Hypertrophic cardiomyopathy mutations in the pliant and light chain-binding regions of the lever arm of human β-cardiac myosin have divergent effects on myosin function
Figures
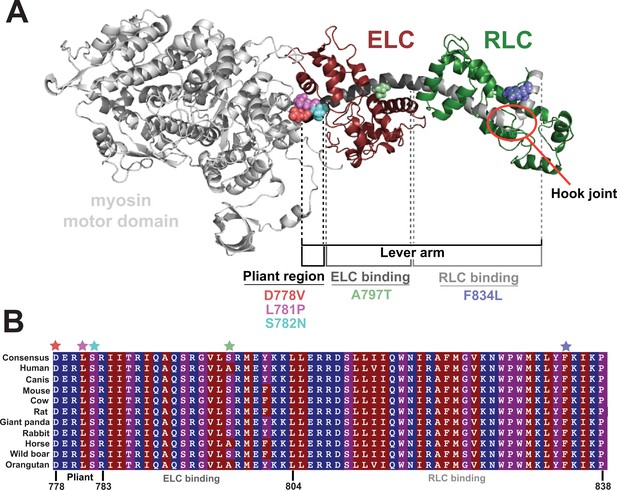
Location of hypertrophic cardiomyopathy-causing mutations along the lever arm.
(A) Homology model of the prestroke β-cardiac myosin subfragment 1 (S1; Homburger et al., 2016) highlighting the five mutations examined in the present study: D778V, L781P, S782N, A797T, and F834L and their locations along the lever arm. The essential light chain (ELC) and regulatory light chain (RLC) are shown in maroon and green, respectively, and the hook joint is highlighted in the red circle. (B) Alignment of β-cardiac myosin lever arms for several model organisms, demonstrating the high degree of conservation of the lever arm across species. Residues are colored by hydrophobicity, where red is most hydrophobic, and blue is most hydrophilic. Mutated residues studied here are indicated with colored stars.
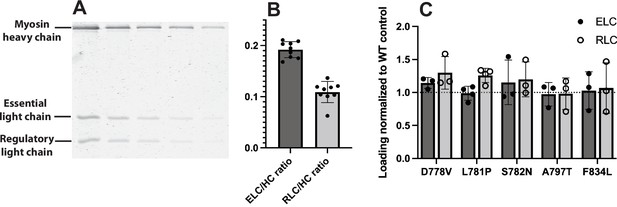
Light chain loading of wild type (WT) vs mutant myosins.
(A) Representative gel used to quantitate light chain loading (L781P 2-hep is shown). For each sample, myosin was titrated in a denaturing SDS-PAGE gel, stained with Coomassie, and scanned for 700 nm fluorescence. See Materials and Methods. (B) Light chain loading across all WT 2-hep samples measured. Mean essential light chain/heavy chain (ELC/HC) = 0.19 ± 0.02, regulatory light chain/heavy chain (RLC/HC) = 0.11 ± 0.02 (mean ± SD). Expected ELC/HC and RLC/HC ratios, if Coomassie staining were unbiased, are 0.168 and 0.146. (C) Light chain loading of mutant 2-hep myosins normalized to WT controls. For each mutant, light chain loading was not statistically different from WT.
-
Figure 2—source data 1
Raw uncropped gel image.
Uncropped representative gel used to quantitate light chain loading (L781P 2-hep is shown) ± annotation.
- https://cdn.elifesciences.org/articles/76805/elife-76805-fig2-data1-v1.pptx
-
Figure 2—source data 2
Ratio of ELC and RLC to myosin 2-hep heavy chain.
(2B) Ratio of pixel density of ELC/RLC over WT 2-hep myosin heavy chain, based on fluorescent imaging of Coomassie stained gels. (2 C) Ratio of LC/HC for mutants over same-day WT LC/HC controls.
- https://cdn.elifesciences.org/articles/76805/elife-76805-fig2-data2-v1.xlsx
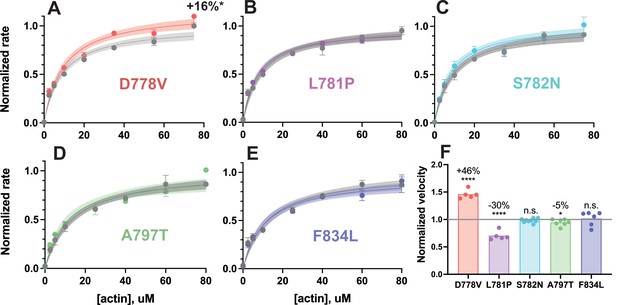
Actin-activated ATPase activity and in vitro motility velocity of 2-hep human β-cardiac myosin lever arm mutants vs wild-type (WT).
