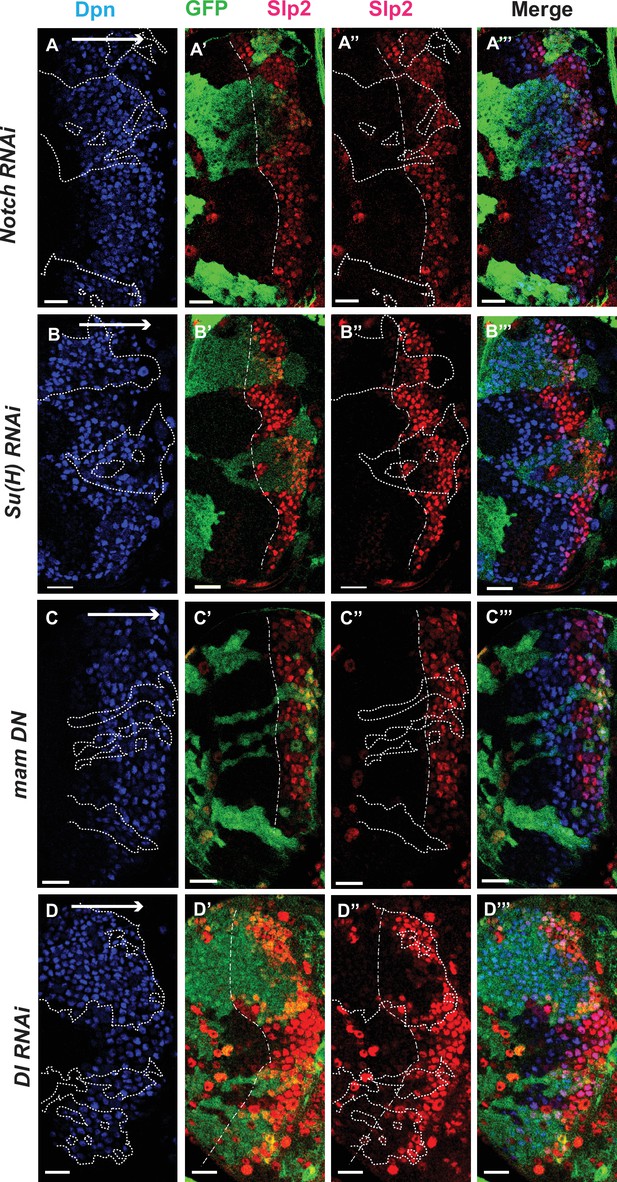
The Notch pathway regulates expression of slp genes in medulla neuroblasts.
White arrow to the right indicates the neuroblast age from young to old. All clones are generated with ay-Gal4 UAS-GFP and clone boundaries are shown in dotted lines. The dash-dotted lines indicate where Slp2 should be turned on, and the curvature is due to the curvature of the Dpn initiation. (A) RNAi knockdown of Notch in GFP marked clones leads to a delay in expression of Slp2 compared to contemporaneous wild type neuroblasts outside the GFP marked clones. While in wild-type brains Slp2 expression is seen in the 4th-5th neuroblast from the lateral edge of the medulla, inside N-RNAi clones Slp2 expression is first noted in the 10th –15th neuroblast (n=12 clones). (B) In Su(H) RNAi clones marked by GFP, Slp2 expression begins at the 7th-10th neuroblast (n=11 clones). (C) In clones expressing the dominant negative mutant variant of Mastermind, Slp2 expression is also delayed and start at the 7th-10th neuroblasts (n=6 clones) (D) Knockdown of Dl, a Notch ligand, caused a similar delay in Slp2 expression, with Slp2 expression being initiated at the 6th to 10th NB (n=5 clones). Clonal data from at least three brains were observed for each mutant to infer delay in Slp2 expression initiation. Scale bar 20 µm.
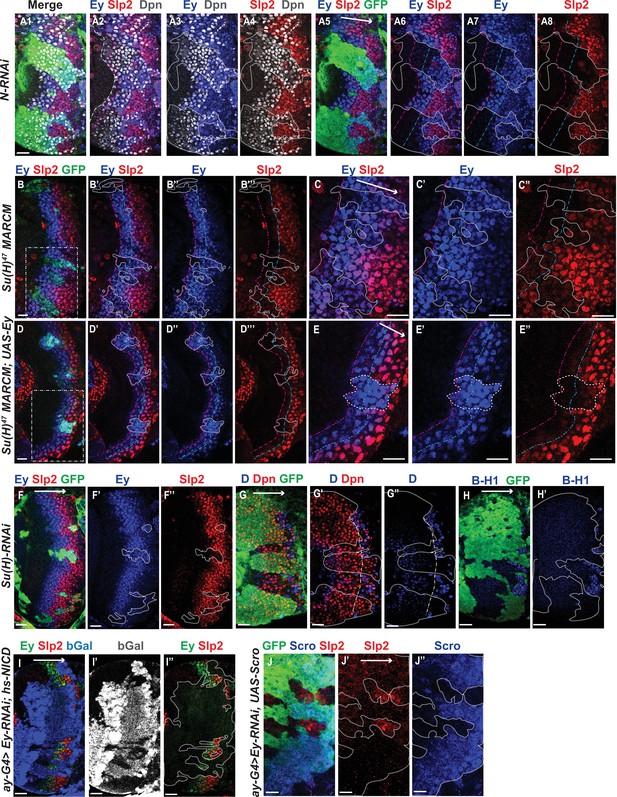
Genetic interactions between Notch, Ey and Scro in the regulation of Slp expression.
In all panels, dotted lines outline clone margins. Dash-dotted lines mark the expected transition front for Dpn or TTFs (white for Dpn, pink for Ey, cyan for Slp2, and yellow for D). The curvature of the dash-dotted lines is determined by the curvature of Dpn or the previous TTF. (A1–A8) In N-RNAi clones marked by GFP in green, Ey is turned on in the 5th-8th NB (6.50±1.05, n=6), and the transition to Slp stage occurs in the 6th –9th Ey +NB (7.67±1.21, n=6). In control regions (not marked by GFP), Ey is turned on in the 2nd –3rd NB (2.50±0.55, n=6), and the transition to Slp stage occurs on the 3rd –4th Ey +NB (3.83±0.41, n=6). The delay of Ey expression and the further delay of Slp transition are significant by t-test (p=4.8 × 10–5 and p=0.0003, respectively). (B-B’’’) In Su(H) mutant clones marked by GFP, Ey expression is delayed and the transition to Slp stage is further delayed: Slp2 is still barely detectable or very weak at the 6th –8th Ey +NBs (6.80±0.84, n=5). In control regions, the transition to Slp2 stage occurs in the 4th Ey +NBs (n=5). The further delay of Ey to Slp transition is significant by t-test (p=0.002). (C-C’’) Magnified view of the rectangle area containing three clones shown in B. (D-D’’’) In Su(H) mutant clones over-expressing Ey, the transition to Slp2 stage is still delayed to the 6th-7th Ey +NBs (6.40±0.55, n=5). In control regions, the transition occurs in the 3rd-4th Ey +NBs (3.80±0.45, n=5), The further delay of Ey to Slp transition is significant by t-test (p=4.50 × 10–5). (E-E”) Magnified view of the rectangle area containing one clone shown in D. (F-H’) The expression of Ey and Slp2 (F-F’’), Dichaete and Dpn (G-G’’), and BarH-1 (H,H’) in Su(H)-RNAi clones marked by GFP. (I-I’’) Supplying active NICD using hsFlp; dpn >FRT-STOP-FRT-NICD (heat shocked for 12 min 3 days before dissection) is not sufficient to activate Slp2 expression in Ey-RNAi clones marked by bGal. (J-J’’) Supplying Scro is not sufficient to activate Slp2 expression in Ey-RNAi clones marked by GFP. Scale bars 20 µm.
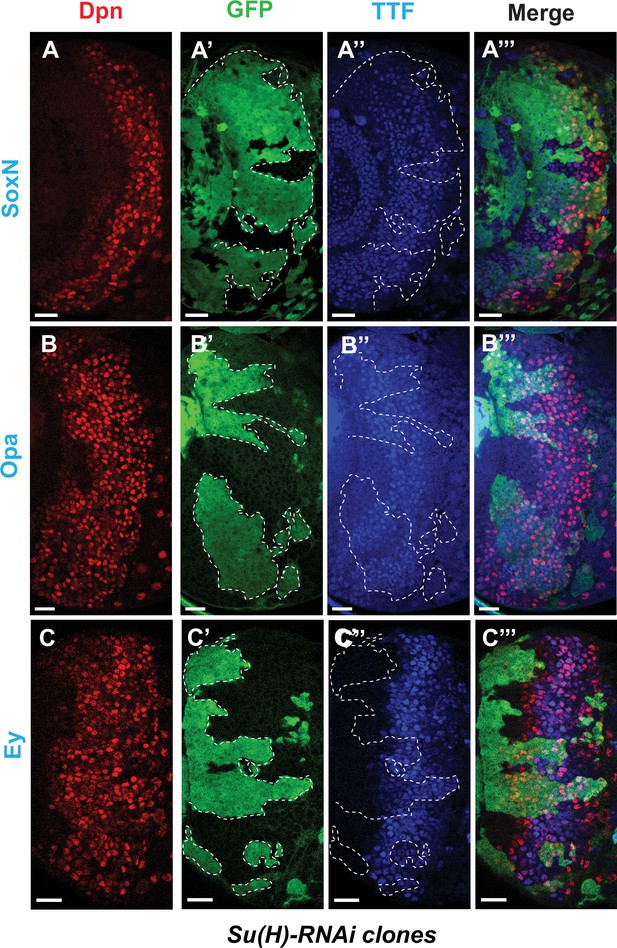
Notch signaling is also involved in early temporal transitions.
(A-A’’’) SoxN expression is not significantly affected in Su(H)RNAi clones marked by GFP. (B-B’’’) The TTF Opa is expressed at two different ‘stripes’. The earlier expression stripe is expanded in width within Su(H) RNAi clones, while Opa expression in the second stripe in older neuroblasts is delayed. (C-C’’’) Expression of Ey is delayed in Su(H) RNAi clones. Representative images of at least four brains are shown. Scale bars 20 µm.
