Metabolic basis for the evolution of a common pathogenic Pseudomonas aeruginosa variant
Figures

Mathematical model built from monoculture growth data is sufficient to explain the rise of LasR loss-of-function strains.
(A) Predicted densities (left y-axis) of mathematical model shown for wild type (WT, dashed line) and LasR– (solid line) strains. Predicted percentages (right y-axis) of LasR– (beige fill) and LasR+ (green fill) strains over the course of evolution regime in LB with passage (Pn) every 2 days. Table shows experimentally measured growth parameters used to create the model as obtained for strains PA14 WT and ∆lasR grown in 5 mL LB cultures on a roller drum. (B) Percentage of LasR– phenotypes observed in n ≥ 4 independent evolution experiments in LB. Different shapes represent independent experiments, and same shapes represent technical replicates. A representative image of the smooth LasR+ and sheen LasR– colonies from Day 6 is shown. (C) lasR alleles detected in the population at Day 4 (4 d) and Day 6 (6 d) by PoolSeq within the lasR coding sequence, which includes the autoinducer binding and DNA binding domains, for a representative experiment (diamond symbols, in B). The percentages of each allele and the sum (i.e. % total) is indicated for each replicate culture. Each color represents a different allele. (D) LasR regulates the production of its cognate autoinducer 3OC12HSL via direct transcriptional control of the gene encoding the LasI synthase. LasI-produced autoinducer activity of evolved pools from a representative experiment (diamond symbols, in B) at days 2 and 6. Activity is presented as a percentage of that produced by unevolved WT monocultures. The levels produced by the engineered ∆lasR control strain is shown for reference (dotted line). **, p = 0.0015 as determined by two-tailed, upaired t-test. (E) Comparison of predicted (dashed line) and observed (solid line) outcomes of competition assays initiated at different initial ratios for which a constitutively tagged WT (att::lacZ) was competed against ∆lasR (beige, gray line) or WT (control, green) competitors for 6 h (final) in planktonic LB cultures with 95% confidence intervals shown for best fit line (quadratic).
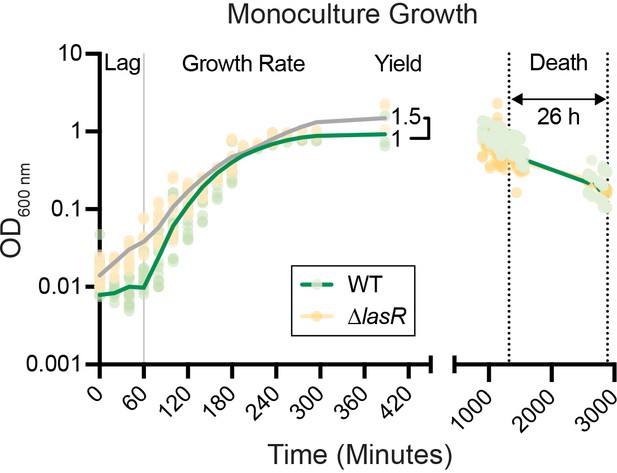
Experimentally determined growth parameters of PA14 wild type and ∆lasR monocultures in LB.
Density measurements of strains PA14 wild type (WT, green) and ∆lasR (beige, gray) monocultures grown in 5 mL LB on a roller drum over time. Lag, growth rate, yield, duration of death, and death rate were determined for use in the mathematical model (Figure 1A).
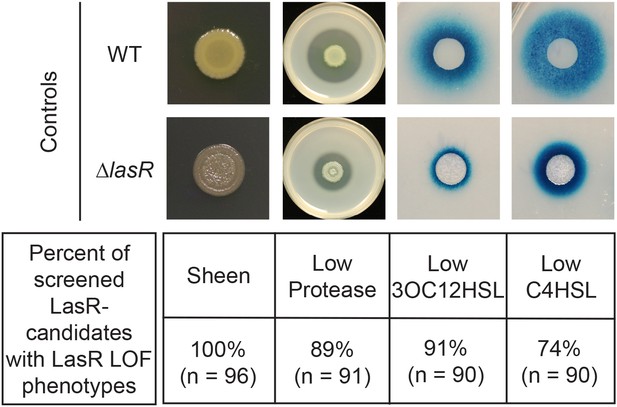
Phenotype analysis of sheen LasR– candidates isolated from the evolution experiments in LB.
LasR loss-of-function (LOF) candidates picked on basis of sheen colony morphology from evolution were screened for phenotypes associated with LasR– strains including low protease activity on milk plates and low levels of acyl homoserine lactone production as measured by ∆lasI∆rhlI bioreporters responsive to LasR-regulated autoinducer 3-oxo-C12-homoserine lactone (3OC12HSL) and RhlR-regulated autoinducer N-butyryl-L-homoserine lactone (C4HSL).
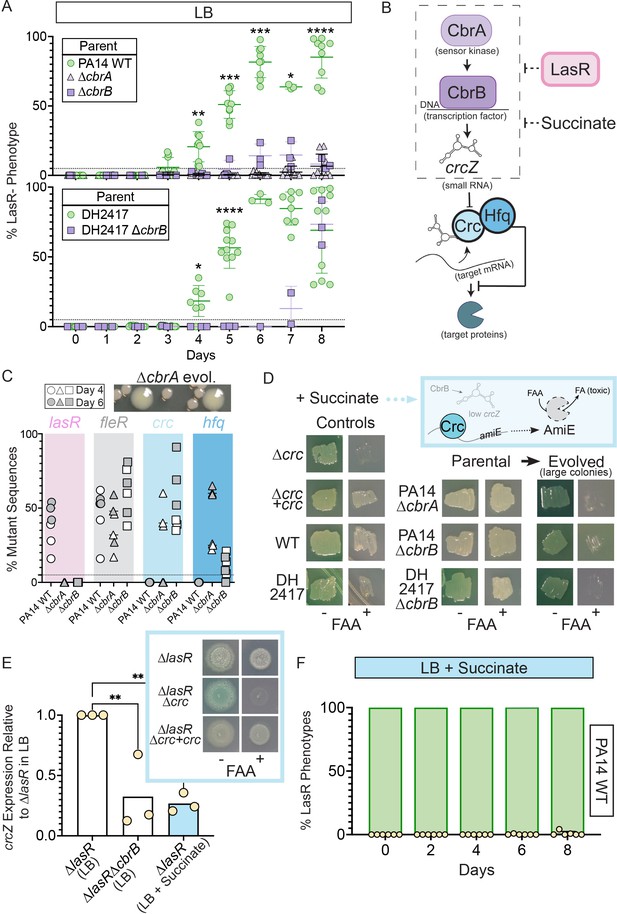
Activity of the carbon catabolite repression system is required for LasR– selection in LB.