(A–E) Representative actin-activated ATPase curves of 2-hep constructs for each lever arm mutant, normalized to their WT controls. Each plot shows a single biological replicate (one of two, see Supplementary file 1), where error bars represent the SD of three technical replicates. Where error bars are not shown, error is smaller than the size of the data point. Mutant data are plotted against their prep-matched WT 2-hep control (gray) and normalized to the WT kcat value. Curves are fit to Michaelis-Menten kinetics, and the shaded areas represent 95% CI of the fits. Only D778V produced a significant difference: the kcat was increased by 16 ± 9% (average of two independent biological replicates compared to WT controls). (F) In vitro motility velocities of lever arm mutants. Each data point represents the average velocity of the mutant 2-hep on a single slide normalized to the WT 2-hep velocity from the same slide. Bars represent the average of the data points. D778V increased the velocity by 46 ± 8% over WT, L781P reduced the velocity by 30 ± 8% compared to WT, A797T reduced the velocity by 5 ± 6%, and S782N and F834L had no significant effects on motility velocity (mean ± SD of the data points shown). **** indicates p≤0.0001, * indicates p≤0.05.
-
Figure 3—source data 1
Rate vs (actin) for WT and mutant 2-hep myosins.
Turnover rate per second vs (actin) of the actin-activated ATPase activity of each mutant 2-hep myosin along with its same day WT control. Raw rates are depicted in the left-hand boxes and rates normalized to WT 2-hep control are shown in the right-hand boxes. Data shown is for all technical replicates of the depicted representative biological replicate for each mutant/WT pair.
- https://cdn.elifesciences.org/articles/76805/elife-76805-fig3-data1-v1.xlsx
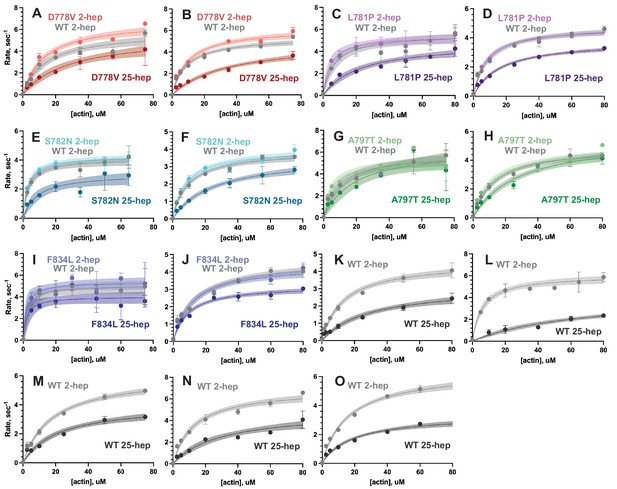
Full actin-activated ATPase results.
(A–O) Each plot shows a single biological replicate of the ATPase results reported here (raw data, not normalized). Data points represent the average of three technical replicates, and error bars show SD (where error bars are not shown, error is smaller than the size of the data point). Curves are fit to Michaelis-Menten kinetics, and shaded areas represent the SE of the fit. Every curve is a separate biological replicate (i.e. each WT 2-hep, WT 25-hep, mutant 2-hep, and mutant 25-hep shown are from a separate batch of cells and protein preparation). Data are shown in the same order presented in Supplementary file 1, where fitting results for each curve are detailed.
-
Figure 3—figure supplement 1—source data 1
Turnover rate per second vs (actin) of the actin-activated ATPase activity of each mutant 2-hep myosin and each mutant 25-hep myosin along with its same day WT 2-hep control.
Raw rates are depicted in the left-hand boxes, rates of mutant 2-hep normalized to WT 2-hep control are shown in the middle boxes, and rates of mutant 25-hep normalized to mutant 2-hep are shown in the right-hand side boxes. For the five WT 2-hep/25-hep biological replicates, the raw rates are shown. Data shown is for all technical replicates of all biological replicates.