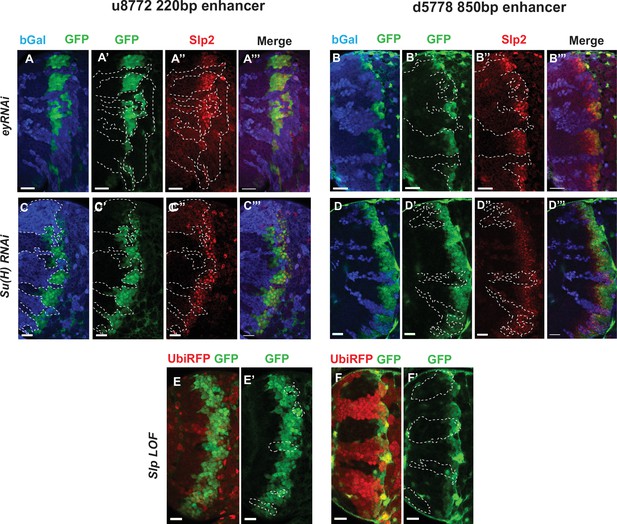
Eyeless and the Notch pathway act through the 220 bp and 850 bp enhancers to regulate expression of slp genes.
(Panels A-A’’’ through D-D’’’) RNAi clones in which Ey or Su(H) is knocked down are marked by Beta-galactosidase (bGal) in blue, while regions unmarked by bGal are populated by wild type neuroblasts. (A-A’’’) The expression of a GFP reporter driven by the 220 bp enhancer and endogenous Slp2 in ey RNAi clones and wild type neuroblasts. Expressions of both the GFP reporter and endogenous Slp2 are similarly abolished in ey knockdown clones (representative data shown, n=5 brains). (B-B’’’) The expression of a GFP reporter driven by the d5778 850 bp enhancer and endogenous Slp2 in ey RNAi knockdown clones and in wild type neuroblasts. Knockdown of Ey leads to loss of both the GFP reporter expression and the endogenous Slp2 protein (representative data shown, n=15 brains). (C-C’’’) The expression of the GFP reporter driven by 220 bp enhancer and endogenous Slp2 in Su(H) RNAi clones and in wild type neuroblasts (representative data shown, n=11 brains). (D-D’’’) The expression of the GFP reporter driven by 850 bp enhancer and of endogenous Slp2 in Su(H) RNAi clones and in wild-type neuroblasts (representative data shown, n=14 brains). In case of both enhancers the expression of the GFP reporter is lost or delayed. (E, E’) The expression of the 220 bp GFP reporter in slp1 and slp2 loss-of-function mitotic clones (‘dark’ regions) and in wild-type neuroblasts (marked by RFP) (representative data shown, n=24 brains). (F,F’) Expression of the 850 bp enhancer GFP reporter in slp LOF clones and wild-type neuroblasts similar to (E,E’) (representative data shown, n=16 brains). Slp1 binding sites were identified in both enhancer elements which indicate that Slp1 may auto-regulate its expression. However, GFP reporter expression driven by these two enhancers is not significantly affected within slp LOF neuroblasts, suggesting that Slp1/2 are not required to initiate their own expression in neuroblasts. Representative images are provided. Scale bar 20 µm.
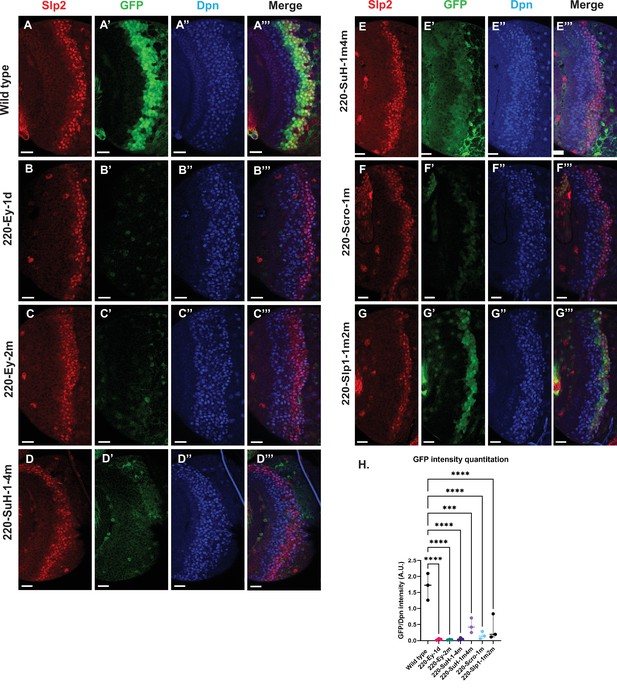
Mutations of predicted binding sites for transcription factors Ey, Su(H) and Scro in the u8772 220 bp enhancer establish their requirement for activating slp1/slp2 mRNA transcription in medulla neuroblasts.
Expression of GFP reporter driven by the wild-type 220 bp enhancer (A-A’’’) was compared against GFP reporters driven by mutated versions of the 220 bp enhancer in which predicted binding sites for specific transcription factors had been mutated: (B-B’’’) with a predicted site for Ey binding (220-Ey-site1 in Table 2) deleted, (C-C’’’) with a different predicted site for Ey binding (site 220-Ey-Site2 in Table 2) mutated, (D-D’’’) with four predicted Su(H) binding sites mutated (sites 220-SuH-Site1 to 4), (E-E’’’) with two Su(H) binding sites mutated (sites 220-SuH-Site1 and 220-SuH-Site4 in Table 2), (F-F’’’) with a predicted site for Scro binding mutated (220-Scro-Site1 in Table 2), (G-G’’’) with two predicted Slp1 binding sites mutated (sites 220-Slp1-Site1 and 220-Slp1-Site2). Loss of binding sites for Ey, Su(H) and Scro in the 220 bp enhancer led to a loss or reduction of GFP reporter expression. Scale bar: 20 µm. (H) Intensity comparisons of mutated and wild type GFP reporters of the u8772 220 bp enhancer. The decrease in intensity of GFP expression for Ey, Scro, Su(H) and Slp1 binding site mutated reporters relative to wild type are all statistically significant. Ordinary one way ANOVA was performed to determine statistical significance between the wild type and the mutated enhancer variants. The differences in GFP expressions were statistically significant for all enhancer variants relative to the wild-type enhancer. Adjusted p values from Dunnett’s multiple comparisons test between wild type and mutants are as follows: for both Ey site mutants, the 220-Scro-1m mutant, and the 220-Slp-1m2m, p values were<0.0001. For the 220-SuH-1–4 m, p value was also <0.0001, but for the 220-SuH-1m4m the p value was 0.0001. Three different sets of observations were used for analyses (n=3). The graph shows the distribution of individual observations about the median and within 95% confidence intervals.
-
Figure 7—source data 1
Quantification and comparison of reporter expression level driven by wild type and mutant forms of u8772 220bp enhancer.
- https://cdn.elifesciences.org/articles/75879/elife-75879-fig7-data1-v1.xlsx
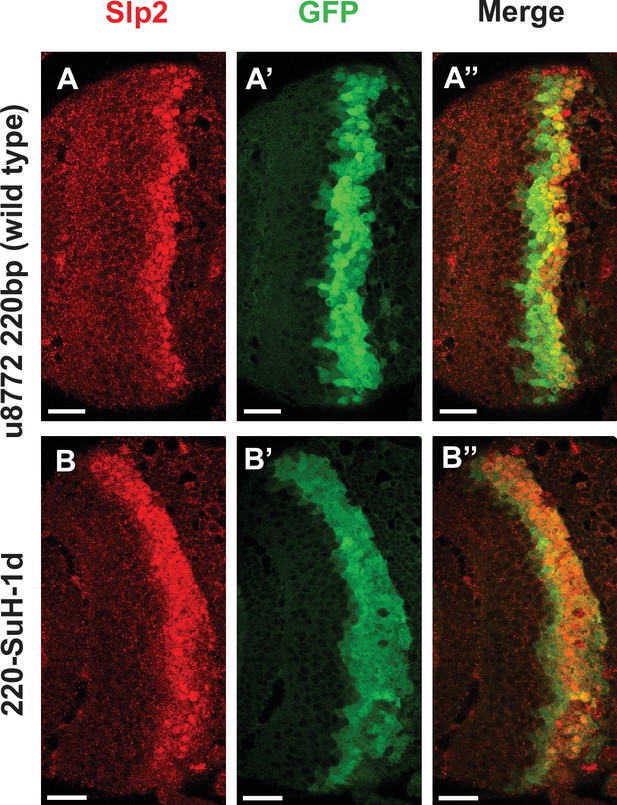
Deletion of one Su(H) site identified by FIMO in the u8772 220 bp enhancer GFP reporter does not abolish GFP expression.
Deletion of the 220-Su(H)-site1 in the u8772 220 bp enhancer (B-B’’) did not change the intensity of GFP reporter expression compared to wild-type u8772 220 bp GFP reporter (A-A’’).
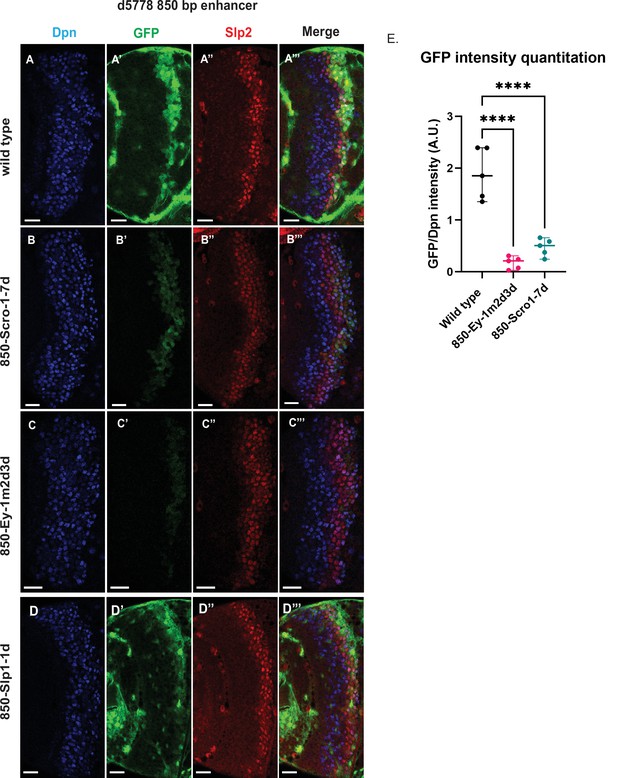
Mutations of putative binding sites for Ey and Scro in the d5778 850 bp enhancer show their requirement for activating Slp1/Slp2 gene expressions in medulla neuroblasts.