(A) The percentage of colonies with LasR– phenotypes enumerated over the course of evolution for ∆cbrA or ∆cbrB mutants (purple triangle and square, respectively) in strains PA14 or a LasR+ cystic fibrosis isolate (DH2417) relative to ‘wild-type‘ comparators. PA14 WT strain data is the same as in Figure 1B (n ≥ 3). Statistical significance was determined between percent LasR– phenotypes in CbrA/B + and cbrA/B mutant pools each day via Two-Way ANOVA with Dunnet’s multiple hypothesis correction. For all panels: *, p < 0.05; **, p < 0.005; ***, p < 0.0005; ****, p < 0.0001. (B) The carbon catabolite repression system promotes the preferential consumption of succinate (and other preferred substrates) through the two-component system CbrAB. CbrA activates its response regulator CbrB which directly induces expression of the small RNA crcZ. crcZ sequesters Crc thereby allowing translation of the target gene to occur. Often the target gene enables the utilization of specific (i.e. less preferred) substrates. In a catabolite repressed state, such as when succinate is present, Crc binds to target mRNA with the RNA binding protein Hfq and blocks translation. CbrB protein levels are higher in strains lacking LasR function, but the mechanism linking these pathways is uncharacterized. (C) Percent total mutant alleles in lasR (pink bar), fleR (gray bar), crc (light blue bar), and hfq (darker blue bar) in a representative experiment (Figure 1B, diamond symbols) for PA14 wild type, ∆cbrA, ∆cbrB evolved populations sequenced on Day 4 (white filled symbol) and six (gray filled symbol). Representative image of the larger colony morphologies observed in the evolved pools from CbrA/B- deficient strains (∆cbrA shown) above. (D) Crc represses amiE encoding an amidase that can turnover the fluoroacetamide (FAA) protoxin to fluoroacetate (FA) mediating cell death. In the presence of succinate, cells with functional Crc survive in the presence of FAA. PA14 WT, PA14 ∆cbrA, PA14 ∆cbrB, and DH2417 WT strains were included as controls. The ∆cbrA and ∆cbrB parental strains used for the evolution experiments and representative colonies that emerged with a larger colony size in these backgrounds were patched (or struck out) onto succinate containing plates in the absence and presence of the FAA protoxin. (E) crcZ expression of PA14 ∆lasR in LB (white bar) and LB with 40 mM succinate (blue bar) measured by qRT-PCR and plotted relative to expression of ∆lasR in LB (n = 4). Inset shows representative image of ∆lasR, ∆lasR∆crc, and ∆lasR∆crc +crc grown on succinate containing plates in the absence and presence of FAA. (F) Percentage of colonies with LasR– phenotypes observed in evolution experiment initiated with strain PA14 WT in LB supplemented with 40 mM succinate (n = 6). P-values of 0.007 and 0.005 for comparison of ∆lasR∆cbrB (LB) and ∆lasR (LB +succinate) relative to ∆lasR grown in LB as determined by ordinary one-way ANOVA with Dunnett’s multiple comparison test.
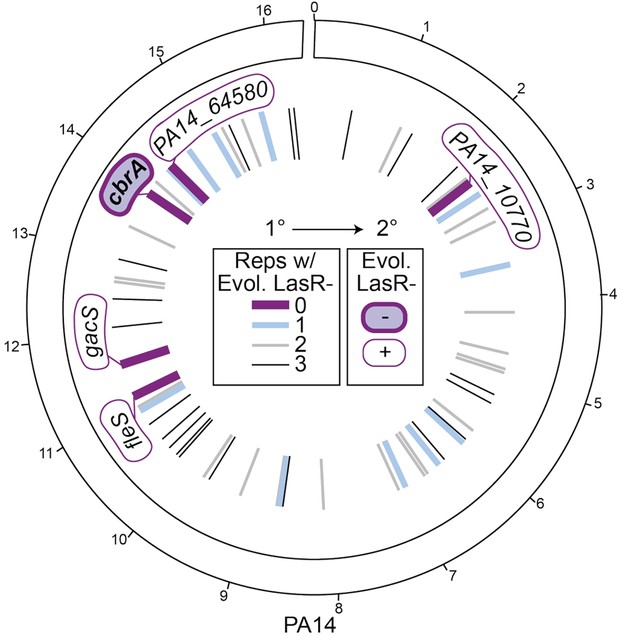
Screen reveals specific requirement of CbrA, and not other sensor kinases encoded in the PA14 genome for LasR– strain selection.
Circos plot of the PA14 genome, with the genomic location of the genes deleted to create the kinase mutant collection indicated by lines in the inner circle (and noted in Supplementary file 2); outer ticks indicate 4 × 105 bp genome increments. The primary screen (1°) was performed in a 96-well plate format in LB with each strain in triplicate. Deletion backgrounds that had zero, one, two, or three replicates containing LasR– phenotypes at the time of plating (i.e. when all wild-type controls had >50% colonies with LasR– phenotypes) are represented as lines colored purple, blue, or gray, respectively, with decreasing line thickness. Deletion mutants that were found to have zero colonies with LasR– phenotypes (thick dark purple lines, gene names indicated) were secondarily screened in 5 mL LB evolution assays (Figure 2—figure supplement 2A); only ∆cbrA (filled in purple) was negative (-) for LasR– phenotypes in the secondary screen (2°).
