Small proline-rich proteins (SPRRs) are epidermally produced antimicrobial proteins that defend the cutaneous barrier by direct bacterial membrane disruption
Figures
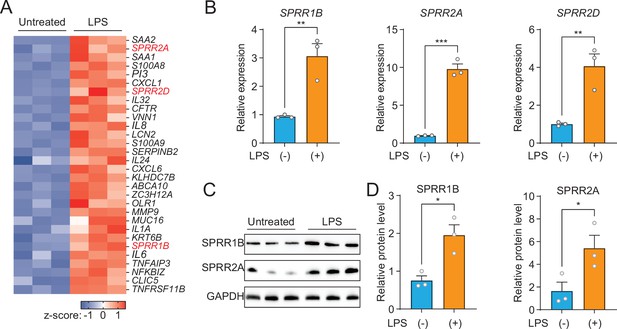
The expression of SPRR family genes are upregulated by lipopolysaccharide (LPS) in human sebaceous gland cells.
(A) Heat map of significantly upregulated genes, represented as Z-scored RPKM (reads per kilo base per million reads). (B) Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis of SPRR1B, SPRR2A, and SPRR2D transcript in the vehicle- and LPS-treated human SZ95 sebocytes. (C) Western blot analysis of SPRR1B and SPRR2A was performed on vehicle- or LPS-treated SZ95 cells. GAPDH was used as the loading control. (D) Quantification of western blot in (C). Means ± standard error of the mean (SEM) are plotted. *p < 0.05, **p < 0.01, and ***p < 0.001 were determined by unpaired t-test.
-
Figure 1—source data 1
The expression of SPRR proteins increases with lipopolysaccharide.
- https://cdn.elifesciences.org/articles/76729/elife-76729-fig1-data1-v3.zip
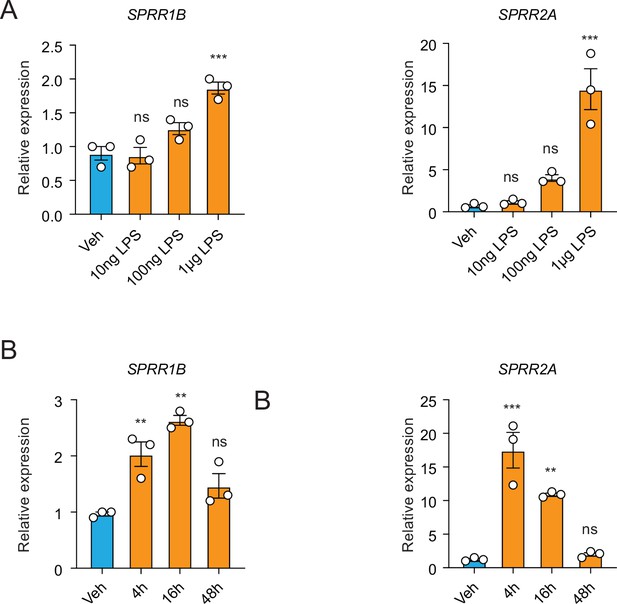
Dose–response and time course analysis of lipopolysaccharide (LPS) treatment on human sebocyte cells.
Quantitative reverse transcription PCR (qRT-PCR) analysis of SPRR1B and SPRR2A transcript in the vehicle- and various LPS-treated human SZ95 sebocytes. (A) SZ95 sebocyte cells were treated with different concentration of LPS for 16 hr. (B) SZ95 cell were treated with 1 μg/ml LPS for different length of time. Means ± standard error of the mean (SEM) are plotted. **p < 0.01, ***p < 0.001; ns, not significant by unpaired t-test.