- https://cdn.elifesciences.org/articles/76805/elife-76805-fig3-figsupp1-data1-v1.xlsx
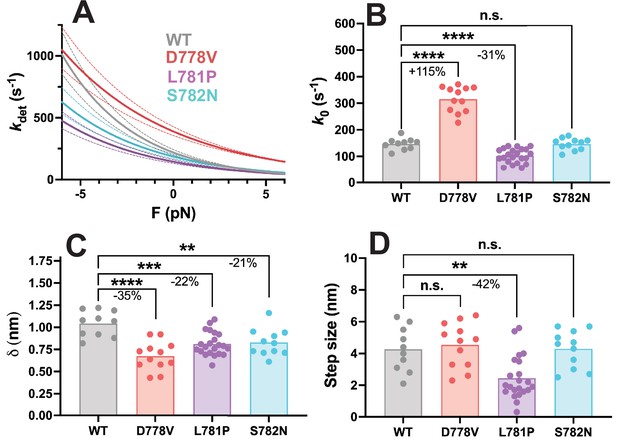
Harmonic force spectroscopy measurements of pliant region mutations in short subfragment 1 (sS1) human β-cardiac myosin.
(A) Detachment rate of sS1 myosin as a function of load force, average of all molecules (wild type [WT]: n=10 molecules, D778V: n=12 molecules, L781P: n=22 molecules, S782N: n=11 molecules). WT curve is shown in gray, D778V curve is shown in red, L781P curve is shown in purple, and S782N curve is shown in cyan. Dotted lines show error propagated from SEM of fitted parameters. (B) k0, the detachment rate at zero load force, for WT and each mutant sS1. D778V increased the detachment rate by 115 ± 9% (mean ± SEM), while L781P reduced the detachment rate by 31 ± 10%, and S782N had no significant effect on detachment rate. (C) δ, the measure of force sensitivity, for WT and each mutant sS1. All the pliant region mutations reduced the force sensitivity: D778V by 35 ± 11%, L781P by 22 ± 8%, and S782N by 21 ± 10% (mean ± SEM). (D) Step size for WT and each mutation. Only L781P affected step size, reducing it by 42 ± 22% (mean ± SEM). Each data point represents the average value for one molecule, and bars show the average of the data points. ** indicates p≤0.01, *** indicates p≤0.001, **** indicates p≤0.0001.
-
Figure 4—source data 1
Detachment rate, δ, and step size for all molecules of WT and pliant region mutant myosins.
Detachment rate k0, δ, and step size (nm) for all molecules of WT and mutant sS1 myosin, as measured by harmonic force spectroscopy. Each row represents an individual molecule. Molecules are show in the same order for each group of measurements.
- https://cdn.elifesciences.org/articles/76805/elife-76805-fig4-data1-v1.xlsx
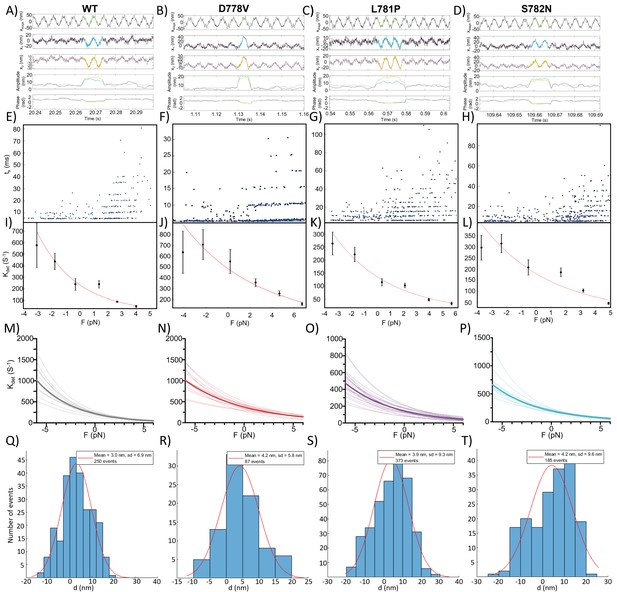
Representative raw data and analysis for harmonic force spectroscopyHFS measurements.