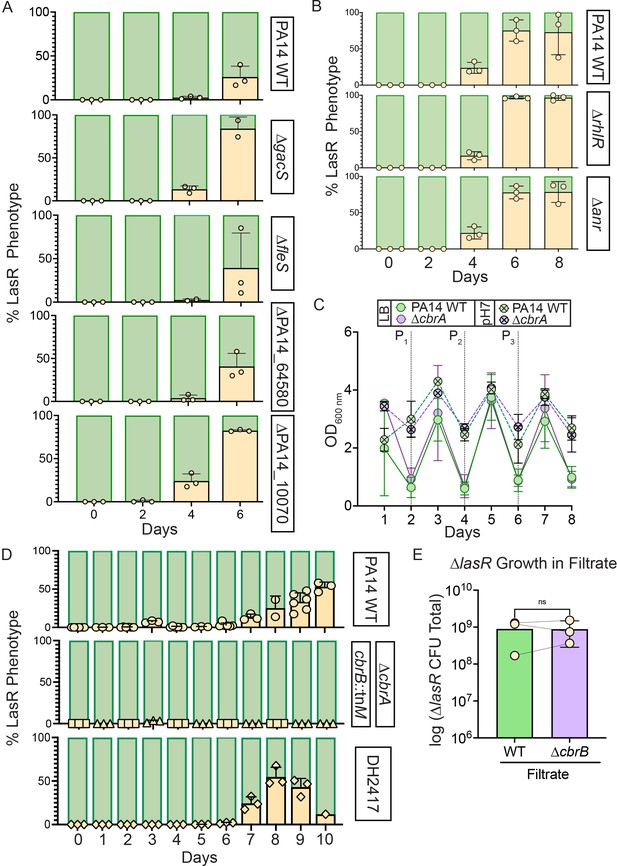
LasR– strains evolve in all tested strain backgrounds except for those deficient in cbrAB, and this is independent of cellular density, lysis, and filtrate toxicity.
(A) Kinase deletion mutant backgrounds that did not evolve LasR– phenotypes in any replicate of a 96-well evolution (Figure 2—figure supplement 1) were secondarily screened for the appearance of LasR– colonies in 5 mL cultures. The colony phenotypes were quantified over time with percentage of LasR– phenotypes indicated (beige). (B) The percent of colonies with LasR– phenotypes observed for LB evolution experiments initiated with strains PA14 ∆rhlR or ∆anr relative to wild type. (C) Optical density for PA14 wild type (WT, green) or ∆cbrA (purple) cultures over course of evolution in LB (circles, n ≥ 9) or LB buffered to pH 7 with HEPES (circle with ‘x’, n ≥ 3). Points represent the average and error bars, standard deviation. Statistical significance between WT and ∆cbrA determined by two-way ANOVA with Šídák’s multiple comparisons test for LB and buffered LB datasets separately. For LB: Day 1, p < 0.0001. For buffered LB: Day 1, p = 0.023 and Day 6, p < 0.009. All other comparisons are non-significant. D. Percentage of colonies with LasR– phenotypes over the course of evolution in buffered LB for PA14 WT (circles), ∆cbrA (triangles) or cbrB::tnM (squares), or DH2417 LasR +clinical isolate (diamonds) (n ≥ 3). E. Density (i.e. total colony forming unit counts) of PA14 ∆lasR after 24 hr of growth in filtrate from saturated PA14 wild type or ∆cbrB cultures. ns, not significant as determined by Student’s paired two-tailed t-test. P-value = 0.9597.
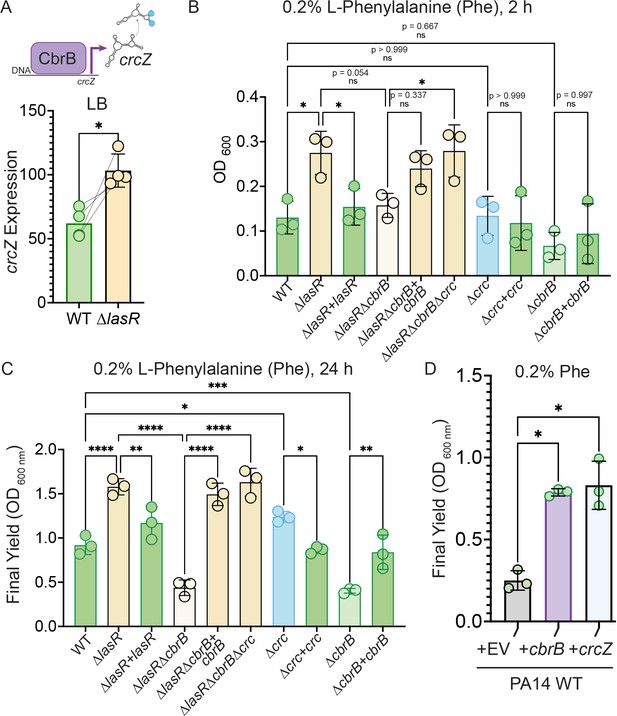
Increased CbrB activity of LasR– strains is necessary and sufficient to promote growth on non-repressive substrates like phenylalanine via Crc.
(A) CbrB promotes the transcription of crcZ, and crcZ thus can be a readout of CbrB transcriptional activity. crcZ expression was measured by qRT-PCR relative to the average expression of the housekeeping genes rpoD and rpsL in cultures of PA14 WT and ∆lasR strains grown to OD600 nm = 1 from four independent experiments. *, p = 0.0334 as determined by Student’s paired two-tailed t-test. (B) Early growth (2 hr) and (C) Final yield (24 hr) on phenylalanine (Phe) as a sole carbon source shows enhanced growth for ∆lasR, cbrB dependence, and the requirement for cbrB is abolished by deletion of crc. Each point is the average of three replicates, repeated three independent days. Statistical significance determined by one-way ANOVA with Šídák’s multiple comparisons test. ns, not significant. *, p < 0.05. **, p < 0.005. ****, p < 0.0001. (D) Final yield on Phe under (0.2%) arabinose-inducing conditions for the PA14 WT strain expressing an empty vector or crcZ, and cbrB overexpression constructs. Each point is the average of three replicates, performed on three separate days. Statistical significance determined by one-way ANOVA with multiple hypothesis correction as above.
