Changes in transmission of Enterovirus D68 (EV-D68) in England inferred from seroprevalence data
Figures
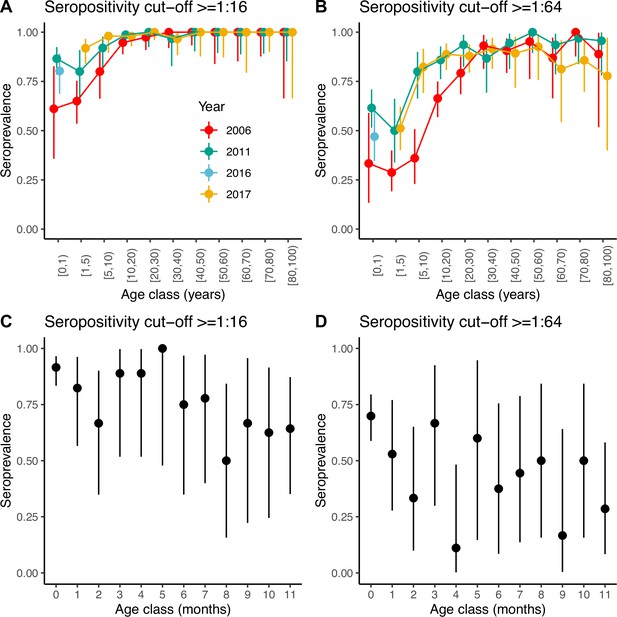
Seroprevalence by age.
Seroprevalence by age class in years for the three serosurveys and for two different cut-offs of neutralising antibody titre used to define positivity: (A) 1:16 and (B) 1:64. Seroprevalence by month during the first year of life for children across the three serosurveys combined and two seropositivity cut-offs: (C) 1:16 and (D) 1:64. Bars are 95% binomial confidence intervals. Note that in (A) and (B) individuals in the [0–1) age class from the 2017 serosurvey were in fact sampled in 2016 and as such, are shown with a different colour.
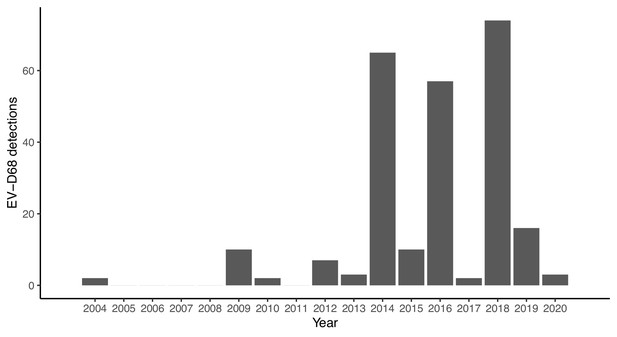
Annual number of Enterovirus D68 (EV-D68) detections reported by Public Health England (now UKHSA) between 2004 and 2020.
The data were extracted from three sources: for the years 2004–2011, from Kadambari et al., 2014; for 2012–2013, from a risk assessment report published by Public Health England, 2018; and for 2014–2020, we digitised data from Figure 2 in this website (Public Health England, 2023).
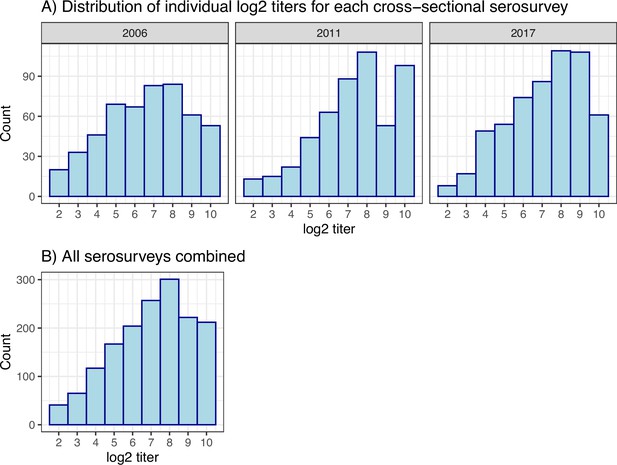
Individual log2 titre distributions.