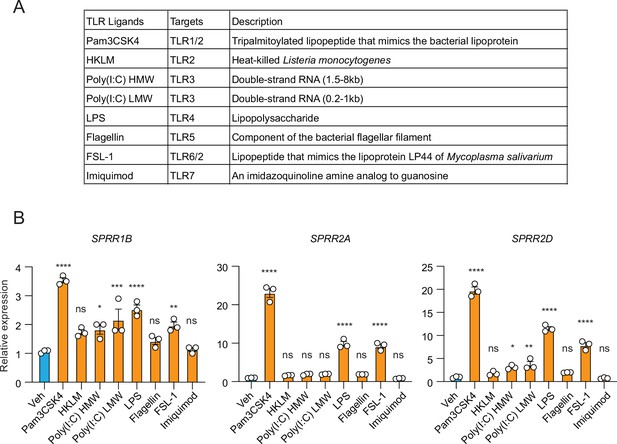
The expression of SPRR family genes are upregulated by Toll-like receptor (TLR)2 and TLR4 agonists in human sebaceous gland cells.
(A) A list of different TLR agonists. (B) Quantitative reverse transcription PCR (qRT-PCR) analysis of SPRR family genes expression in the vehicle and various TLR agonists treated SZ95 cells. Means ± standard error of the mean (SEM) are plotted. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant by one-way analyses of variance (ANOVAs).
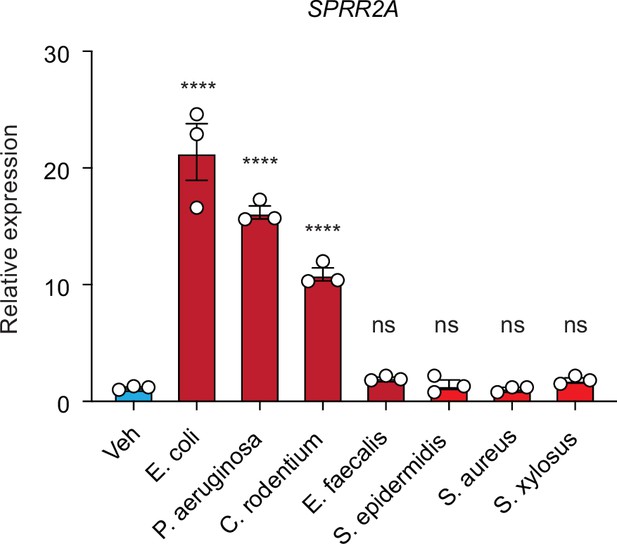
Gram-negative bacteria can trigger Sprr2a gene expression in human sebaceous gland cells.
Quantitative reverse transcription PCR (qRT-PCR) analysis of Sprr2a transcript in the vehicle and various heat-inactivated bacteria treated human SZ95 sebocyte cells. Means ± standard error of the mean (SEM) are plotted. ****p < 0.0001; ns, not significant by one-way analyses of variance (ANOVAs).
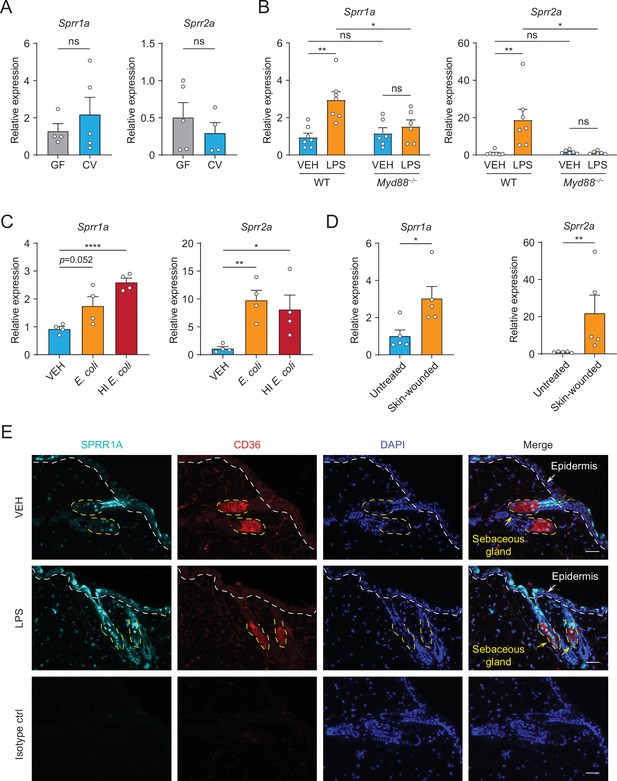
The expression of SPRR family genes are upregulated by lipopolysaccharide (LPS) in mice.