(A–D) Detection of single actin-myosin interaction events (representative) from the change in the amplitude and the phase (region marked with cyan color for bead 1 [x1] and yellow color for bead 2 [x2]) of the oscillating trapped beads for wild type (WT) (A), D778V (B), L781P (C), and S782N (D). Stage oscillation is shown in the top panel of (A–H). Detachment rates (one point corresponds to one event) obtained from HFS measurements of one representative molecule of WT (E), D778V (F), L781P (G), and S782N (H) at different assistive (<0), zero and resistive (>0) load forces. (I–L) Representative fitting using Arrhenius equation (corrected for harmonic force, equation 1) of the detachment rates obtained at different load forces for one molecule each of WT (I), D778V (J), L781P (K), and S782N (L).(M–P) Detachment rate curves obtained for every molecule of WT (M), D778V (N), L781P (O), and S782N (P), where the thick line shows the average of all of the molecules (same as Figure 3A). (Q – T) Fitting of histogram of displacements (obtained from each event) of one representative molecule of WT (Q), D778V (R), L781P (S), and S782N (T).
-
Figure 4—figure supplement 1—source data 1
(I–L) Data fof representative fitting using Arrhenius equation (corrected for harmonic force, equation 1) of the detachment rates (Kdet) obtained at different load forces for one molecule each of WT (I), D778V (J), L781P (K), and S782N (L).
(Q–T) Data of displacement measurements binned in the histogram of displacements (obtained from each event) of one representative molecule of WT (Q), D778V (R), L781P (S), and S782N (T).
- https://cdn.elifesciences.org/articles/76805/elife-76805-fig4-figsupp1-data1-v1.xlsx
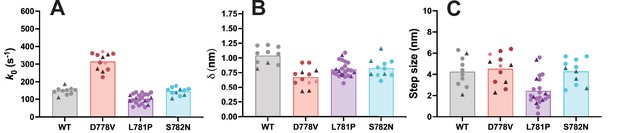
Harmonic force spectroscopy measurements across unique protein preparations.
(A–C) Plots show the same data from Figure 3B–D with data points (representing unique molecules) from independent protein preparations shown in different colors and with different shapes. Wild type (WT), L781P, and S782N molecules come from two unique protein preparations, and D778V molecules come from three unique protein preparations.
-
Figure 4—figure supplement 2—source data 1
Detachment rate, delta, and step size for all molecules of WT and pliant region mutant myosins across separate myosin preps.
Data matches data from Figure 4—source data 1, with the addition of the column entitled ‘prep number; which denotes the myosin prep from which each molecule originated.
- https://cdn.elifesciences.org/articles/76805/elife-76805-fig4-figsupp2-data1-v1.xlsx
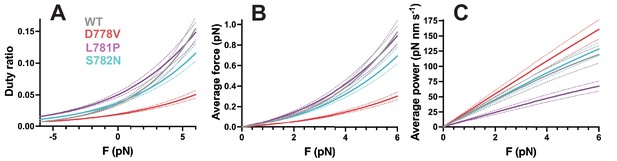
Effects of pliant region mutations on ensemble duty ratio, average force, and average power.
Calculation of (A) duty ratio, (B) average force, and (C) average power output at resistive forces based on measured detachment rate k0, actin-activated ATPase rate kcat, force-dependent detachment rate kdet(F), and step size (see equations 2-6). Wild type (WT) curves are shown in gray, D778V curves are shown in red, L781P curves are shown in purple, and S782N curves are shown in cyan, where dotted lines in the same color show error, propagated from the SEM of the individual molecules.
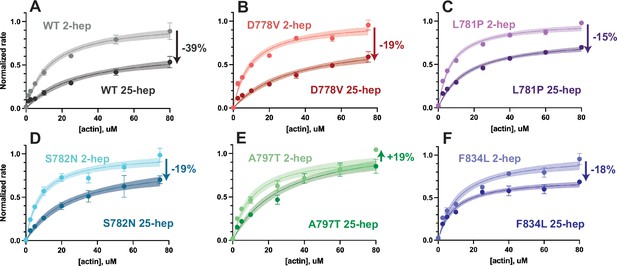
Actin-activated ATPase rates of 2-hep vs 25-hep β-cardiac myosin constructs.