(A) For each cross-sectional serosurvey. (B) For all three surveys combined.
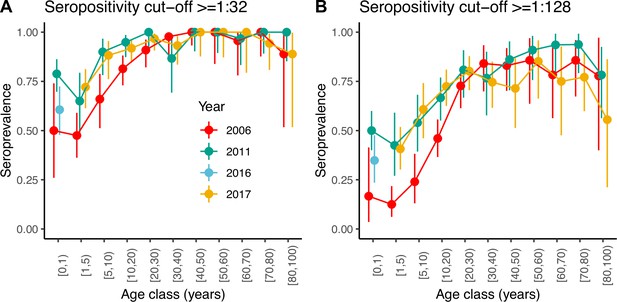
Seroprevalence by age class in years for the three serosurveys and two different cut-offs of neutralising antibody titre used to define positivity: (A) 1:32 and (B) 1:128.
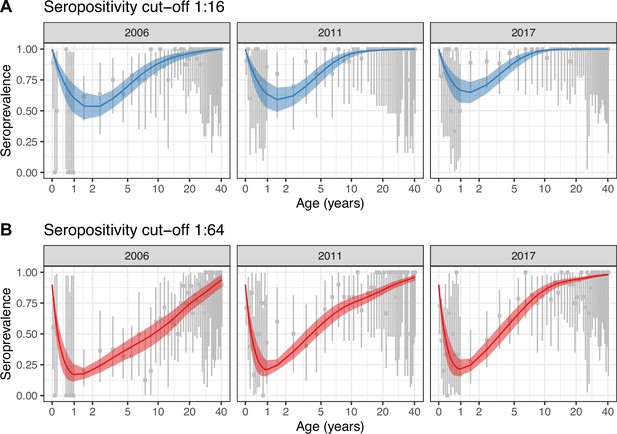
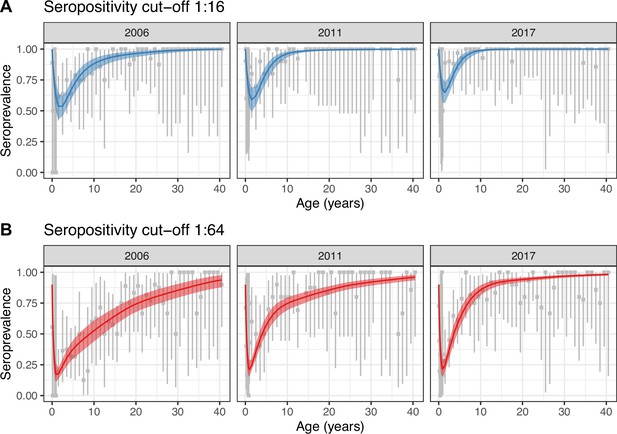
Best model fit to data.
Observed (gray) and estimated (blue and red) seroprevalence by age for the three cross-sectional serosurveys (2006, 2011, and 2017). Model fit is shown for the random walk model (Model 2) using the two seropositivity cut-offs, (A) 1:16 and (B) 1:64. Gray intervals indicate the 95% binomial confidence intervals around the observed proportion of seropositivity. The line and ribbons correspond to the median and 95% credible intervals of the posterior estimates of seroprevalence. Note that the age axis is log-transformed to better show the data in the younger age classes. The same plot with the age axis on a natural scale is shown in Figure 2—figure supplement 2. Note also that the data for <1 yo shown for 2017 were actually collected in 2016.