(A–D) Quantitative reverse transcription PCR (qRT-PCR) analysis of Sprr1a and Sprr2a gene expression in mouse dorsal skin tissue. (A) Germ-free mice (GF) compared to conventionally raised mice (CV). (B) Phosphate-buffered saline (PBS)-treated mouse skin compared to LPS intradermal injection mouse skin in WT or MYD88−/− mouse. (C) Mouse skin intradermally injected by vehicle (PBS), E. coli or heat-inactivated (HI) E. coli. (D) Untreated mouse skin compared to wounded skin abraded in a crosshatch pattern by a 15-blade scalpel. Means ± standard error of the mean (SEM) are plotted. *p < 0.05, **p < 0.01, ****p < 0.0001; ns, not significant by unpaired t-test. (E) Immunofluorescence staining of SPRR1A expression in mouse skin. CD36 was used as marker of sebocyte cells. Nuclei are stained with 4',6-diamidino-2-phenylindole (DAPI) (blue). Epidermis and sebaceous gland indicated with an arrow. White dashed line separates the epidermis and dermis. Yellow dashed line indicates the outline of SG. Scale bar, 50 μm.
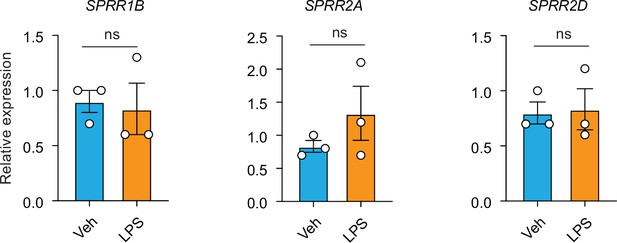
Lipopolysaccharide (LPS) cannot induce the expression of SPRR family genes in human keratinocyte cells.
hTERT immortalized human keratinocyte cells were treated with LPS for 16 hr. Quantitative reverse transcription PCR (qRT-PCR) analysis of SPRR1B, SPRR2A, and SPRR2D transcript in the vehicle- and LPS-treated human hTERT keratinocytes. Means ± standard error of the mean (SEM) are plotted. ns, not significant by unpaired t-test.
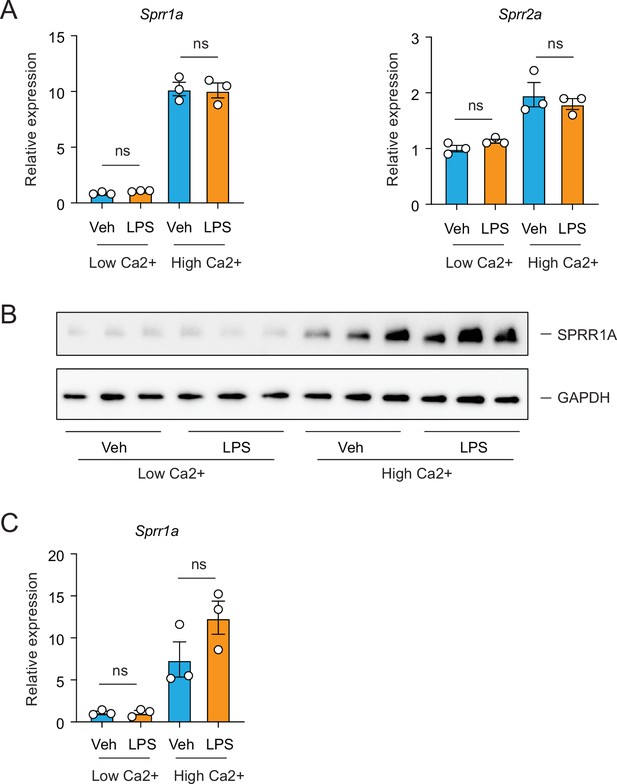
Lipopolysaccharide (LPS) cannot trigger the expression of Sprr family genes in primary mouse keratinocytes.