A parallel comparison was made for GFP reporter expression driven by the wild-type 850 bp enhancer (A-A’’’) to the GFP reporter expression driven by versions of the 850 bp enhancer with binding sites for specific transcription factors Scro, Ey, and Slp1 mutated (B-B’’’, C-C’’’, and D-D’’’ respectively). For the Ey site mutant 850-Ey-1m2d3d shown in (B-B’’’) three potential binding sites for Ey were mutated (850-Ey-Site1 in Table 2 was mutated as indicated, 850-Ey-Site2 and 850-Ey-Site3 were deleted). For the Scro sites mutant 850-Scro-1-7d, seven of the potential Scro binding sites (850-Scro-Site1 through 850-Scro-Site7 in Table 2) were deleted. Loss of binding sites for Ey obliterated GFP reporter expression in neuroblasts, while loss of potential Scro binding sites resulted in a noticeable reduction of GFP expression from the d5778 850 bp reporter. (E) Quantitation of d5778 850 bp reporter intensities of the wild type and the factor binding site mutated variants shows that the reduction of GFP intensity compared to wild type for Ey binding site and Scro binding site mutated reporters is statistically significant. Scale bars: 20 µm. Ordinary one way ANOVA was performed to compare differences in GFP expression between the wild type d5778 850 bp enhancer and the transcription factor binding site mutants. Adjusted p values from Dunnett’s multiple comparisons test between the wild type d5778 850 bp enhancer and both the Ey and Scro sites mutants were less than 0.0001. Five different sets of observations were used for analyses (n=5). The graph shows the distribution of individual observations about the median and within 95% confidence intervals.
-
Figure 8—source data 1
Quantification and comparison of reporter expression level driven by wild type and mutant forms of d5778 850bp enhancer.
- https://cdn.elifesciences.org/articles/75879/elife-75879-fig8-data1-v1.xlsx
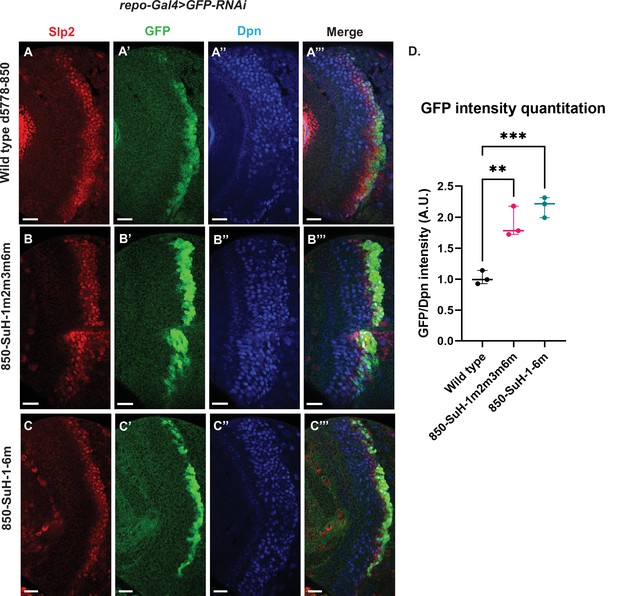
Mutation of potential Su(H) binding sites in the d5778 850 bp enhancer does not decrease GFP reporter expression in neuroblasts.
To measure GFP intensities in medulla neuroblasts unimpeded by GFP fluorescence from glial cells, UAS-GFP-RNAi; d5778 850 bp >GFP wild type reporter or UAS-GFP-RNAi; d5778 850 Su(H) binding site mutant GFP reporters were mated with a UAS-Dcr2; Repo-Gal4 expressing line, and phenotypes in the resulting progeny were observed. Mutation of four Su(H) binding sites in the 850-SuH-1m2m3m6m mutant (B-B’’’) and all six possible Su(H) binding sites in the 850-SuH-1–6 m mutant (C-C’’’) showed no loss of GFP expression relative to the wild type d5778 850 bp GFP reporter (A-A’’’). Instead, GFP expression was upregulated in Su(H) site mutants of the d5778 850 bp reporter. This indicates that Su(H) does not activate slp1/slp2 expression through the d5778 850 bp enhancer directly. Scale bars: 20 µm. (D) Three sets of observations were used for statistical analyses (n=3). Ordinary one-way ANOVA was used to compare relative GFP intensities between Su(H) site mutants and the wild type d5778 850 bp enhancer. Adjusted p values by Dunnett’s multiple comparisons test between the wild type d5778 850 bp reporter and the Su(H) site mutant reporters are as follows: for the 850-SuH-1m2m3m6m reporter p=0.0019, and the 850-SuH-1–6 m reporter is p=0.0005. Difference of means of each sample relative to wild type are statistically significant as p<0.005, below the significance threshold of p<0.05. The graph shows the distribution of individual observations about the median and within 95% confidence intervals.
-
Figure 9—source data 1
Quantification and comparison of reporter expression level driven by wild type and Su(H)-site-mutated forms of d5778 850bp enhancer after glial expression is removed by RNAi.
- https://cdn.elifesciences.org/articles/75879/elife-75879-fig9-data1-v1.xlsx
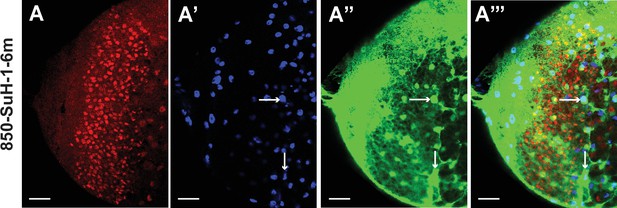
Mutation of Su(H) binding sites in the d5778 850 bp enhancer causes intensified GFP expression in glia.
(A-A’’’) The expression of 850-SuH-1–6 m mutant reporter is very strong in glial cells marked by Repo, preventing us to assess the expression in neuroblasts (marked by Dpn). The 850-SuH-1m2m3m6m mutant reporter has a similar pattern (not shown). Arrows point to examples of cell bodies of glia marked by Repo. Scale bars 20 µm.
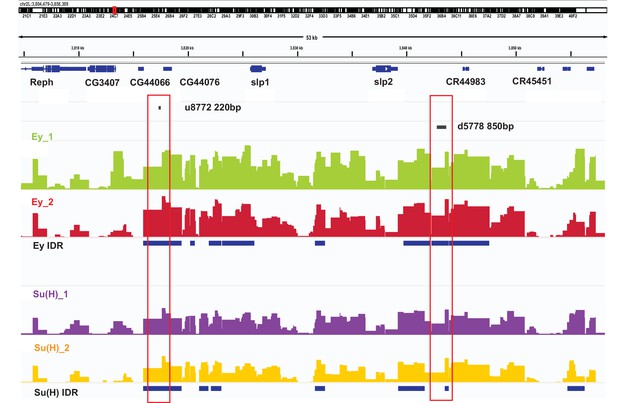
DamID-seq demonstrates reproducible binding of Ey and Su(H) to the slp1/2 enhancers in vivo.
Prominent reproducible peaks of Ey and Su(H) binding are seen in neighborhood of the slp1 and slp2 gene loci including at the genomic locations of identified 220 bp and 850 bp enhancers in both replicates of Ey-Dam and Su(H)-Dam experiments. Peaks at the u8772 220 bp and the d5778 850 bp enhancers have passed IDR <0.05 cut-off supporting their reproducibility.
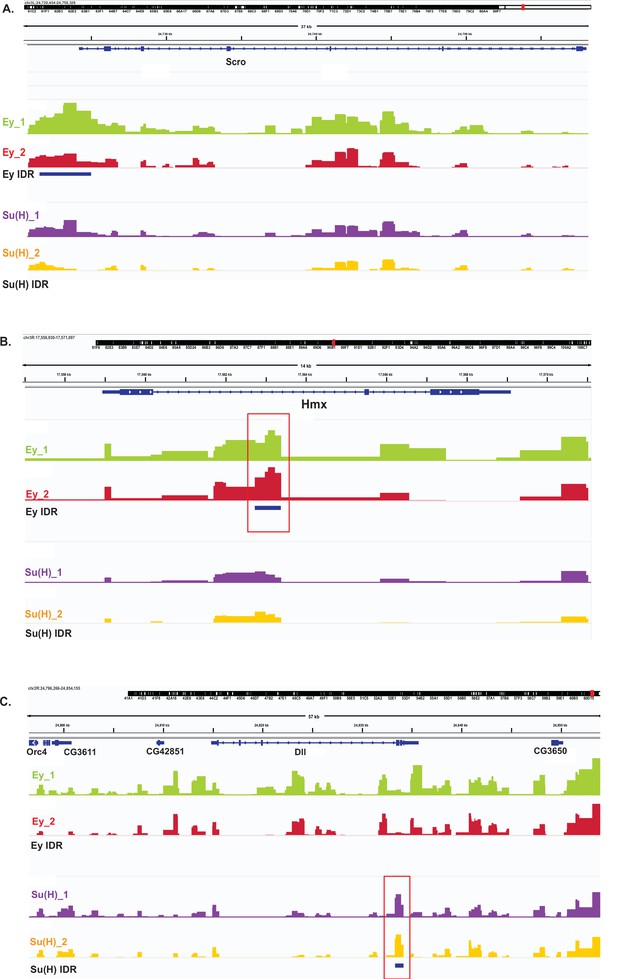
Examples of DamID-seq profiles on selected genes.