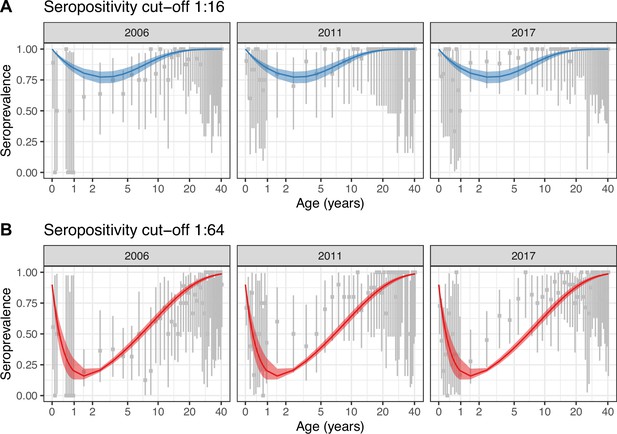
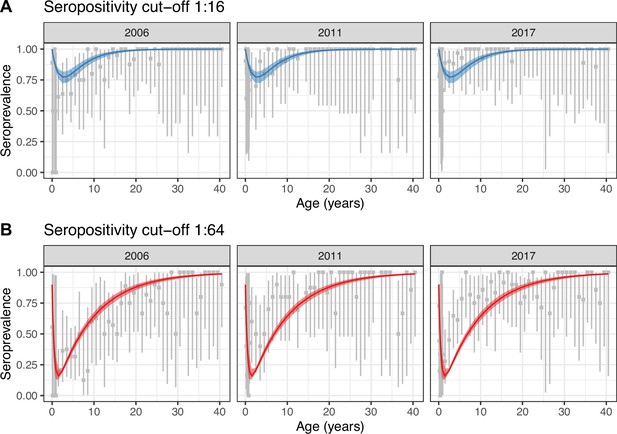
Constant model fit to data.
Observed (gray) and estimated (blue and red) seroprevalence by age for the three cross-sectional serosurveys (2006, 2011, and 2017). Model fit is shown for the constant model (Model 1) using the two seropositivity cut-offs, (A) 1:16 and (B) 1:64. Gray intervals indicate the 95% binomial confidence intervals around the observed proportion of seropositivity. The line and ribbons correspond to the median and 95% credible intervals of the posterior estimates of seroprevalence. Note that the age axis is log-transformed to better show the data in the younger age classes. The same plot with the age axis on a natural scale is shown in Figure 2—figure supplement 3. Note also that the data for <1 yo shown for 2017 were actually collected in 2016.
Same as Figure 2, with x-axis on a natural scale.
Same as Figure 2—figure supplement 1, with x-axis on a natural scale.
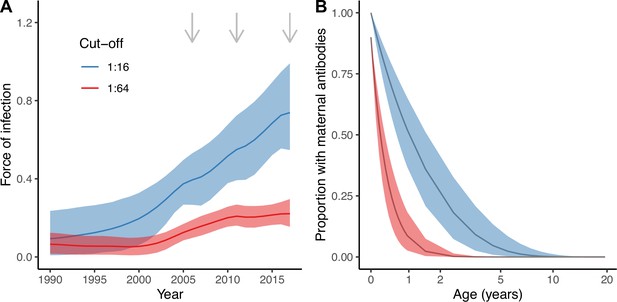
Estimates from the best model (Model 2).
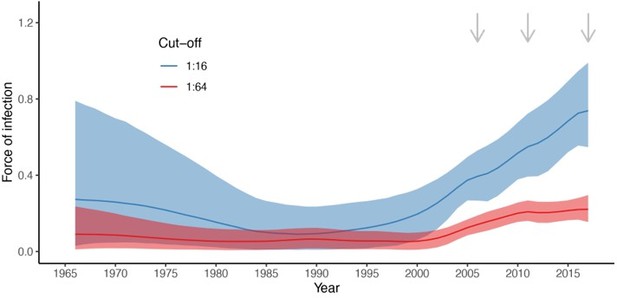
Estimated force of infection (FOI) over time (A) and estimated proportion of individuals with detectable maternal antibodies through age (B) for the two seropositivity cut-offs considered. Median and 95% credible intervals are shown for both. In (A), gray arrows indicate the years for which there is cross-sectional seroprevalence data.