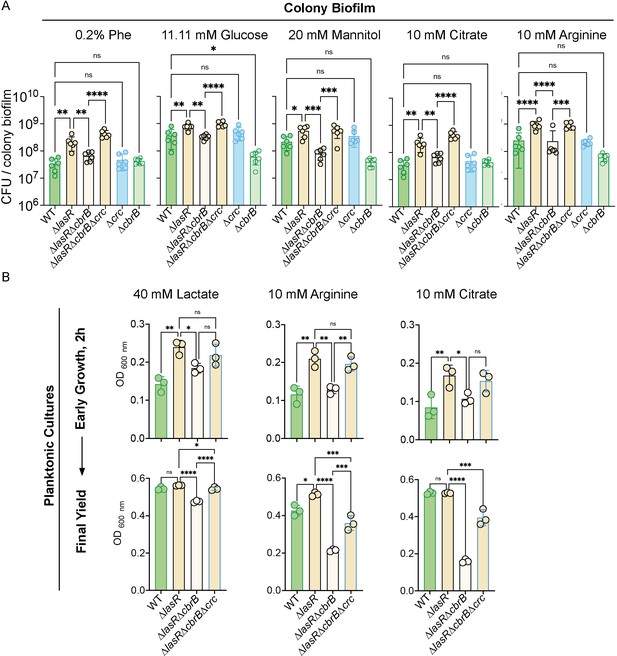
∆lasR strains have CbrB-dependent growth advantages that can be restored via loss of crc.
(A) Colony biofilm colony forming units (CFUs) were enumerated for strains PA14 WT, ∆lasR, ∆lasR∆cbrB, ∆lasR∆cbrB∆crc, ∆crc, and ∆cbrB after 16 hr on M63 minimal medium containing 0.2% phenylalanine, 20 mM mannitol, or 0.2% glucose, which have been well studied in the context of carbon catabolite repression (i.e. the CbrAB pathway) in addition to 10 mM citrate and 10 mM arginine. Bottom of y-axis set to starting inoculum density. P-values: ns, not significant; *, p < 0.05; **, p <; ***, p < 0.0007; ****, p < 0.0001 as determined by ordinary one-way ANOVA with Šídák’s multiple comparisons test. Each data point of colony count (CFU) was collected on a separate day (n = 6). (B). Early planktonic growth (2 hr post inoculation, top) and final yields on different carbon sources (bottom): 10 mM arginine (post 8 hr), 40 mM lactic acid (post 8 hr), and 10 mM citrate (post 7 hr). Each point displays the average growth (OD 600 nm) across three independent experiments with three replicates per day. P-values: ns, not significant; *, p < 0.05; **, p <; ***, p < 0.0007; ****, p < 0.0001 as determined by ordinary one-way ANOVA with Šídák’s multiple comparisons test.
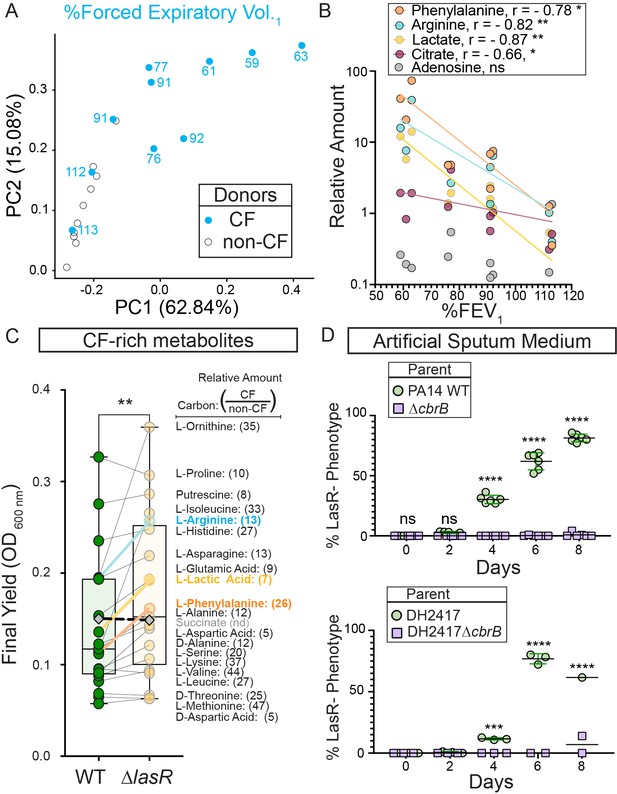
CbrB-dependent growth advantages may contribute to lasR mutant selection in distinct nutrient profiles of progressive cystic fibrosis airways.
(A) The first two dimensions (PC1 and PC2) of a principal component analysis of log normalized metabolite counts from bronchoalveolar lavage (BAL) samples collected from cystic fibrosis (CF, blue filled) and non-cystic fibrosis (non-CF, gray open) donors explain 62.84% and 15.08% of the variation in the data, respectively. PC1 separates the metabolite data by relative lung function as measured by percent forced expiratory volume in 1 s (%FEV1) for samples from people with CF. The %FEV1 is overlayed for CF-donor samples with text. Samples from non-CF donors group more closely with CF-donor samples that have high lung function. (B) Spearman correlation analysis of the relative phenylalanine (orange), arginine (aqua), lactate (yellow), citrate (magenta), and adenosine (gray) metabolite counts in the BAL samples relative to %FEV1 with exact p-values of 0.010, 0.005, 0.002, 0.041, and 0.714, respectively. Best fit semilog lines shown for visual clarity. (C) Comparison of the final yield measured after 24 hr for strains PA14 wild type and ∆lasR across a subset of carbon sources in BIOLOG growth assays for which the metabolite was found to be in higher abundance in CF-donor relative to non-CF donor BAL samples. Bold font indicates carbon sources analyzed in Figure 3 and Figure 3—figure supplement 1. Number in parenthesis refers to the ratio of the average counts for each metabolite in CF relative to non-CF samples. **, p-value = 0.003 as determined by two-tailed paired t-test comparing growth across CF-enriched metabolites between the wild type and ∆lasR strains. Succinate (gray diamond, black dashed line) was not detected (nd) in the BAL samples and thus not included in the statistical analysis, but the growth data is shown for reference. (D) Observed percentage of colonies with LasR– phenotypes over the course of evolution from strains (top) PA14 WT or (bottom) CF isolate (both green circles) with ∆cbrB (purple squares) derivatives in artificial sputum medium (ASM), which was designed to recapitulate the CF lung nutritional profile. ns, not significant (p-value > 0.9); ***, p = 0.0008; ****, p < 0.0001 as determined by ordinary two-way ANOVA with Šídák’s multiple comparisons test.