The IDR tracks are showing peaks that have passed the IDR <0.05 cut-off. (A) IDR Peaks for Ey but not Su(H) are observed in the vicinity of the scro gene locus. (B) IDR Peaks for Ey but not Su(H) are observed within the intron of Hmx gene locus. (C) IDR Peaks for Su(H) but not Ey are observed in the dll gene locus.
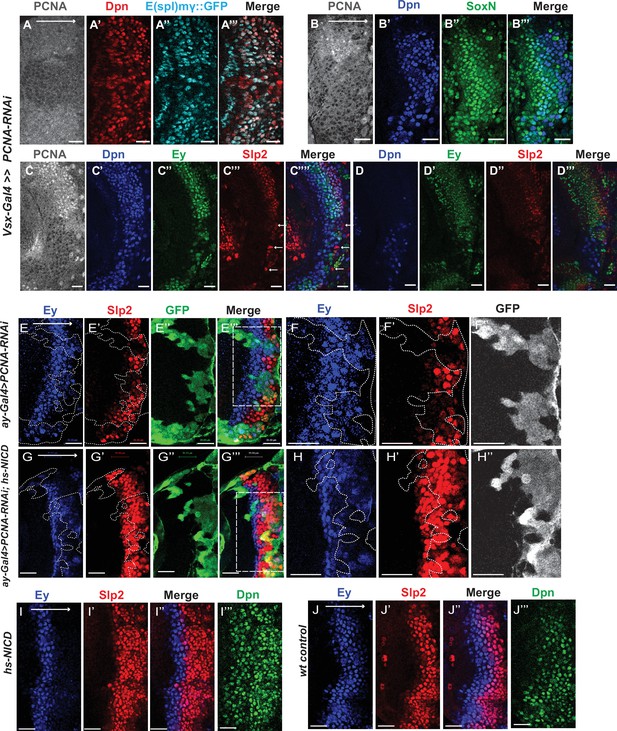
Cell cycle progression is required for the precise timing of Slp expression and this is mediated in part through Notch signaling.
(A-A’’’) The expression of PCNA, Dpn (red), and E(spl)mγGFP (cyan) in vsx-Gal4 driving UAS-DCR2 and UAS-PCNA-RNAi (VDRC51253) optic lobes (n=5). The affected region is indicated by loss of PCNA staining. (B-B’’’) The expression of Dpn and SoxN in vsx-Gal4 driving UAS-DCR2 and UAS-PCNA-RNAi optic lobes (n=5). (C-D’’’) The expression of Dpn, Ey, and Slp2 in vsx-Gal4 driving UAS-DCR2 and UAS-PCNA-RNAi optic lobes (n=9) showing the surface neuroblast layer view (C-C’’’’) and the deep progeny layer view (D-D’’’) of the same brain. (C-C’’’’) In affected region marked by loss of PCNA staining, Ey expression is reduced and Slp2 expression is lost in Dpn +neuroblasts. Some glia express Slp2 but not Dpn are indicated by white arrows. (D-D’’’) Dpn expressing neuroblasts are observed in deep layers in the affected region, but they don’t express Ey or Slp2. (E-E’’’) Larvae of genotype ywhsFLP; ay-Gal4 UAS-GFP /UAS-PCNA-RNAi; UAS-DCR2/+ were heat shocked at 37°C for 8 min 70 hours before dissection at 3rd instar larval stage. Ey expression (blue) is minimally affected in clones marked by GFP (green), while Slp2 (red) expression is delayed (n=17 clones). (F-F’’) Magnified view of the boxed region in E’’’. (G-G’’’) Larvae of genotype ywhsFLP; ay-Gal4 UA-SGFP /UAS-PCNA-RNAi; UAS-DCR2/dpn >FRT-STOP-FRT-NICD were heat shocked at 37 °C for 8 min 70 hr before dissection at 3rd instar larval stage. Ey expression (blue) is minimally affected in clones marked by GFP (green), while Slp2 (red) expression initiation is rescued (n=10 clones). (H-H’’) Magnified view of the boxed region in G’’’. (I-J’’’) The expression of Ey, Slp2, and Dpn in ywhsFLP; dpn >FRT-STOP-FRT-NICD (I-I’’’) and yw control (J-J’’’) optic lobes (n=5 each) (heat shocked 37°C for 8 min 70 hr before dissection). Scale bars 20 µm.
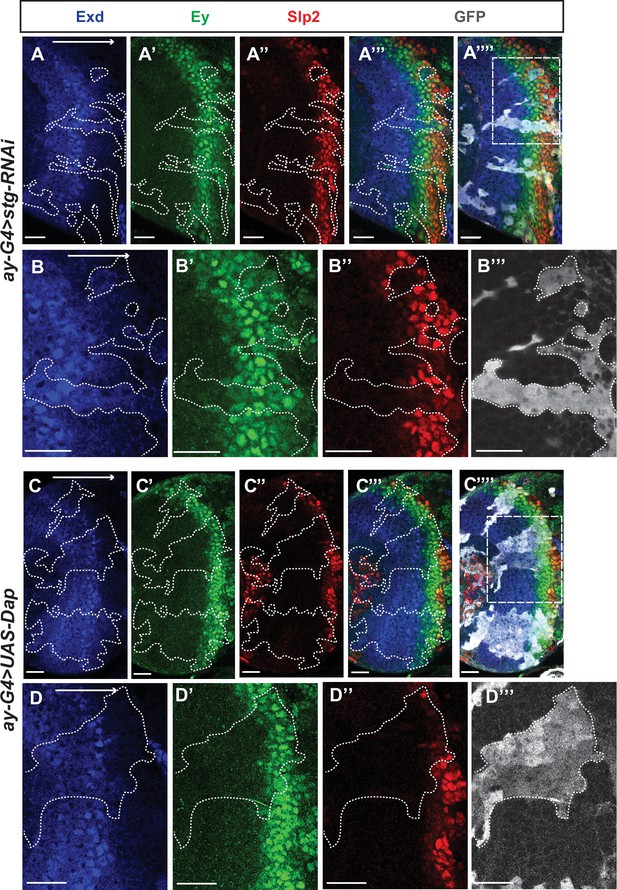
Cell cycle progression is required for the temporal cascade progression.
(A-A’’’) Larvae of genotype ywhsFLP; ay-Gal4 UAS-GFP /+; UAS-DCR2/UAS-stgRNAi (VDRC 17760) were heat shocked at 37°C for 8 min 70 hr before dissection at 3rd instar larval stage. Exd expression (co-factor of Hth, and always expressed in the same pattern) is normal in clones marked by GFP (gray), Ey expression is slightly delayed, while Slp2 expression is severely delayed (11 clones). (B-B’’’) Magnified view of the boxed region in A’’’’. (C-C’’’’) Larvae of genotype ywhsFLP; ay-Gal4 UAS-GFP /+; UAS-DCR2/UAS-Dap (BDSC 83338) were heat shocked at 37°C for 8 min 70 hr before dissection at 3rd instar larval stage. Exd expression is normal in clones marked by GFP (gray), Ey expression is slightly delayed, while Slp2 expression is severely delayed (8 clones). (D-D’’) Magnified view of the boxed region in C’’’’.
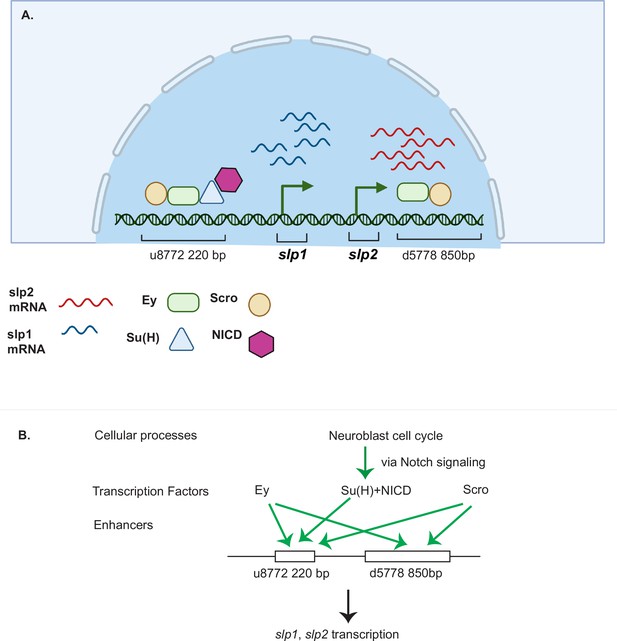
A coordinated action of temporal patterning factors Eyeless (Ey) and Scarecrow (Scro) and the Notch signaling pathway regulates the expressions of Slp1 and Slp2 in medulla neuroblasts.
(A) A schematic summarizing the main findings of this study. In medulla neuroblasts, transcription of the slp1 and slp2 genes is regulated by other temporal patterning factors Ey and Scro and by the Notch pathway. Ey and Scro both activate slp1/2 transcription by binding to the identified enhancers u8772 220 bp and d5778 850 bp. Su(H) activates slp1/2 transcription by binding to the u8772 220 bp enhancer. (B) The cell cycle in neuroblasts is required for continued activation of Notch signaling in medulla neuroblasts. We show that the cell cycle influences expression of slp1 and slp2 genes via its effects on the Notch signaling pathway; Slp1/2 expression is lost in cell cycle arrested neuroblasts due to a failure of Notch signaling. Restoration of Notch signaling rescues Slp1/2 expression in cell cycle arrested neuroblasts. Cellular processes like the neuroblast cell cycle cooperate with temporal patterning and Notch signaling to attain precise developmental outcomes. Graphic A created using BioRender.
Tables
Fragments screened or cloned for identifying enhancers of slp1/2 transcription in medulla neuroblasts.