Primary mouse keratinocytes were isolated from 3- to 5-day-old neonatal mice through dispase digestion. With 0.05 mM CaCl2 (Low Ca2+), keratinocytes can proliferate but will not differentiate into a stratified layer. Under 1 mM CaCl2 (High Ca2+) condition, keratinocytes will initiate differentiation process. (A) Quantitative reverse transcription PCR (qRT-PCR) analysis of Sprr1a and Sprr2a transcript in the vehicle- and LPS-treated primary mouse keratinocyte cells cultured under Low Ca2+ or High Ca2+ condition. (B) Western blot analysis of SPRR1A protein was performed on vehicle- or LPS-treated primary mouse keratinocytes under both Low Ca2+ and High Ca2+ condition. (C) Quantification of the western blot in (B). Means ± standard error of the mean (SEM) are plotted. ns, not significant by unpaired t-test.
-
Figure 2—figure supplement 2—source data 1
Lipopolysaccharide cannot trigger the expression of SPRR proteins in primary mouse keratinocytes.
- https://cdn.elifesciences.org/articles/76729/elife-76729-fig2-figsupp2-data1-v3.zip
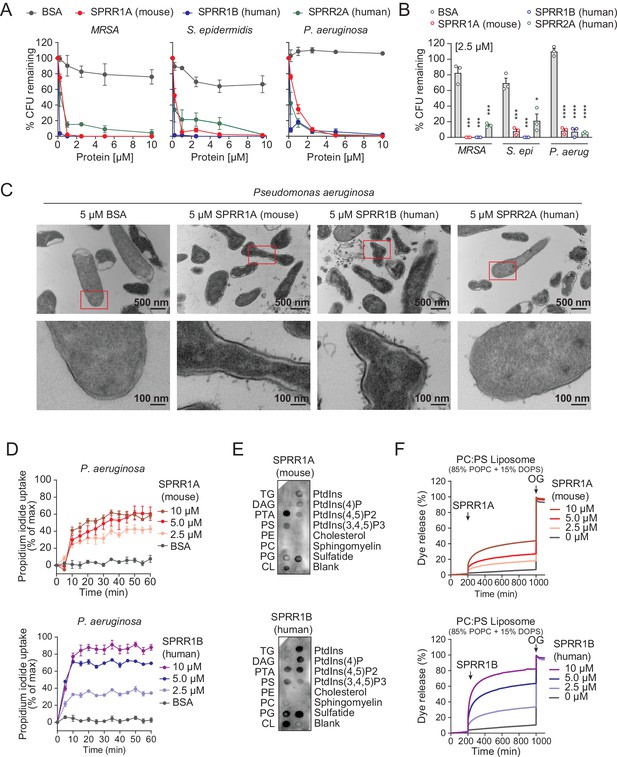
SPRR family proteins exert bactericidal activity against various skin commensal and pathogenic bacteria by membrane disruption.
(A) Increasing concentrations of purified recombinant SPRR proteins were added to mid-logarithmic phase methicillin-resistant Staphylococcus aureus (MRSA), S. epidermidis, P. aeruginosa for 2 hr and surviving bacteria were quantified by dilution plating. (B) 2.5 μM of SPRR proteins was added to midlogarithmic phase bacteria for 2 hr and surviving bacteria were quantified by dilution plating. Remaining colony-forming units (CFUs) are expressed as a percentage of untreated bacteria control. (C) Transmission electron microscopy of P. aeruginosa after incubation with 5 μM purified recombinant SPRR proteins. Bovine Serum Albumin (BSA) was used as negative control. Examples of cell surface damage and cytoplasmic leakage are indicated with red rectangular box. Upper scale bars, 500 nm. Lower scale bars, 100 nm. (D) Propidium iodide (PI) uptake by P. aeruginosa in the presence of increasing concentrations of mouse SPRR1A and human SPRR1B protein. (E) Membranes displaying various lipids were incubated with 1 μg/ml SPRR proteins and detected with specific antibody. (F) Carboxyfluorescein (CF)-loaded liposomes were treated with increasing concentrations of mouse SPRR1A and human SPRR1B protein, and dye efflux was monitored over time and was expressed as a percentage of total efflux in the presence of the detergent octyl glucoside (OG). All assays were performed in triplicate. Means ± standard error of the mean (SEM) are plotted. *p < 0.05, ***p < 0.001, ****p < 0.0001; ns, not significant by two-tailed t-test.