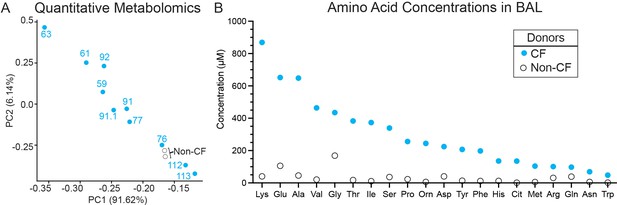
Quantitative amino acid analysis.
(A) The first two components (PC1 and PC2) of a principal component analysis of log normalized amino acid concentrations measured in bronchoalveolar lavage (BAL) fluid collected from cystic fibrosis (CF, blue filled circles) and non-cystic fibrosis (Non-CF, black open circles) donors by the Biocrates AbsoluteIDQ p180 Kit explain 91.62% and 6.14% of the variation in the data, respectively. As with the relative metabolite counts measured by LC/MS used in Figure 4A, PC1 of the amino concentrations measured by the Biocrates AbsoluteIDQ p180 Kit separates BAL samples by the respective percent forced expiratory volume in 1 s (%FEV1) (overlayed text). (B) Average amino acid concentrations (µM) measured from BAL samples collected from cystic fibrosis (CF, blue) and non-cystic fibrosis (Non-CF, black) donors. See Supplementary file 5 for data by sample.

CbrAB activity contributes to the positive selection of LasR– strains in complex media.
LasR– strain fitness relative to wild type is shown across growth phases, including lag, exponential growth, stationary, and death phases. Relative fitness of the LasR– strain (dotted black line) is calculated from the experimentally determined monoculture growth data of strains PA14 wild type (WT) and ∆lasR over time. Values above one indicate a LasR– strain fitness advantage over the WT strain during that growth phase. Circled insets show representative cartoons of LasR- (beige) and LasR+ (green) cells at each growth phase to indicate dividing or lysing cells (burst cells) across growth stages. The heights of the peaks or valleys of the relative fitness lines can be altered by several modulating factors including those that contribute to the positive and negative selection of LasR– strains. Other modulating factors reported or suggested in the literature include inter- and intra- species competition, extracellular protease, immunoclearance, and oxygenation which are likely condition dependent. In the absence of CbrA or CbrB (CbrAB-, dotted purple line) or in the presence of succinate (one CbrAB repressive substrate), the relative growth of LasR– strains is lower resulting in a reduction in the observed selection. This could be partially relieved in the CbrAB- background through disruption of Crc or Hfq function (blue dotted line), restoring activity through the pathway.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Pseudomonas aeruginosa) | PA14 | NCBI | Accession: GCF_000014625.1 | Reference file |
| Strain, strain background (Saccharomyces cerevisiae) | Saccharomyces cerevisiae | PMID:16820502 | Cloning yeast | |
| Strain, strain background (Escherichia coli) | DH5a | Invitrogen | Electrocompetent cells | |
| Strain, strain background (Escherichia coli) | S17 λpir | PMID:8226632 | Electrocompetent cells made in lab | |
| Strain, strain background (P. aeruginosa) | PA14 WT | PMID:7604262 | Hogan Laboratory reference strain | |
| Strain, strain background (P. aeruginosa) | DH2417; NC-AMT0101-1-2 | PMID:16687478 | Chronic CF lung infection isolate with functional LasR allele | |
| Strain, strain background (P. aeruginosa) | PA14 WT | PMID:33771779 | Laub Lab; strain background of kinase clean deletion mutants | |
| Genetic reagent (P. aeruginosa) | PA14 ∆lasR | PMID:15554963 | ||
| Genetic reagent (P. aeruginosa) | PAO-MW1qsc102 | PMID:10570171; PMID:11544214 | AHL-sensing bioreporter; PAO1 lasIrhlI mutant with Tn5-B22, which contains promoterless lacZ located within PA1896 (hypothetical protein) at chromosomal location of 2,067,716. Responsive to 3OC12-HSL but not C4-HSL. | |
| Genetic reagent (P. aeruginosa) | PAO-MW1qsc131 | PMID:10570171; PMID:11544214 | AHL-sensing bioreporter; PAO1 lasIrhlI mutant with Tn5-B22, which contains promoterless lacZ, under phzC promoter control. Responsive to either 3OC12-HSL or C4-HSL but requires both for full activation. | |
| Genetic reagent (P. aeruginosa) | PA14 WT att::lacZ | PMID:31980538 | PA14 WT with constitutive expression of lacZ for competition assays | |
| Genetic reagent (P. aeruginosa) | PA14 ∆cheA | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆chpA | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆creC | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆uhpB | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆bfiS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆bphP | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_10770 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_11630 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆rocS1 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆narX | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆wspE | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_19340 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆mxtR | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆cpxS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆gtrS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_24340 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆rocS2 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_26810 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆sagS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆copS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆pfeS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆bqsS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_30700 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_30840 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆czcS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_32570 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_36420 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆ercS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆exaD | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆ercS’ | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆parS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆kdpD | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_43670 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_45590 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_45870 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_46370 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_46980 