Table summarizing names and chromosomal locations of enhancer fragments screened or cloned. Those with names beginning with GMR are Janelia FlyLight Gal4 lines. Enhancer fragments with names beginning in u, i, or d are enhancers from the study (Fujioka and Jaynes, 2012).
| Screened fragments | Start base | End base | Expression in medulla neuroblasts |
|---|---|---|---|
| GMR35H02 | 3817265 | 3821014 | Yes |
| GMR79H09 | 3830094 | 3833128 | No |
| GMR35G02 | 3821714 | 3825360 | No |
| GMR79H08 | 3832497 | 3833703 | No |
| u8772 | 3816967 | 3818532 | Yes |
| u8781 | 3816967 | 3817608 | Yes |
| u8172 | 3817605 | 3818532 | No |
| u8166 | 3817605 | 3819171 | Mainly in neurons |
| u5534 | 3820168 | 3822240 | unspecific broad expression |
| u5547 | 3820168 | 3821009 | weak expression |
| u3925 | 3821751 | 3823158 | No |
| u3725 | 3822001 | 3823158 | No |
| u2316 | 3823356 | 3824032 | No |
| u1609 | 3824033 | 3824739 | No |
| i1523 | 3827196 | 3827972 | No |
| i2330 | 3827970 | 3828686 | No |
| i4053 | 3829629 | 3830998 | No |
| i4060 | 3829629 | 3831819 | No |
| i5882 | 3831440 | 3833928 | No |
| d2445 | 3839285 | 3841364 | No |
| d5778 | 3842530 | 3844660 | Yes |
| d6383 | 3843114 | 3845187 | No |
| Cloned fragments | Start base | End base | Expression in medulla neuroblasts |
| 35H02-L1.5 | 3817261 | 3818808 | Yes |
| 35H02-M1.0 | 3818785 | 3819757 | No |
| 35H02-R2.2 | 3818872 | 3821014 | No |
| slpf1-220bp (u8772 220 bp) | 3817298 | 3817517 | Yes |
| slpf2-592bp | 3817782 | 3818373 | No |
| slpf3-123bp | 3818686 | 3818808 | No |
| d5778 850 bp | 3842770 | 3843619 | Yes |
List of predicted binding sites and mutagenesis strategy.
Sequences of 21 bp fragments including the underlined binding sites are shown. The critical invariant bases are in bold. For sites on the reverse strand (-), the reserve strand sequence is shown. For deletions, all 21bps are deleted. Mutated bases are shown as lower-case letters. * indicates that the binding site is predicted by the FIMO pipeline.
| Site Name | Strand | Original Sequence | Mutate to | Constructs (Size in bp) |
|---|---|---|---|---|
| 220-Ey-Site1 | + | TAATTCCGTTTAAATGATTTC | Deleted | 220-Ey-1d (199 bp) |
| 220-Ey-Site2* | - | TTTTCACGGCTCAGTGGCCTT | TTTTCACGGCTatGTGGCCTT | 220-Ey-2m (220 bp) |
| 220-SuH-Site1* | + | GGTGAGCGATAGTTTTCACAT | GGTGAGCGActaggagacgAT | 220-SuH-1d has the first site deleted. 220-SuH-1–4 m (220 bp) has all four sites mutated. 220-SuH-1m4m (220 bp) has site 1 and site 4 mutated. |
| 220-SuH-Site2 | + | TTTGAGCTTTGCACGTAATCA | TTTGAGCTTaGactGTAATCA | |
| 220-SuH-Site3 | - | AAATTCATTTCCACTTGATTA | AAATcCAcTTCCACTTGATTA | |
| 220-SuH-Site4 | - | CTGATATTTTTCACGGCTCAG | CTGActcgcTTtACGGCTCAG | |
| 220-Scro-Site1 | + | CACGTAATCAAGTGGAAATGA | CACGTAATCgtGTGGAAATGA | 220-Scro-1m (220 bp) |
| 220-Slp1-Site1 | + | TTCACATGTTTTGTTCGAGCA | TTCACATGTaccaagCGAGCA | 220-Slp1-1m2m has both sites mutated (220 bp) |
| 220-Slp1-Site2 | - | CTCTTGTGTTGGTCAAATTCA | CTCTTGgagTcGatcAATTCA | |
| 850-Ey-Site1* | + | TTGTGCCCTGTGTGTGTGAAA | aTGTGgCtTGTGTGTGTGAAA | 850-Ey-1m2d3d has site 1 mutated, and sites 2 and 3 deleted (808 bp). |
| 850-Ey-Site2 | + | TGATTCCAATTGACTGATTTC | Deleted | |
| 850-Ey-Site3 | - | ATGTGAGCCACTTGTGTGGCA | Deleted | |
| 850-SuH-Site1 | - | TTTTTGTTTCACACACACAGG | TaTcTGTcgCACACACACAGG | 850-SuH-1m2m3m6m has sites 1, 2, 3 and 6 mutated (850 bp). 850-SuH-1–6 m has sites 1–6 mutated (850 bp). |
| 850-SuH-Site2 | - | CGTCTGTTTCCCAACACCGTA | CGTCTGTaTCCgtACACCGTA | |
| 850-SuH-Site3* | - | TCCAAAGTTCCCACAGTGCGC | TCCAAAGaTCCgACAGTGCGC | |
| 850-SuH-Site4 | + | GCATTTTTAACAACTTTAAGC | GCATTTcTAAgtACTTTAAGC | |
| 850-SuH-Site5 | + | GTCGACCTTGTCAAGTACCTT | GTCGACCaTGTCAAGTACCTT | |
| 850-SuH-Site6 | - | AGCAGCGTTTTCTCAACTTTG | AGCAGCGTTaTCTgAACTTTG | |
| 850-Scro-Site1* | + | TTGATGTTAAGTGTCTGCCAC | Deleted | 850-Scro-1-7d has all 7 sites deleted (724 bp) |
| 850-Scro-Site2 | - | CATAGTGTCCTAAATGCTGTT | Deleted | |
| 850-Scro-Site3 | + | CAAGCCAAAACTAATGGGTAT | Deleted | |
| 850-Scro-Site4,5 | - | TCGTTAAAGG GGTTAAATAGG | Deleted | |
| 850-Scro-Site6* | + | TTGTCAAGTACCTTGTTAGCT | Deleted | |
| 850-Scro-Site7 | + | CGTTTAATTGACAATTGAACC | Deleted | |
| 850-Slp1-Site1* | + | TGACTTTGTTTTTGCTACTCA | Deleted | 850-Slp1-1d (829 bp) |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti SoxN (Rabbit polyclonal) | Claude Desplan | N/A | IF 1:250 |
| Antibody | anti- Ey (Rabbit polyclonal) | Claude Desplan | N/A | IF 1:500 |
| Antibody | anti-Slp1 (Rabbit polyclonal) | Claude Desplan | N/A | IF 1:500 |
| Antibody | anti-Slp2 (Guinea-pig polyclonal) | Claude Desplan | N/A | IF 1:500 |
| Antibody | anti-Scro (Guinea-pig polyclonal) | Claude Desplan | N/A | IF 1:100 |
| Antibody | Anti-D (Rabbit polyclonal) | Claude Desplan | N/A | IF 1:500 |
| Antibody | Anti-Dpn (Guinea pig polyclonal) | Chris Doe | N/A | IF 1:500 |
| Antibody | Anti-Dpn (Rat monoclonal) | Abcam | Antibody#: Ab195173 | IF 1:500 |
| Antibody | Anti-B-H1 (Rat polyclonal) | Tiffany Cook | N/A | IF 1:200; Tiffany Cook |
| Antibody | Anti-GFP (Sheep polyclonal) | AbD Serotec | 4745–1051 | IF 1:500 |
| Antibody | Anti-Repo (mouse monoclonal) | Developmental Studies Hybridoma Bank | 8D12 anti-Repo | IF 1:50 |
| Antibody | Anti-PCNA (mouse monoclonal) | Abcam | ab29 | IF 1:10 |
| Antibody | Anti-betaGalactosidase (Chicken polyclonal) | Abcam | Antibody#: Ab9361 | IF 1:500 |
| Antibody | Cy5 AffiniPure Anti-RatIgG (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 712-175-153 | IF!:500 |
| Antibody | Cy3 AffiniPure Anti-RatIgG (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 712-165-153 | IF 1:500 |
| Antibody | Cy3 AffiniPure Anti-Guinea Pig IgG (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 706-165-148 | IF 1:500 |
| Antibody | Alexa Fluor 647 AffiniPure Anti-Guinea Pig (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 706-605-148 | IF 1:500 |
| Antibody | Alexa Fluor 488 AffiniPure Anti-Guinea Pig IgG (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 706-545-148 | IF 1:500 |
| Antibody | Alexa Fluor 647 AffiniPure Anti-Goat IgG (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 705-605-147 | IF 1:500 |
| Antibody | DyLight 405 AffiniPure Anti-Mouse IgG (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 715-475-151 | IF 1:500 |
| Antibody | Alexa Fluor 647 AffiniPure Anti-Rabbit IgG (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 711-605-152 | IF 1:500 |
| Antibody | Cy5 AffiniPure Anti-Mouse IgG (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 715-175-151 | IF 1:500 |
| Antibody | Alexa Fluor 647 AffiniPure Anti-Chicken IgY (IgG) (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 703-605-155 | IF 1:500 |
| Antibody | Alexa Fluor 488 AffiniPureDonkey Anti-Mouse IgG (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 715-545-151 | IF 1:500 |
| Antibody | Alexa Fluor 488 AffiniPure Anti-Sheep (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 713-545-147 | IF 1:500 |
| Antibody | DyLight 405 AffiniPure Anti-Rat IgG (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 712-475-153 | IF 1:500 |
| Antibody | DyLight 405 AffiniPure Anti-Rabbit IgG (Donkey polyclonal) | Jackson ImmunoResearch Laboratories Inc | Catalog_#: 711-475-152 | IF 1:500 |
| Antibody | anti-Rabbit IgG, Alexa Fluor  555 conjugate (Donkey polyclonal) | Life Technologies | Catalog_#: A-31572 | IF 1:500 |
| Antibody | Anti-Opa (Rabbit polyclonal) | J. Peter Gergen | N/A | IF 1:100; J. Peter Gergen |
| Antibody | Anti-Erm (Rat polyclonal) | Claude Desplan | N/A | 1:100 |
| Recombinant DNA Reagent | pJR12 plasmid | Rister et al., 2015 PMID:26785491 DOI: 10.1126/science.aab3417 | N/A | Jens Rister |
| Recombinant DNA Reagent | pCFD5 plasmid | Addgene | Addgene Plasmid #73,914 | Fillip Port |
| Recombinant DNA Reagent | UAS-LT3-Dam plasmid | Andrea Brand | N/A | |