-
Figure 3—source data 1
SPRR proteins bind to negatively charged lipids.
- https://cdn.elifesciences.org/articles/76729/elife-76729-fig3-data1-v3.zip
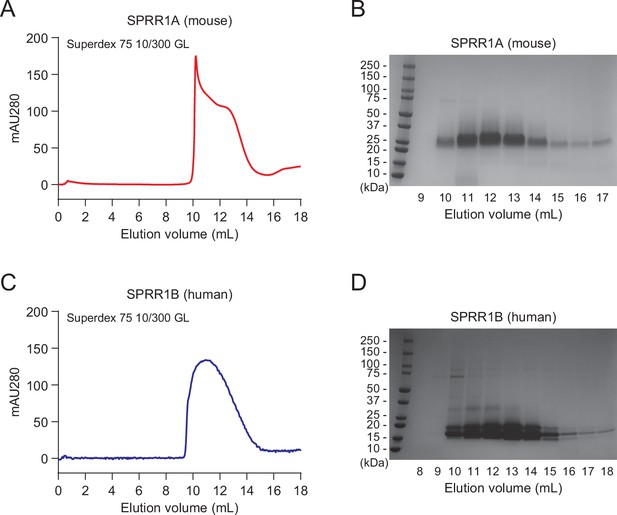
Recombinant expression and purification of SPRR proteins.
Recombinant mouse SPRR1A protein (A) and human SPRR1B protein (C) were expressed using a baculovirus expression system and purified by size-exclusion chromatography (Superdex 75 10/300 GL) (Hu et al., 2021). (B, D) Fractions from (A) and (C) were visualized by Coomassie blue staining following sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE).
-
Figure 3—figure supplement 1—source data 1
Coomassie blue staining of fractions in (A) and (C) resolved by SDS-PAGE.
- https://cdn.elifesciences.org/articles/76729/elife-76729-fig3-figsupp1-data1-v3.zip
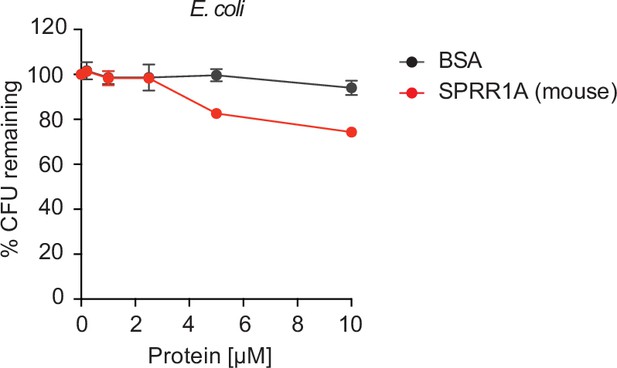
SPRR1A protein was resistant to Gram-negative bacteria Escherichia coli.
Increasing concentrations of purified recombinant mouse SPRR1A protein was added to mid-logarithmic phase E. coli for 2 hr and surviving bacteria were quantified by dilution plating. Remaining colony-forming units (CFUs) are expressed as a percentage of untreated bacteria control. BSA was used as negative control. Means ± standard error of the mean (SEM) are plotted.