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_48160 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆phoQ | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_49420 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆fleS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆pirS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆gacS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆tctE | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆pprA | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆colS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_57170 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆roxS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆rcsC | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆pvrS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆pilS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆cbrA | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆pmrB | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆retS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_64580 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆aruS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆ntrB | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_68230 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆amgS | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆algZ | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆phoR | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆kinB | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆PA14_72740 | PMID:33771779 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆rhlR | PMID:30936375 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆anr | PMID:31527114 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆cbrB | This paper | PA14 WT with in-frame deletion of cbrB (PA14_62540) | |
| Genetic reagent (P. aeruginosa) | PA14 cbrB::TnM | PMID:22911607; PMID:16477005 | cbrB MAR2xT7 transposon insertion mutant | |
| Genetic reagent (P. aeruginosa) | DH2417∆cbrB; NC-AMT0101-1-2∆cbrB | This paper | CF clinical isolate NC-AMT0101-1-2 (DH2417) with in-frame deletion of cbrB (PA14_62540) | |
| Genetic reagent (P. aeruginosa) | PA14 ∆crc | This paper | PA14 WT with in-frame deletion of crc (PA14_70390) | |
| Genetic reagent (P. aeruginosa) | PA14 ∆crc + crc | This paper | PA14 ∆crc with complementation of crc (PA14_70390) at the native locus | |
| Genetic reagent (P. aeruginosa) | PA14 ∆lasR + lasR | PMID:31980538 | ||
| Genetic reagent (P. aeruginosa) | PA14 ∆cbrB + pMQ70 cbrB (plasmid) | This paper | PA14 ∆cbrB expressing arabinose inducible pMQ70 cbrB expression vector | |
| Genetic reagent (P. aeruginosa) | PA14 ∆lasR∆cbrB | This paper | PA14 ∆lasR with in-frame deletion of cbrB (PA14_62540) | |
| Genetic reagent (P. aeruginosa) | PA14 ∆lasR∆cbrB + cbrB | This paper | PA14 ∆lasR∆cbrB with complementation of cbrB (PA14_62540) at the native locus | |
| Genetic reagent (P. aeruginosa) | PA14 ∆lasR∆cbrB∆crc | This paper | PA14 ∆lasR∆cbrB with in-frame deletion of crc (PA14_70390) | |
| Genetic reagent (P. aeruginosa) | PA14 ∆lasR∆crc | This paper | PA14 ∆lasR with in-frame deletion of crc (PA14_70390) | |
| Genetic reagent (P. aeruginosa) | PA14 ∆lasR∆crc + crc | This paper | PA14 ∆lasR∆crc with complementation of crc (PA14_70390) at the native locus | |
| Genetic reagent (P. aeruginosa) | PA14 + pMQ72 EV | This paper | PA14 WT expressing pMQ72 empty vector | |
| Genetic reagent (P. aeruginosa) | PA14 + pMQ70 cbrB | This paper | PA14 WT expressing arabinose inducible pMQ70 cbrB expression vector | |
| Genetic reagent (P. aeruginosa) | PA14 + pMQ72 crcZ | This paper | PA14 WT expressing arabinose inducible pMQ72 crcZ expression vector | |
| Recombinant DNA reagent | pMQ30 EV | PMID:16820502 | ||
| Recombinant DNA reagent | pMQ72 EV | PMID:16820502 | ||
| Recombinant DNA reagent | pMQ70 EV | PMID:16820502 | ||
| Recombinant DNA reagent | pMQ30_cbrB_KO | This paper | pMQ30 plasmid for knocking out cbrb | |
| Recombinant DNA reagent | pMQ30_crc_KO | This paper | pMQ30 plasmid for knocking out crc | |
| Recombinant DNA reagent | pMQ30_cbrB_KON | This paper | pMQ30 plasmid for complementing cbrb at the native locus | |
| Recombinant DNA reagent | pMQ30_crc_KON | This paper | pMQ30 plasmid for complementing cbrb at the native locus | |
| Recombinant DNA reagent | pMQ72_crcZ | This paper | pMQ72 plasmid backbone with arabinose inducible crcZ expression | |
| Recombinant DNA reagent | pMQ70_cbrB | This paper | pMQ70 plasmid backbone with arabinose inducible cbrB expression | |
| Sequence-based reagent | pMQ72 crcZ OE 1 F | This paper | PCR Primers; construct design | gtttctccatacccgtttttttgggctagcGCACAACAACAATAACAAGCAACGACGAAG |
| Sequence-based reagent | pMQ72 crcZ OE 2 R | This paper | PCR Primers; construct design | ctagaggatccccgggtaccgagctcgaattcgaaatggtgtaaggcgaaggaaaaacgg |
| Sequence-based reagent | pMQ70 cbrB OE 1 F | This paper | PCR Primers; construct design | ctctctactgtttctccatacccgtttttttgggctagcgAGACGAGCgaattcACGTCGAGAGAGCtgaatacatggcac |
| Sequence-based reagent | pMQ70 cbrB OE 2 R | This paper | PCR Primers; construct design | ttgcatgcctgcaggtcgactctagaggatccccgggtacGTAACAGGTTGCAGGGTaccGTtacgagtcggccgaggcccc |
| Sequence-based reagent | pMQ30 cbrB KO 1 F | This paper | PCR Primers; construct design | taacaatttcacacaggaaacagctatgaccatgattacgaattcAGGAAGTGCTGATGTGGAACC |
| Sequence-based reagent | pMQ30 cbrB KO 2 R | This paper | PCR Primers; construct design | GTAACAGGTTGCAGGGTGTTTATTCAGCTCTCTCGACGTGCT |
| Sequence-based reagent | pMQ30 cbrB KO 3 F | This paper | PCR Primers; construct design | CACGTCGAGAGAGCTGAATAAACACCCTGCAACCTGTTACC |
| Sequence-based reagent | pMQ30 cbrB KO 4 R | This paper | PCR Primers; construct design | aggtcgactctagaggatccccgggtaccgagctcgaattcCAGGGAGTGCTGGTTGTTACCGATGACTtc |
| Sequence-based reagent | pMQ30 crc KO 1 F | This paper | PCR Primers; construct design | taacaatttcacacaggaaacagctatgaccatgattacgaattcTGGAATACAGGCGCAGCAac |
| Sequence-based reagent | pMQ30 crc KO 2 R | This paper | PCR Primers; construct design | TAGAAAAGCCGGCGCATGCGCTGGCTTTTTCGTGTCTGACGGGGCAAATGGCCCCCAAAATCACGTGCG |
| Sequence-based reagent | pMQ30 crc KO 4 R | This paper | PCR Primers; construct design | ctgcaggtcgactctagaggatccccgggtaccgagctcgaattcttggctgaccgccgagtacggcatgc |