| Recombinant DNA Reagent | pJR12-hsNICD plasmid | This paper | N/A | See section on ’Plasmid constructs’ in ‘Materials and methods’ |
| Recombinant DNA Reagent | UAS-LT3-DamEy | This paper | N/A | See section on ’Plasmid constructs’ in ‘Materials and methods’ |
| Recombinant DNA Reagent | UAS-LT3-DamSu(H) | This paper | N/A | See section on ’Plasmid constructs’ in ‘Materials and methods’ |
| Recombinant DNA Reagent | CoinFlp plasmid | Addgene | Addgene Plasmid #52,889 | Iswar Hariharan |
| Recombinant DNA Reagent | GMR35H02 BAC | BACPAC resources | CH321-94O18 | |
| Recombinant DNA Reagent | Su(H) cDNA. | Drosophila Genomicsa Resource Center | GH10914 | |
| Recombinant DNA Reagent | Notch cDNA | Drosophila Genomics Resource Center | LD34134 | |
| Recombinant DNA Reagent | CH321-86A18 (BAC) | BACPAC resources | BAC encoding dpn enhancer | |
| Sequence-based reagent | 220-Ey-1d gBlock | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | 220-Ey-2m gBlock | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | 220-SuH-1–4 m | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | 220-SuH-1m4m | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | 220-Scro-1m | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | 220-Slp1-1m2m | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | 850-Ey-1m2d3d | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | 850-SuH-1m2m3m6m | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | 850-SuH-1–6 m | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | 850-Scro-1-7d | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | 850-Slp1-1d | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | Ey gene block | This study | N/A | See Supplementary file 4 for sequence |
| Sequence-based reagent | SlpR2-2FP | This study | N/A | 5’TTAGGCGCGCCAGTGCGTGTCTGCCCTTTCATTTTG 3’ |
| Sequence-based reagent | SlpR2-2RP | This study | N/A | 5’ ATATATGCGGCCGCCCCAGCTAGCTCCCTTCACTCTTCT 3’ |
| Sequence-based reagent | SlpL1-5FP | This study | N/A | 5’ TTAGGCGCGCCGAATCGAAATGCTTCCCCGCCTCG 3’ |
| Sequence-based reagent | SlpL1-5RP | This study | N/A | 5’ ATATATGCGGCCGCTGAACGTGCAACATCAAAGGCCGC 3’ |
| Sequence-based reagent | SlpM1-FP | This study | N/A | 5’ TTAGGCGCGCCGCGGCCTTTGATGTTGCACGTTCA 3’ |
| Sequence-based reagent | SlpM1-RP | This study | N/A | 5’ ATATATGCGGCCGCGGACAGTTCGGAATGTGCCTCGA 3’ |
| Sequence-based reagent | Slpf1-FP | This study | N/A | 5’ TTAGGCGCGCCGAATCGAGTGGTGAGCGATAG 3’ |
| Sequence-based reagent | Slpf1-RP | This study | N/A | 5’ ATATATGCGGCCGCTCTGATATTTTTCACGGCTCA 3’ |
| Sequence-based reagent | Slpf2-FP | This study | N/A | 5’ TTAGGCGCGCCTTCGACCTTGTAGTGGCAAG 3’ |
| Sequence-based reagent | Slpf2-RP | This study | N/A | 5’ ATATATGCGGCCGCCGGAGATCGGAAGGTTAGTG 3’ |
| Sequence-based reagent | Slpf3-FP | This study | N/A | 5’ TTAGGCGCGCCTCTCCTTGTTGCTCCTCACA 3’ |
| Sequence-based reagent | Slpf3-RP | This study | N/A | 5’ ATATATGCGGCCGCTGAACGTGCAACATCAAAGG 3’ |
| Sequence-based reagent | d5778FP | This study | N/A | 5’ TTAGGCGCGCCTGGTCTTTTACGTTAATCTGGGCAGCT 3’ |
| Sequence-based reagent | d5778RP | This study | N/A | 5’ ATATATGCGGCCGCACATTACGCATTGCATTCCTCCTCCTT 3’ |
| Sequence-based reagent | d5778-850FP | This study | N/A | 5’ TTAGGCGCGCCCATTAACTCGAGTCTGGTTTCCGAT 3’ |
| Sequence-based reagent | d5778-850RP | This study | N/A | 5’ ATATATGCGGCCGCCGTACATATTCTCCAGGAGTTCGGTC 3’ |
| Sequence-based reagent | pJRLPseq3 | This study | N/A | 5’ AGATGGGTGAGGTGGAGTACG 3’ |
| Sequence-based reagent | pJR12-TATA-seq | This study | N/A | 5’ AGCTGCGCTTGTTTATTTGCTTAG 3’ |
| Sequence-based reagent | SuHDamFP | This study | N/A | 5’ ATATATGCGGCCGCAAATGAAGAGCTACAGCCAATTTAATTTAAACGCCGCC 3’ |
| Sequence-based reagent | SuHDamRP | This study | N/A | 5’ AAAATACTCGAGTCAGGATAAGCCGCTACCATGACTATTCCATTGC 3’ |
| Sequence-based reagent | DamseqFP1 | This study | N/A | 5’ TGAGGGGAGACATAGTACTGGT 3’ |
| Sequence-based reagent | DamseqFP2 | This study | N/A | 5’ GAAGGTCTGGTTGAGCGCCATA 3’ |
| Sequence-based reagent | pCFD5-u8772-gRFP1 | This study | N/A | 5’ GCGGCCCGGGTTCGATTCCCGGCCGATGCATCGAAATTTCCTGGTATTCGGTTTTAGAGCTAGAAATAGCAAG 3’ |
| Sequence-based reagent | pCFD5-u8772-gRRP1 | This study | N/A | 5’ CGCCTCGCCCAAATGCATTTTGCACCAGCCGGGAATCGAACCC 3’ |
| Sequence-based reagent | pCFD5-u8772-gRFP2 | This study | N/A | 5’ AAATGCATTTGGGCGAGGCGGTTTTAGAGCTAGAAATAGCAAG 3’ |
| Sequence-based reagent | pCFD5-u8772-gRRP2 | This study | N/A | 5’ TTAGTTTGATTGTTTCGAAGTGCACCAGCCGGGAATCGAACCC 3’ |
| Sequence-based reagent | pCFD5-u8772-gRFP3 | This study | N/A | 5’ CTTCGAAACAATCAAACTAAGTTTTAGAGCTAGAAATAGCAAG 3’ |
| Sequence-based reagent | pCFD5-u8772-gRRP3 | This study | N/A | 5’ ATTTTAACTTGCTATTTCTAGCTCTAAAACAAACTGGAAGTCATTGACCCTGCACCAGCCGGGAATCGAACCC 3’ |
| Sequence-based reagent | pCFD5-d5778-gRFP1 | This study | N/A | 5’ GCGGCCCGGGTTCGATTCCCGGCCGATGCAATAAGTCCTTGGGTAATACGGTTTTAGAGCTAGAAATAGCAAG 3’ |
| Sequence-based reagent | pCFD5-d5778-gRRP1 | This study | N/A | 5’ AACATTTATCTAGGACATCTTGCACCAGCCGGGAATCGAACCC 3’ |
| Sequence-based reagent | pCFD5-d5778-gRFP2 | This study | N/A | 5’ AGATGTCCTAGATAAATGTTGTTTTAGAGCTAGAAATAGCAAG 3’ |
| Sequence-based reagent | pCFD5-d5778-gRRP2 | This study | N/A | 5’ TGTTGGCAAGCGGCGCTTCATGCACCAGCCGGGAATCGAACCC 3’ |
| Sequence-based reagent | pCFD5-d5778-gRFP3 | This study | N/A | 5’ TGAAGCGCCGCTTGCCAACAGTTTTAGAGCTAGAAATAGCAAG 3’ |
| Sequence-based reagent | pCFD5-d5778-gRRP3 | This study | N/A | 5’ ATTTTAACTTGCTATTTCTAGCTCTAAAACTTTCGATATCCCAGCTCCTTTGCACCAGCCGGGAATCGAACCC 3’ |
| Sequence-based reagent | u8772crdelFP | This study | N/A | 5’ TTGCAAATACTTTTTATTCAAGGAATCGAC 3’ |
| Sequence-based reagent | u8772crdelRP | This study | N/A | 5’ AATCTCAAGTTTGGTGTTTGTAATTTTTGG 3’ |
| Sequence-based reagent | d5778crdelFP | This study | N/A | 5’ CTATTGAAGGGCGGACATATTAGACAACAATTGGATCGCTTG 3’ |
| Sequence-based reagent | d5778crdelRP | This study | N/A | 5’ CTGCATTCCATCCCGTCGCATCCTTGTC 3’ |
| Sequence-based reagent | pJRGFPdelFP1 | This study | N/A | 5’ TTAGAGATGCATCTCAAAAAAATGGTGGGCATAATAGTGTTGTTTATATATATCAAAAATAACAAC 3’ |
| Sequence-based reagent | pJRGFPdelRP1 | This study | N/A | 5’ CCACCGGTCGCCACCGACGTCAGC GGCCGGCCGC 3’ |
| Sequence-based reagent | pJRGFPdelFP2 | This study | N/A | 5’ GTCGCGGCCGGCCGCTGACGTCGGTGGCGACCGGTGGATCGTTTAAACAGGCC 3’ |
| Sequence-based reagent | pJRGFPdelRP2 | This study | N/A | 5’ CAATAACTCGAGGAGCGCCGGAGT ATAAATAGAGGCGCTTCGTCTACG 3’ |
| Sequence-based reagent | pJRGFPdelseqFP | This study | N/A | 5’ CCATTATAAGCTGCAATAAACAAGTTAACAAC 3’ |
| Sequence-based reagent | pJRGFPdelseqRP | This study | N/A | 5’ GTCGCTAAGCGAAAGCTAAGC 3’ |
| Sequence-based reagent | U63seqfwd | This study | N/A | 5’ ACGTTTTATAACTTATGCCCCTAAG 3’ |
| Sequence-based reagent | pCFD5seqrev | This study | N/A | 5’ GCACAATTGTCTAGAATGCATAC 3’ |
| Sequence-based reagent | dpnenFP | This study | N/A | 5’ TTAGGCGCGCCCTTCGCTTTTGCCTG GTCGGCTCATCGG 3’ |
| Sequence-based reagent | dpnenRP | This study | N/A | 5’ ATATATGCGGCCGCACGCCTCGTCCTGGCACCCTC 3’ |
| Sequence-based reagent | NICDFP | This study | N/A | 5’ TATTTAACCGGTTATTATCAAATGTAGATGGCCTCGGAACCCTTG 3’ |
| Sequence-based reagent | NICDRP | This study | N/A | 5’ ATAATAGTTTAAACATGAGTACGCAAAGAAAGCGGGCAC 3’ |
| Sequence-based reagent | hscFP1 | This study | N/A | 5’ TATTTAACCGGTTATTATCAAATGTAGATGGCCTCGGAACCCTTG 3’ |
| Sequence-based reagent | hscRP1 | This study | N/A | 5’ GGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCGAATTCCAAAATGAGTACGCAAAGAAAGCGGGCAC 3’ |
| Sequence-based reagent | hscFP2 | This study | N/A | 5’ GTGCCCGCTTTCTTTGCGTACTCATTTTGGAATTCGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCC 3’ |
| Sequence-based reagent | hscRP2 | This study | N/A | 5’ ATAATAGTTTAAACGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCCCCGC 3’ |
| Genetic reagent (D. melanogaster) | UAS-ey -RNAi | Bloomington Drosophila Stock Centre | BDSC 32486; Symbol: CG1464; Flybase ID:FBgn0005558 | |