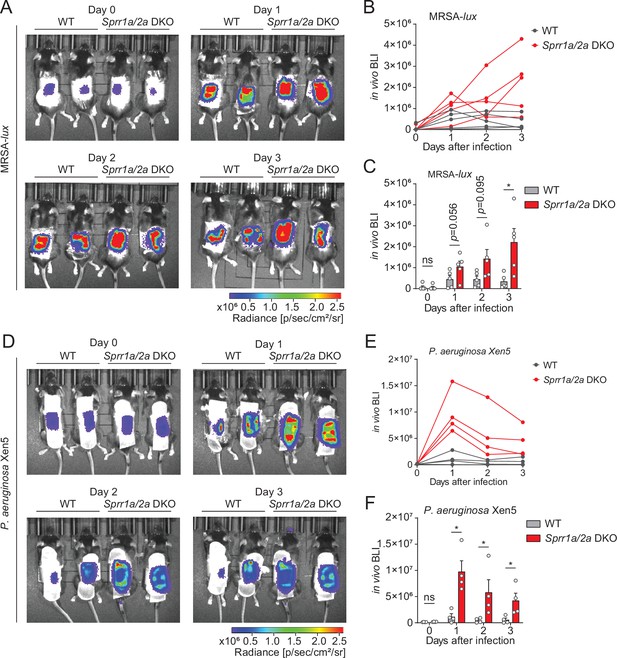
SPRR family proteins protect against skin methicillin-resistant Staphylococcus aureus (MRSA) and P. aerugonisa infection.
(A–C) WT and Sprr1a−/−;Sprr2a−/− mice were epicutaneously challenged with MRSA (1 × 106 CFU) on the shaved dorsal skin for three consecutive days. (A) Representative in vivo bioluminescent imaging (BLI) photographs from days 0 to 3. Quantification of MRSA total flux (photons/s) for each mouse (B) or plotted as means ± standard error of the mean (SEM) (C). (D–F) WT and Sprr1a−/−;Sprr2a−/− mice were superficially abraded in a crosshatch pattern by a 15-blade scalpel to introduce skin wounding. One day post crosshatch wounding, WT and Sprr1a−/−;Sprr2a−/− mice were topically applied with 1 × 106 CFU P. aerugonisa on the back skin for 3 days. (D) Representative in vivo bioluminescent imaging (BLI) signals from days 0 to 3. Quantification of P. aerugonisa total flux (photons/s) for each mouse (E) or plotted as means ± SEM (F). *p < 0.05, ns, not significant by unpaired t-test.
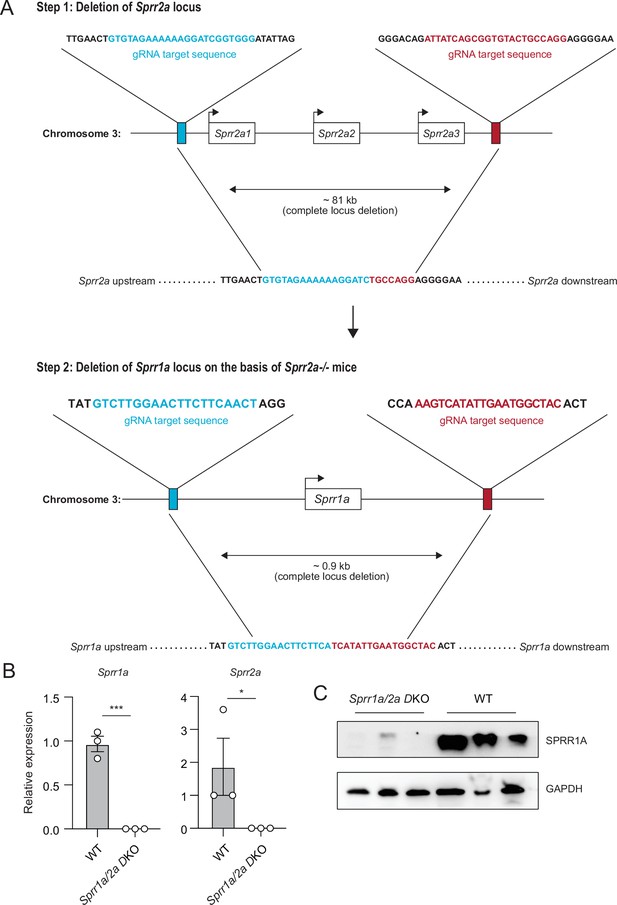
Generation and validation of Sprr1a−/−;Sprr2a−/− mice by CRISPR/Cas9 genomic targeting.