(A–F) Representative actin-activated ATPase curves for each mutant, normalized to the mutant 2-hep control kcat. (A) is reproduced from Vander Roest et al., 2021. Each data point represents the average of three technical replicates of one biological replicate (one of two, see Figure 3—figure supplement 1 and Supplementary file 1), and error bars represent SDs. Where error bars are not shown, error is smaller than the size of the data point. Curves are fitted to Michaelis-Menten kinetics, and shaded areas represent the 95% CI of the fits. Arrows with percentages on each graph show the percent change from the 2-hep to the matched 25-hep (average of both biological replicates, not just the representative curve shown). See Supplementary file 1 for full results.
-
Figure 6—source data 1
Rate vs (actin) for wild type (WT) and mutant 2- vs 25-hep myosin.
Turnover rate per second vs (actin) of the actin-activated ATPase activity of each mutant 25-hep and 2-hep myosin pair. Raw rates are depicted in the left-hand boxes and 25-hep myosin rates normalized its mutant 2-hep myosin control are shown in the right-hand boxes. Data shown is for all technical replicates of the depicted representative biological replicate for each mutant 2-hep/25-hep pair.
- https://cdn.elifesciences.org/articles/76805/elife-76805-fig6-data1-v1.xlsx
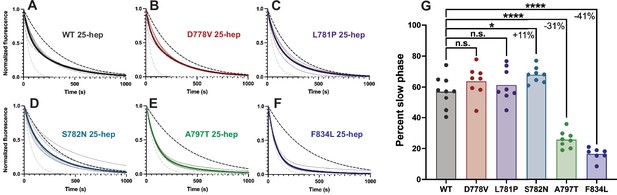
Single ATP turnover kinetics for 25-hep myosin lever arm mutants compared to wild type (WT).
(A–F) Single mant-ATP turnover curves for WT 25-hep and each mutant. Thin curves show curves fitted to a biexponential decay and normalized to the fitted Y0=1.0 and plateau=0.0 from each replicate, while thick lines show average fitted curves across all replicates. Note that in several cases, the average curve obscures individual replicate curves underneath. The dotted black line represents a simulated single-exponential decay with the slow rate of the average curve, and the dotted gray line represents a simulated single-exponential decay with the fast rate of the average curve. (G) Percent slow phase of multiple replicates of each mutant. Pairwise comparisons show that only A797T and F834L significantly reduce the super relaxed state (SRX) state, resulting in a 31 ± 4% and 41 ± 4% (mean ± SEM) reduction in slow phase, respectively. S782N resulted in an 11 ± 4% (mean ± SEM) increase in SRX. * indicates p≤0.05, **** indicates p≤0.0001.
-
Figure 7—source data 1
Fast and slow rates and percent slow turnover for all 25-hep single turnover experiments.
Each row represents an individual technical replicate, where rows are matched by replicate across the three data groups.
- https://cdn.elifesciences.org/articles/76805/elife-76805-fig7-data1-v1.xlsx
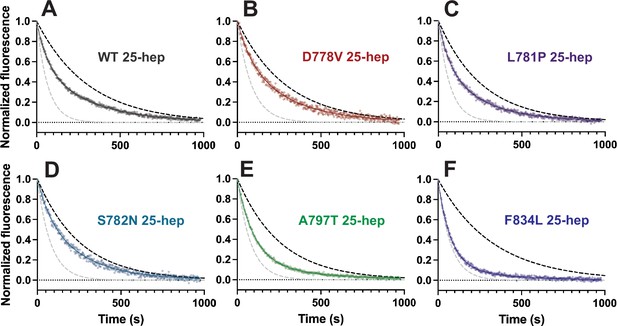
Representative single ATP turnover results for 25-hep myosins.
(A–F) Representative single ATP turnover data (semi-transparent data points) for each wild type (WT) and mutant 25-hep, fitted to a biexponential decay (solid line) and normalized to the fitted Y0=1.0 and fitted plateau=0.0. The gray dotted line shows a simulated single-exponential decay with the fast rate of the shown fitted curve, and the black dotted line represents a simulated single-exponential decay with the slow rate of the shown fitted curve.
-
Figure 7—figure supplement 1—source data 1
Representative single turnover data for a single technical replicate of each WT and mutant 25-hep.
Time (seconds) vs normalized fluorescence, where Y0=1.0 of the fitted curve and 0.0 corresponds to the plateau value of the fitted curve. Raw (non-normalized) fluorescence prior to normalization is also shown.