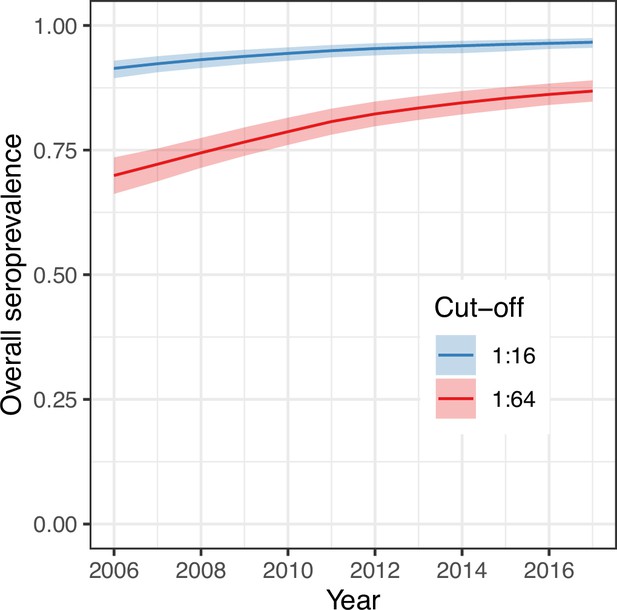
Overall (age-weighted) seroprevalence per year for the best model using the two different seropositivity cut-offs.
Median and 95% credible intervals are shown.
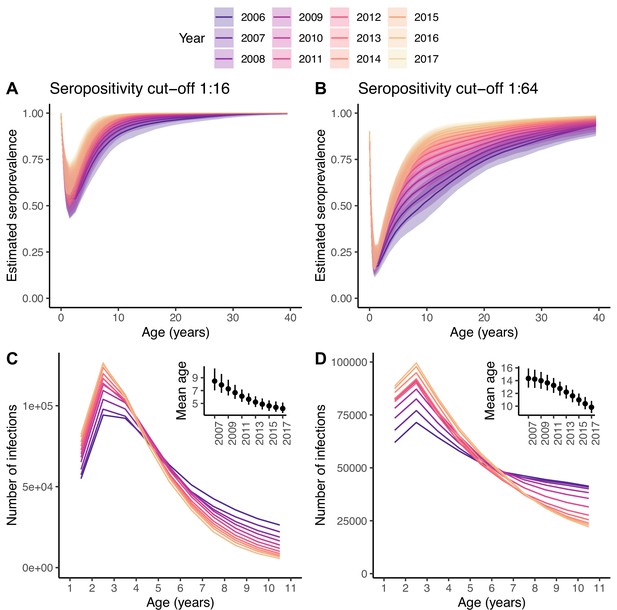
Changes in the number of infections.
Seroprevalence (A, B) and reconstructed number of infections (C, D) for each age class and year under the best model and for the two seropositivity cut-offs, 1:16 (A, C) and 1:64 (B, D). The insets in (C, D) show the mean age at infection for each year. In (A, B), the median and 95% credible intervals are shown, whereas in (C, D) only the median is shown, for clarity of the plots.
Additional files
-
Supplementary file 1
Supplementary results.
(a) Proportion of seropositive individuals in the 25–40 years old group. This proportion is used to fix the proportion of individuals born with maternal antibodies and is assumed constant across the study. (b) Model comparison for the two different datasets, corresponding to two different seropositivity cut-offs. For each model comparison, the first row corresponds to the model with the largest expected log pointwise density (ELPD), which measures the model expected predictive accuracy. For each model, its log-likelihood and the difference in the Bayesian leave-one-out (LOO) estimate of the ELPD compared to the best model are shown. (c) Model parameter estimates. Median and 95% credible intervals are presented.
- https://cdn.elifesciences.org/articles/76609/elife-76609-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76609/elife-76609-transrepform1-v1.docx