| Sequence-based reagent | pMQ30 crc KO 3 F | This paper | PCR Primers; construct design | TTTGAGCTCGGGTATCATACACGCACGTGATTTTGGGGGCCATTTGCCCCGTCAGACACGAAAAAGCCAG |
| Sequence-based reagent | cbrB check F | This paper | PCR primer, KO check | GCGTCTGCTCCCTGGCCAAG |
| Sequence-based reagent | cbrB check R | This paper | PCR primer, KO check | GTGGCGCTGGTGGCGACATC |
| Sequence-based reagent | crc check F | This paper | PCR primer, KO check | GCTCGATGGCGAAACGAATG |
| Sequence-based reagent | crc check R | This paper | PCR primer, KO check | GCGCTGGTGTTGACCATCATC |
| Sequence-based reagent | crcZ RT 1 F | PMID:31911486 | PCR Primers | GCACAACAACAATAACAAGCAACG |
| Sequence-based reagent | crcZ RT 2 R | PMID:31911486 | PCR Primers | AGTTTTATTCTTCTTCCGACTGGCT |
| Sequence-based reagent | rpsL RT1F | PMID:31911486 | PCR Primers | GTAAGGTATGCCGTGTACG |
| Sequence-based reagent | rpsL RT 2 R | PMID:31911486 | PCR Primers | CACTACGCTGTGCTCTTG |
| Sequence-based reagent | rpoD RT 1 F | PMID:30936375 | PCR Primers | CGCCGAGATCAAGGAAATCA |
| Sequence-based reagent | rpoD RT 2 R | PMID:30936375 | PCR Primers | TACTTCTTGGCGATGGAAATCA |
| Commercial assay, kit | NEBuilder HiFi DNA Assembly | Biolabs | Cat. #: E2621L | Gibson cloning |
| Commercial assay, kit | Master Pure Yeast DNA purification kit | Lucigen | Cat. No.: MPY80200 | |
| Commercial assay, kit | RNAeasy Mini kit | QIAGEN | Cat. No.: 74,104 | |
| Commercial assay, kit | Turbo DNA-free kit | Thermo Fisher Scientific | AM1907 | |
| Commercial assay, kit | RevertAid H Minus First Strand cDNA synthesis | Thermo Scientific | Cat. No.: EP0451 | cDNA synthesis with IDT random hexamer |
| Commercial assay, kit | SsoFast EvaGreen Supermix | BIO-RAD | Cat.#: 1725201 | |
| Commercial assay, kit | Zymoprep Yeast Plasmid Miniprep II | Zymo Research | Cat. No.: D2004 | Yeast cloning |
| Commercial assay, kit | Biocrates AbsoluteIDQ p180 kit | biocrates | Amino acid. quantification | |
| Chemical compound, drug | Brain Heart Infusion | BD | SKU:211,059 | BBL Brain Heart Infusion |
| Chemical compound, drug | Agar | BD | SKU:214,510 | Difco Agar, granulated (2 Kg pail) |
| Chemical compound, drug | XGAL; 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside | Research Products International | B71800-5.0 | Stock dissolved in DMSO |
| Chemical compound, drug | Fluoroacetamide | Aldrich | 128341–5 G | dissolved in water, filter sterilized |
| Chemical compound, drug | Lactamide | Tokyo Chemical Industry | Product Number: L0001 | |
| Chemical compound, drug | Mannitol | Sigma | M8429-500G | D-Mannitol |
| Chemical compound, drug | Succinate | Sigma | S9512-500G | Succinic acid, to pH7 with NaOH |
| Chemical compound, drug | Phenylalanine | Sigma-Aldrich | P2126-100G | L-Phenylalanine |
| Chemical compound, drug | Arginine | Sigma | A5131-100G | L-arginine monohydrochloride |
| Chemical compound, drug | Lactate | Fisher Scientific | S326-500 | Sodium lactate syrup, 60% w/w |
| Chemical compound, drug | Glucose | VWR Chemicals | BDH9230-2.5KG | Dextrose, anhydrous |
| Chemical compound, drug | Citrate | FisherScientific | A104-500 | Citric acid, monohydrate |
| Chemical compound, drug | Gentamicin | Research Products International | G38000-10.0 | Gentamicin Sulfate |
| Chemical compound, drug | Nalidixic acid | Research Products International | N42000-25.0 | |
| Chemical compound, drug | Carbinicillin | Goldbio | C-103–25 | Carbenicillin (Disodium) |
| Chemical compound, drug | Sucrose | Fisher BioReagents | BP220-212; 2.5 kg | D-Sucrose |
| Chemical compound, drug | HEPES | SIGMA | H3375-100G | buffer |
| Chemical compound, drug | Tryptone | Fisher Bioreagents | BP1421-500 | LB |
| Chemical compound, drug | NaCl; sodium chloride | Fisher Chemical | S271-3 | LB |
| Chemical compound, drug | Yeast extract | Fisher Bioreagents | BP1422-500 | LB |
| Chemical compound, drug | Ammonium sulfate | Fisher Chemical | A702-500 | M63 |
| Chemical compound, drug | Potassium phosphate monobasic | Fisher Chemical | P382-500 | M63 |
| Chemical compound, drug | Potassium phosphate dibasic, anhydrous | Fisher Chemical | P288-500 | M63 |
| Chemical compound, drug | Magnesium Sulfate | Fisher Scientific | M63-500 | M63 |
| Chemical compound, drug | Milk | BD | 232,100 | Difco Skim Milk |
| Chemical compound, drug | Yeast Nitrogen Base without amino acids | Research Products International | Y20040-500.0 | Yeast cloning |
| Chemical compound, drug | Yeast Synthetic Drop-out Medium Supplements without uracil | Sigma | Y1501-20G | Yeast cloning |
| Chemical compound, drug | Peptone | Fisher bioreagents | BP1420-500 | YPD |
| Chemical compound, drug | Sodium phosphate dibasic anhydrous | Fisher Chemicals | S374-500 | ASM |
| Chemical compound, drug | Sodium phosphate monobasic | Fisher Chemicals | S369-500 | ASM |
| Chemical compound, drug | Potassium Nitrate | Fisher Chemicals | M-12636 | ASM |
| Chemical compound, drug | Potassium Sulfate | Fisher Chemicals | P304-500 | ASM |
| Chemical compound, drug | L-(+)-Lactic acid | Sigma | L1750-10G | ASM; 1 M stock; pH to 7 with NaOH |
| Chemical compound, drug | Calcium chloride dihydrate | Sigma | C7902-500G | ASM |
| Chemical compound, drug | Magnesium Chloride Hexahydrate | Fisher chemical | M33-500 | ASM |
| Chemical compound, drug | FeSO4*7H2O | Sigma-Aldrich | F-8048 | ASM; Ferrous sulfate heptahydrate; Filter sterilized |
| Chemical compound, drug | N-acetylglucosamine | Fisher Scientific | AAA1304718 | ASM |
| Chemical compound, drug | DPPC | Sigma | P0763-250MG | ASM; 1,2-dipalmitoyl-sn-glycero-3-phosphocholine; dissolved in chloroform |
| Chemical compound, drug | Tryptophan | Sigma-Aldrich | T0254-25G | ASM, L-Tryptophan |
| Chemical compound, drug | Mucin | Sigma | M2378-100G | ASM; Mucin from porcine stomach, Type II |
| Peptide, recombinant protein | Phusion | New England BioLabs | M0530L | High-Fidelity DNA polymerase |
| Software, algorithm | MATLAB | MathWorks | ||
| Software, algorithm | breseq | PMID:24838886 | Version 0.35.4 | |
| Software, algorithm | bcl2fastq | Illumina | RRID:SCR_015058 | v2.20.0422 |
| Software, algorithm | GraphPad Prism 9 | GraphPad | Version 9.2.0 |
Additional files
-
Supplementary file 1
Unfixed mutations observed in Pool-Seq data from evolved populations.