| Genetic reagent (D. melanogaster) | UAS-Su(H)-RNAi | Vienna Drosophila Stock Centre | VDRC 103597 Symbol: CG3497; Flybase ID: FBgn0004837 | |
| Genetic reagent (D. melanogaster) | UAS-N-RNAi | Bloomington Drosophila Stock Centre | BDSC 7078; Symbol: CG3936; Flybase ID: FBgn0004647 | |
| Genetic reagent (D. melanogaster) | UAS-Dl-RNAi | Vienna Drosophila Stock Centre | VDRC 32788; Symbol: CG3619; Flybase ID: FBgn0000463 | |
| Genetic reagent (D. melanogaster) | UAS-PCNA-RNAi | Vienna Drosophila Stock Centre | VDRC 51253; Symbol: CG9193; Flybase ID: FBgn0005655 | |
| Genetic reagent (D. melanogaster) | UAS-mam-DN | Justin Kumar | Symbol: CG8118; Flybase ID: FBgn0002643 | Justin Kumar |
| Genetic reagent (D. melanogaster) | UAS-stg-RNAi | Vienna Drosophila Stock Centre | VDRC 17760; Symbol: CG1395 Flybase ID: FBgn0003525 | |
| Genetic reagent (D. melanogaster) | UAS-dap | Bloomington Drosophila Stock Centre | BDSC 83338; Symbol: CG1772 Flybase ID: FBgn0010316 | |
| Genetic reagent (D. melanogaster) | UAS-scro-RNAi | Bloomington Drosophila Stock Centre | BDSC 33890; Symbol: CG17594 Flybase ID: FBgn0287186 | |
| Genetic reagent (D. melanogaster) | UAS-Ey | Bloomington Drosophila Stock Centre | BDSC56560 | |
| Genetic reagent (D. melanogaster) | UAS-Scro | FlyORF | F000666 | |
| Genetic reagent (D. melanogaster) | GMR35H02-Gal4 | Bloomington Drosophila Stock Centre (Pfeiffer et al., 2008) | BDSC 49923 | |
| Genetic reagent (D. melanogaster) | GMR41H10-Gal4 (SoxN-Gal4) | Bloomington Drosophila Stock Centre (Pfeiffer et al., 2008) | N/A | No longer available from BDSC |
| Genetic reagent (D. melanogaster) | UAS-GFP-nls | Bloomington Drosophila Stock Centre | BDSC 4776 | |
| Genetic reagent (D. melanogaster) | UAS-EGFP-RNAi | Bloomington Drosophila Stock Centre | BDSC 9931 | |
| Genetic reagent (D. melanogaster) | ayGal4 “y w hsFLP; act >y +> Gal4 UAS-GFP / CyO” | Ito et al., 1997 PMID:9043058 DOI: 10.1242/dev.124.4.761 | N/A | |
| Genetic reagent (D. melanogaster) | “y w hs FLP; act >y +> Gal4 UAS GFP / CyO; UASDCR2/TM6B” | Zhu et al., 2022 PMID:35273186 DOI: 10.1038/s41467-022-28915-3 | N/A | |
| Genetic reagent (D. melanogaster) | ayGal4 UASlacZ | Bloomington Drosophila Stock Centre | BDSC 4410 | |
| Genetic reagent (D. melanogaster) | “y,w, hsFLP, UASCD8GFP; FRT40A tubGal80; tubGal4/TM6B” | Liqun Luo | N/A | |
| Genetic reagent (D. melanogaster) | “y,w,; UbiRFPnls FRT40A/CyO” | Bloomington Drosophila Stock Centre | BDSC 34500 | |
| Genetic reagent (D. melanogaster) | FRT40A slpS37A /SM6-TM6B | Sato and Tomlinson, 2007 PMID:17215299 DOI: 10.1242/dev.02786 | N/A | Andrew Tomlinson |
| Genetic reagent (D. melanogaster) | E(spl)mγGFP | Almeida and Bray, 2005 PMID:16275038 DOI: 10.1016 /j.mod.2005.08.004 | N/A | |
| Genetic reagent (D. melanogaster) | “VsxGal4;Dpn-LacZ/CyO; UAS-Dcr2/TM6B” | Erclik et al., 2017 PMID:28077877 DOI: 10.1038/nature20794 | N/A | |
| Genetic reagent (D. melanogaster) | dpn-Gal4 (GMR13C02) | Bloomington Drosophila Stock Centre | BDSC47859 | |
| Genetic reagent (D. melanogaster) | repo-Gal4 | Bloomington Drosophila Stock Centre | BDSC7415 | |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; Sp/CyO; u8772 220 GFP/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; Sp/CyO; d5778 850 GFP/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 220-Ey-1d/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 220-Ey-2m/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 220-SuH-1–4 m/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 220-SuH-1m4m/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 220-Scro-1m/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 220-Slp1-1m2m/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 850-Ey-1m2d3d/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 850-SuH-1m3m6m/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 850-SuH-1m2m3m6m/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 850-SuH-1–6 m/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 850-Scro-1-7d/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; 850-Slp1-1d/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; UAS-DamEy/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w, hsFlp; Sp/CyO; UAS-SuHDam/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; Sp/CyO; UAS-Dam/ Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “Dcr2; tubG80ts; SoxNG4” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “Dcr2; Su(H)RNAi/CyO; u8772-220-GFP/ Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “Dcr2; Su(H)RNAi/CyO; d5778-850-GFP/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; Sp/CyO; UAS-eyRNAi/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; Sp/CyO; UAS-eyRNAi/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; ayGal4 UASlacZ/CyO; UAS-eyRNAi/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; ayGal4 UASlacZ/CyO; hsNICD/Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; Ubi RFPnls FRT40A; u8772-220-GFP/Sm6-Tm6B “ | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; Ubi RFPnls FRT40A; d5778-850-GFP/Sm6-Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; UAS Dc2; dpn-Gal4 /Sm6-Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; UAS Dc2; repo-Gal4 /Sm6-Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; UAS GFP RNAi; d5778-850-GFP /Sm6-Tm6B” | This study | N/A | See section on ‘Fly stocks’ and ‘Making of transgenic fly stocks’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; UAS GFP RNAi; 850-SuH-1m3m6m/Sm6-Tm6B” | This study | N/A | See section on ‘Fly stocks’ and ‘Making of transgenic fly stocks’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; UAS GFP RNAi; 850-SuH-1m2m3m6m/Sm6-Tm6B” | This study | N/A | See section on ‘Fly stocks’ and ‘Making of transgenic fly stocks’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; UAS GFP RNAi; 850-SuH-1–6 m/Sm6-Tm6B” | This study | N/A | See section on ‘Fly stocks’ and ‘Making of transgenic fly stocks’ in ‘Materials and Methods’ |
| Genetic reagent (D. melanogaster) | “y,w,hsFlp; ayGal4 UAS PCNA RNAi; hsNICD/ Sm6-Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ and ‘Making of transgenic fly stocks’ in ‘Materials and methods’ |
| Genetic reagent (D. melanogaster) | “VsxGal4; E(spl) mγGFP/ CyO; UAS Dc2 /Tm6B” | This study | N/A | See section on ‘Fly stocks and genetics’ in ‘Materials and methods’ |
| Genetic reagent (D. melanogaster) | y, w, nos-Cas9 | Bloomington Drosophila Stock Centre | BL54591 | See section on ‘Fly stocks’ and ‘Making of transgenic fly stocks’ in ‘Materials and methods’ |
| Genetic reagent (D. melanogaster) | “y, w; PBAC{y[+]-attP-9A}VK00027” | Bloomington Drosophila Stock Centre | BL9744 | See section on ‘Fly stocks’ and ‘Making of transgenic fly stocks’ in ‘Materials and methods’ |
| Sequence-based reagent | Stellaris-Quasar 570-conjugated slp2 mRNA probes | This study | Symbol: CG2939-RA; Flybase ID: FBtr0077500 | See section ‘Fluorescence in situ hybridization’ in ‘Materials and methods’ |
| Sequence-based reagent | Stellaris-Quasar 670- conjugated slp1 mRNA probes | This study | Symbol: CG16738-RA; Flybase ID: FBtr0077499 | See section ‘Fluorescence in situ hybridization’ in ‘Materials and methods’ |
| Peptide, recombinant protein | AscI | New England Biolabs | Catalog_#: R0558S | See section ‘Plasmid constructs ’ in ‘Materials and methods’ |
| Peptide, recombinant protein | NotI-HF | New England Biolabs | Catalog_#: R3189S | See section ‘Plasmid constructs ’in ‘Materials and methods’ |
| Peptide, recombinant protein | XhoI | New England Biolabs | Catalog_#: R0146S | See section ‘Plasmid constructs ’in ‘Materials and methods’ |
| Peptide, recombinant protein | AgeI-HF | New England Biolabs | Catalog_#: R3552S | See section ‘Plasmid constructs ’ in ‘Materials and methods’ |
| Peptide, recombinant protein | PmeI | New England Biolabs | Catalog_#: R0560S | See section ‘Plasmid constructs ’ in ‘Materials and methods’ |