(A) Schematic diagram of two-step strategy using CRISPR/Cas9-mediated gene targeting to delete the entire Sprr2a and Sprr1a locus. (B) Quantitative reverse transcription PCR (qRT-PCR) analysis of Sprr1a and Sprr2a expression in the skin of WT and Sprr1a−/−;Sprr2a−/− mice. Values were normalized to Gapdh expression. Means ± standard error of the mean (SEM) are plotted. *p < 0.05, ***p < 0.001 by unpaired t-test. (C) Western blot analysis of SPRR1A protein in the skin of WT and Sprr1a−/−;Sprr2a−/− mice. GAPDH was used as the loading control.
-
Figure 4—figure supplement 1—source data 1
Western blot analysis of SPRR1A protein in the skin of WT and Sprr1a−/−;Sprr2a−/− mice.
- https://cdn.elifesciences.org/articles/76729/elife-76729-fig4-figsupp1-data1-v3.zip
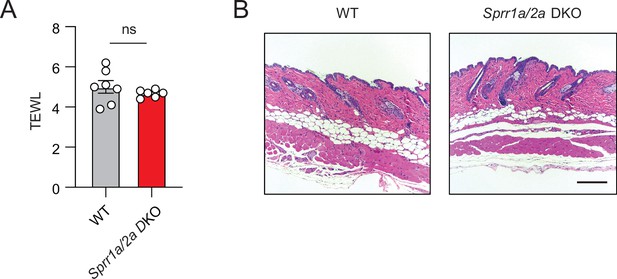
Sprr1a−/−;Sprr2a−/− mice does not show signs of inflammation or impaired skin barrier.
(A) Transepidermal water loss (TEWL) of WT and Sprr1a−/−;Sprr2a−/− mice skin was analyzed. Means ± standard error of the mean (SEM) are plotted, ns, not significant by unpaired t-test. (B) Representative H&E staining of mouse skin from WT and Sprr1a−/−;Sprr2a−/−. Scale bar, 200 μm.
Tables
Primers for qRT-PCR gene expression analysis.
| Gene | Species | Sequence, 5′→3′ |
|---|---|---|
| Sprr1a | Mus musculus | Forward: GCCCTGCACTGTACCTCCTC |
| Reverse: GTGGCAGGGATCCTTGGTTTT | ||
| Sprr2a | Mus musculus | Forward: CCTTGTCCTCCCCAAGTG |
| Reverse: AGGGCATGTTGACTGCCAT | ||
| Gapdh | Mus musculus | Forward:CACTGCCACCCAGAAGACTGT |
| Reverse: GGAAGGCCATGCCAGTGA | ||
| SPRR1B | Homo sapiens | Forward: TATTCCTCTCTTCACACCAG |
| Reverse: TCCTTGGTTTTGGGGATG | ||
| SPRR2A | Homo sapiens | Forward: CCTGAGCACTGATCTGCCTT |
| Reverse: GACATGGCTCTGGGCACTTT | ||
| SPRR2D | Homo sapiens | Forward: GAGCTAAGAAAAGGAAGTCCTCA |
| Reverse: TTATTCAGGGAGTGAACGATAAAT | ||
| GAPDH | Homo sapiens | Forward: GGATTTGGTCGTATTGGG |
| Reverse: GGAAGATGGTGATGGGATT |
| Gene | Protein | Mean-FPKM | Stdeve |
|---|---|---|---|
| Sprr1a | SPPR1 | 66.30906576 | 26.04283 |
| Sprr2a | SPPR2 | 6.587556046 | 3.645252 |
| Lyz1 | LYSOZYME | 36.73892838 | 32.33246 |
| Lyz2 | LYSOZYME | 255.6400659 | 234.8976 |
| Defb1 | DEFENSIN | 26.85869015 | 7.757099 |
| Defb6 | DEFENSIN | 163.2368575 | 64.78706 |
| Cramp1 | CATHELICIDIN | 7.629683869 | 0.813505 |
| S100a8 | S100 | 2.002705491 | 0.908623 |