- https://cdn.elifesciences.org/articles/76805/elife-76805-fig7-figsupp1-data1-v1.xlsx
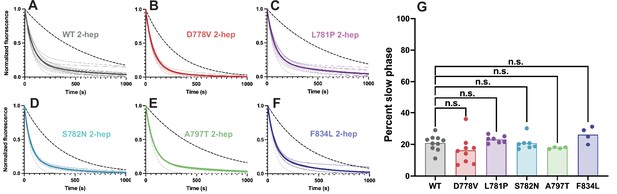
Single ATP turnover kinetics for wild type (WT) vs mutant 2-hep myosins.
(A–F) Single mant-ATP turnover curves for WT 2-hep and each mutant. Thin curves show curves fitted to a biexponential decay and normalized to the fitted Y0=1.0 and plateau=0.0 from each replicate, while thick lines show average fitted curves across all replicates. Average curves obscure individual replicate curves underneath in some cases. The dotted black line represents a simulated single-exponential decay with the slow rate of the average curve, and the dotted gray line represents a simulated single-exponential decay with the fast rate of the average curve. (G) Percent slow phase of multiple replicates of each WT and mutant 2-hep. For all mutations, the slow phase is similarly low as compared to the WT control.
-
Figure 7—figure supplement 2—source data 1
Fast and slow rates and percent slow turnover for all 2-hep single turnover experiments.
Each row represents an individual technical replicate, where rows are matched by replicate across the three data groups.
- https://cdn.elifesciences.org/articles/76805/elife-76805-fig7-figsupp2-data1-v1.xlsx
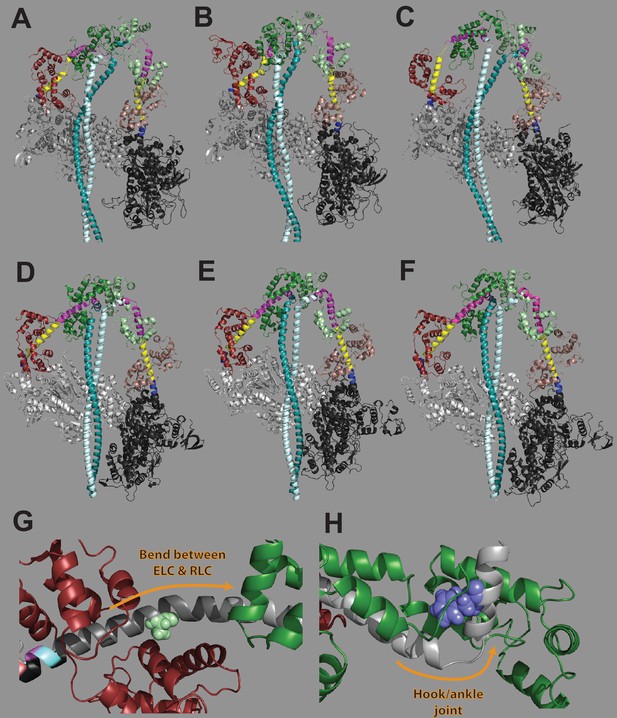
Contributions of lever arm position to the folded state structure.
(A–F) Homology modeled structures of human β-cardiac myosin in the interacting heads motif (IHM) structural state from (A) (Alamo et al., 2017), (B) (Nag et al., 2017), and (C) (Robert-Paganin et al., 2018) and cryo-EM solved structures of smooth muscle myosin in the IHM structural state from (D) (Scarff et al., 2020), (E) (Yang et al., 2020), and (F) (Heissler et al., 2021). In each structure, the pliant region is colored yellow, the essential light chain (ELC)-binding region is colored blue, and the regulatory light chain (RLC)-binding region is colored pink. The myosin heads are in gray, ELCs are in dark red and brown, RLCs are in green, and subfragment 2 tail regions are in cyan. Homology modeled structures (A–C) show a markedly different lever arm structure as compared to experimentally determined structures (D–F). (G) A797 (light green) is located in the region where the lever arm bends between the ELC and RLC. This region has been previously implicated in formation of the IHM (Houdusse and Cohen, 1996). (H) F834 (blue) is located very near to the hook or ankle joint at the end of the lever arm. This joint has been previously shown to be important in the formation of the IHM (Brown et al., 2011). (G) and (H) are from a homology modeled structure of human β-cardiac myosin in the prestroke state (Farman et al., 2014; see Figure 1A).