| Peptide, recombinant protein | ZraI | New England Biolabs | Catalog_#: R0659S | See section ‘Plasmid constructs ’ in ‘Materials and methods’ |
| Commercial assay or kit | NEB Builder HiFi DNA Assembly kit | New England Biolabs | E2621L | See section ‘Plasmid constructs ’in ‘Materials and methods’ |
| Commercial assay or kit | Expand High Fidelity PCR System, dNTPack | Roche | 04738268001 | See section ‘Plasmid constructs ’in ‘Materials and methods’ |
| Commercial assay or kit | Platinum SuperFi II PCR mastermix | Invitrogen | 12368010 | See section ‘Plasmid constructs ’in ‘Materials and methods’ |
| Commercial assay or kit | REDExtract-N-Amp PCR ReadyMix | Sigma-Aldrich | R4775-1.2ML | See section ‘Plasmid constructs ’ in ‘Materials and methods’ |
| Commercial assay or kit | DNeasy Blood and Tissue Kit | Qiagen | 69,504 | See section ‘Plasmid constructs ’ in ‘Materials and methods’ |
| Peptide, recombinant protein | DpnI | New England Biolabs | Catalog_#: R0176S | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Peptide, recombinant protein | DpnII | New England Biolabs | Catalog_#: R0543S | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Peptide, recombinant protein | AlwI | New England Biolabs | Catalog_#: R0513S | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Peptide, recombinant protein | Quick Ligation kit | New England Biolabs | Catalog_#: M2200S | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Peptide, recombinant protein | T4 DNA ligase | New England Biolabs | Catalog_#: M0202S | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Peptide, recombinant protein | T4 DNA polymerase | New England Biolabs | Catalog_#: M0203S | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Peptide, recombinant protein | Klenow fragment | New England Biolabs | Catalog_#: M0210S | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Peptide, recombinant protein | Klenow 3’->5’ exo-enzyme | New England Biolabs | Catalog_#: M0201S | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Peptide, recombinant protein | NEBNext High-Fidelity 2 X PCR master mix | New England Biolabs | Catalog_#: M0541S | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Peptide, recombinant protein | Advantage 2 cDNA polymerase | Clontech | Catalog_#: A63880 | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Peptide, recombinant protein | RNase A | Roche | Catalog_#: 11119915001 | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Commercial assay or kit | Qubit dsDNA HS assay kit | Invitrogen | Catalog_#: Q32851 | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Commercial assay or kit | Agencourt AMPure XP beads | Beckman-Coulter | Catalog_#: A63880 | See section ‘DamID-seq’ in ‘Materials and methods’ |
| Sequence-based reagent | AdRt oligo | Marshall and Brand, 2015 PMID:27490632DOI: 10.1038/nprot.2016.084 | N/A | 5’ CTAATACGACTCA CTATAGGGCAG CGTGGTCGC GGCCGAGGA 3’ |
| Sequence-based reagent | AdRb oligo | Marshall and Brand, 2015 PMID:27490632 DOI: 10.1038/nprot.2016.084 | N/A | 5’ TCCTCGGCCG 3’ |
| Sequence-based reagent | DamID_PCR oligo | Marshall and Brand, 2015 PMID:27490632 DOI: 10.1038/nprot.2016.084 | N/A | 5’ GGTCGCGG CCGAGGATC 3’ |
| Sequence-based reagent | NGS_PCR1 | Marshall and Brand, 2015 PMID:27490632 DOI: 10.1038/nprot.2016.084 | N/A | 5’ AATGATACGGC GACCACCGA*G 3’ *=phosphorothioate linkage |
| Sequence-based reagent | NGS_PCR2 | Marshall and Brand, 2015 PMID:27490632 DOI: 10.1038/nprot.2016.084 | N/A | 5’ CAAGCAGAAGA CGGCATACGA*G 3’ *=phosphorothioate linkage |
| Sequence-based reagent | NGS_adaptors | Marshall and Brand, 2015 PMID:27490632 DOI: 10.1038/nprot.2016.084 | N/A | |
| Software, algorithm | FIMO based web-application | Bart Deplancke | Site: https://biss.epfl.ch | |
| Software, algorithm | FIMO | Grant et al., 2011 PMID:21330290 DOI: 10.1093/bioinformatics/btr064 | Site: https://meme-suite.org/meme Versions 4.11.1–5.4.1 | Accessed between 2016 to 2022; See section ‘Bioinformatic analysis’ in ‘Materials and methods’. |
| Software, algorithm | The MEME suite | Bailey et al., 2015 PMID:25953851 DOI: 10.1093/nar/gkv416 | Site: https://meme-suite.org/meme Versions 4.11.1–5.4.1 | Accessed between 2016 to 2022; See section ‘Bioinformatic analysis’ in ‘Materials and methods’. |
| Software, algorithm | TOMTOM | Gupta et al., 2007 PMID:17324271 DOI: 10.1186/gb-2007-8-2-r24 | Site: https://meme-suite.org/meme Versions 4.11.1–5.4.1 | Accessed between 2016 to 2022; See section ‘Bioinformatic analysis’ in ‘Materials and methods’. |
| Software, algorithm | JASPAR | Sandelin et al., 2004 PMID:14681366 doi: 10.1093/nar/gkh012 | Site: https://jaspar.genereg.net/ releases 6, 7, 8,9. | Accessed between 2016 to 2022; See section ‘Bioinformatic analysis’ in ‘Materials and methods’. |
| Software, algorithm | Fly Factor Survey | Zhu et al., 2011 PMID:21097781 DOI: 10.1093/nar/gkq858 | Site: https://mccb.umassmed.edu/ffs | Accessed between 2016 to 2022; See section ‘Bioinformatic analysis’ in ‘Materials and Methods’. |
| Software, algorithm | Sequence Manipulation Suite Version 2 | Stothard, 2000 PMID:10868275 DOI: 10.2144/00286ir01 | Site: https://www.bioinformatics.org/sms2 | Accessed between 2016 to 2022; See section ‘Bioinformatic analysis’ in ‘Materials and Methods’. |
| Software, algorithm | Primer3web version 4.1.0 | Site: https://bioinfo.ut.ee/primer3 | Primer3web version 4.1.0 | |
| Software, algorithm | damidseq_pipeline | Marshall and Brand, 2015 PMID:26112292 DOI: 10.1093/bioinformatics/btv386 | Site: http://owenjm.github.io/damidseq_pipeline Version: v1.4.6 | See section ‘DamID-seq data analysis’ in ‘Materials and Methods’. |
| Software, algorithm | Irreproducibility Discover Rate (IDR) | Nathan Boley | Site: https://github.com/nboley/idr Version: 2.0.3 | See section ‘DamID-seq data analysis’ in ‘Materials and Methods’. |
| Software, algorithm | Integrated Genomics Viewer (IGV) | Robinson, J.T., et al. (2012) PMID:21221095 doi: 10.1038/nbt.1754 | IGV_Linux_2.11.0 | See section ‘DamID-seq data analysis’ in ‘Materials and Methods’. |
| Software, algorithm | DAVID | Site: https://david.ncifcrf.gov | Releases 6.8 and Dec 2021 | See section ‘DamID-seq data analysis’ in ‘Materials and Methods’. |
| Software, algorithm | FIJI | Schindelin et al., 2012 PMID:22743772 doi: 10.1038/nmeth.2019 | FIJI version 2.3.051 | See section ‘Image analysis’ in ‘Materials and Methods’. |
| Software, algorithm | GraphPad Prism | GraphPad | GraphPad Prism version 9.2.0 | See section ‘DamID-seq data analysis’ in ‘Materials and Methods’. |
| Software, algorithm | Adobe Photoshop | Adobe | Adobe Photoshop 2022 version: 23.4.1 | |
| Software, algorithm | Adobe Illustrator | Adobe | Adobe Illustrator 2022 Version 26.3.1 |
Additional files
-
Supplementary file 1
Sequences deleted in the slp double enhancer deletion line.
- https://cdn.elifesciences.org/articles/75879/elife-75879-supp1-v1.docx
-
Supplementary file 2
List of primers used for experiments described.
- https://cdn.elifesciences.org/articles/75879/elife-75879-supp2-v1.docx
-
Supplementary file 3
Predicted transcription factor binding sites by the web-based FIMO analysis.
- https://cdn.elifesciences.org/articles/75879/elife-75879-supp3-v1.xlsx
-
Supplementary file 4
List of gene blocks used for reporter assays and for cloning Ey coding sequence into the pUAST-Dam-attB plasmid.
- https://cdn.elifesciences.org/articles/75879/elife-75879-supp4-v1.docx
-
Supplementary file 5
Gene lists that have IDR peaks for both Su(H) and Ey, Ey only, and Su(H) only from the DamID-seq data.
- https://cdn.elifesciences.org/articles/75879/elife-75879-supp5-v1.xlsx
-
Supplementary file 6
DAVID analysis of gene lists that have IDR peaks for both Su(H) and Ey, Ey only, and Su(H) only from the DamID-seq data.
- https://cdn.elifesciences.org/articles/75879/elife-75879-supp6-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/75879/elife-75879-transrepform1-v1.pdf