Fixed mutations (i.e. those mutations present at 100% across all samples at all time points) were excluded from this list as potential background differences from reference strain. All unfixed mutations are listed with unique mutation ID that consists of gene name, mutation position on the genome and the nature of the mutation. ‡ symbol indicates multiple changes within the same codon. Gene identifiers, gene name and product name indicated in Columns 2–5 (i.e. B - E) for each unique mutation ID. Columns 6 through 14 (i.e. F - N) represent the difference in the fractions of sequenced reads for the listed variants from Day 4 to Day 6 (i.e. Day 6 fraction - Day 4 fraction) for each strain and replicate. Columns 14 - end (i.e. N - AE) represent the fraction of the sequenced reads with the listed variant. Columns are labeled by gene knockout_day_replicate_fraction or change.
- https://cdn.elifesciences.org/articles/76555/elife-76555-supp1-v2.xlsx
-
Supplementary file 2
Evolution screen for spontaneous LasR- strains in kinase deletion mutant backgrounds.
Number of replicates (three possible) with no LasR- phenotypes observed at time of plating in LB microtiter evolution assay for each clean deletion mutant. The locus tag, gene length, start and stop genomic position, gene name indicated were used to generate Circos plot shown in Figure 2—figure supplement 1. Deletion mutants that displayed no LasR- phenotypes across all three replicates were followed up in a secondary 5 mL LB evolution assay.
- https://cdn.elifesciences.org/articles/76555/elife-76555-supp2-v2.xlsx
-
Supplementary file 3
Bronchioalveolar lavage (BAL) fluid donor characteristics from people with and without cystic fibrosis (CF) used for metabolomics analyses.
Samples are grouped according to CF status with non-CF donors denoted ‘HV’ with age group, gender, CFTR genotype, and percent forced expiratory volume in 1 s at the time of encounter listed. nd, not determined.
- https://cdn.elifesciences.org/articles/76555/elife-76555-supp3-v2.docx
-
Supplementary file 4
Relative metabolite counts in bronchioalveolar lavage fluid from people with cystic fibrosis (CF) and non-CF comparators by LC/MS.
Values listed for each biochemical are normalized and imputed as described in the methods section. The percent forced expiratory volume in 1 s is noted for samples from people with CF as part of the column title. For Non-CF samples, column labels are listed numerically with ‘Non-CF’ indicator. Counts are shown per sample, as the average across CF (n = 10) and non-CF (n = 10) samples and as ratio of the averages between CF and non-CF as well. Information on (sub)pathway, biochemical identifiers, and methodology shown per biochemical.
- https://cdn.elifesciences.org/articles/76555/elife-76555-supp4-v2.xlsx
-
Supplementary file 5
Amino acid and biogenic amine concentrations in bronchioalveolar lavage (BAL) fluid from people with cystic fibrosis (CF) and non-CF comparators.
Values listed are micromolar concentrations for specified metabolites quantified in the BAL samples from people with and without CF (non-CF) using the Biocrates AbsoluteIDQ p180 Kit (n = 10 and 2, respectively). The percent forced expiratory volume in 1 s is noted for samples from people with CF as part of the column title. In addition to the concentrations listed per sample for each amino acid, the average is shown for samples from people with and without CF as well as a ratio of the averages.
- https://cdn.elifesciences.org/articles/76555/elife-76555-supp5-v2.xlsx
-
Supplementary file 6
Strains and plasmids used in this study.
List of strains and plasmids used in this study with internal lab strain identifier, short description of strain/use including gene name and gene number, and source listed.
- https://cdn.elifesciences.org/articles/76555/elife-76555-supp6-v2.docx
-
Supplementary file 7
Supplemental methods for the choice of parameters for modeling wild type and LasR- frequencies in passaged cultures.
Description of the terms for lag, growth, and death for both wild type strain PA14 and derived lasR mutants, and estimates of the number of lasR mutants in the initial population.
- https://cdn.elifesciences.org/articles/76555/elife-76555-supp7-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76555/elife-76555-transrepform1-v2.docx
-
Source code 1
Matlab script for deterministic mathematical model of LasR+ (wild type) strain PA14 and lasR mutant growth following density and percentage of each strain over the course of evolution regime as described in methods.
Model parameters include growth rate, carrying capacity/concentration of saturated culture, killing rate (lysis-growth), difference in lag time (hours), and killing time at the end (hours) for each strain. Code output used to generate Figure 1A.
- https://cdn.elifesciences.org/articles/76555/elife-76555-code1-v2.zip