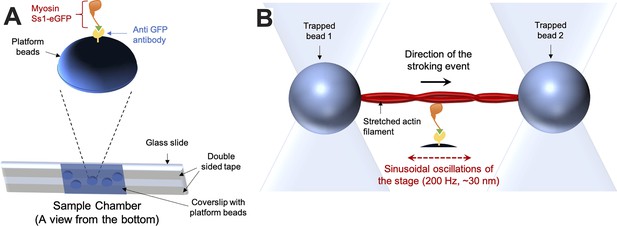
Technical details of the dual-beam optical trap experiment.
(A) The sample chamber (bottom) is shown in an orientation suitable for an inverted microscope. On top, the typical arrangement of the protein complexes on top of a platform bead is depicted. (B) A typical stroking event—as expected during actin-myosin interaction in a harmonic force spectroscopic (HFS) setup—is depicted. In a standard HFS experiment, the stage oscillates, resulting in a variety of assistive or resistive external load forces applied to the stroking myosin.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | MYH7 | NCBI Gene | Gene ID: 4625 | |
| Gene (H. sapiens) | MYL3 | NCBI Gene | Gene ID: 4634 | N-terminal FLAG-TEV tag added |
| Gene (H. sapiens) | MYL2 | NCBI Gene | Gene ID: 4633 | N-terminal His-TEV tag added |
| Strain, strain background (Escherichia coli) | BJ5183-AD-1 | Agilent | 200,157 | |
| Strain, strain background (E. coli) | Rosetta (DE3) pLysS | Sigma-Aldrich | 70,956 | |
| Cell line (H. sapiens) | HEK 293T | ATCC | CRL-3216 | |
| Cell line (H. sapiens) | C2C12 | ATCC | CRL-1772 | |
| Transfected construct (H. sapiens) | pAdEasy-1-MYH7 sS1 | 10.1073/pnas.1309493110 | ||
| Transfected construct (H. sapiens) | pAdEasy-1-MYH7 2hep | 10.1038/nsmb.3408 | ||
| Transfected construct (H. sapiens) | pAdEasy-1-MYH7 25hep | 10.1038/nsmb.3408 | ||
| Biological sample (Bos taurus) | Bovine cardiac acetone powder | Pelfreez | 57,195 | |
| Recombinant DNA reagent | pShuttle-CMV- MYH7 sS1 | 10.1073/pnas.1309493110 | ||
| Recombinant DNA reagent | pShuttle-CMV- MYH7 2hep | 10.1038/nsmb.3408 | ||
| Recombinant DNA reagent | pShuttle-CMV- MYH7 25hep | 10.1038/nsmb.3408 | ||
| Sequence-based reagent | D778V S | This paper | Mutagenesis primer | GGAGGAAATGAGGGTCGAGAGGCTGAGCC |
| Sequence-based reagent | D778V AS | This paper | Mutagenesis primer | GGCTCAGCCTCTCGACCCTCATTTCCTCC |
| Sequence-based reagent | L781P S | This paper | Mutagenesis primer | GATGATGCGGCTCGGCCTCTCGTCCCT |
| Sequence-based reagent | L781P AS | This paper | Mutagenesis primer | AAGGACGAGAGGCCGAGCCGCATCATC |
| Sequence-based reagent | S782N S | This paper | Mutagenesis primer | TGAGGGACGAGAGGCTGAACCGCATCATC |
| Sequence-based reagent | S782N AS | This paper | Mutagenesis primer | GATGATGCGGTTCAGCCTCTCGTCCCTCA |
| Sequence-based reagent | A797T S | This paper | Mutagenesis primer | GTACTCCATTCTGGTGAGCACACCTCGGG |
| Sequence-based reagent | A797T AS | This paper | Mutagenesis primer | CCCGAGGTGTGCTCACCAGAATGGAGTAC |
| Sequence-based reagent | F834L S | This paper | Mutagenesis primer | CTGGATGAAGCTCTACTTAAAGATCAAGCCGCTG |
| Sequence-based reagent | F834L AS | This paper | Mutagenesis primer | CAGCGGCTTGATCTTTAAGTAGAGCTTCATCCAG |
| Software, algorithm | FAST | 10.1016 /j.celrep.2015.04.006 | Filament tracking and velocity measurement software |
Full numbers of biological and technical replicate.
| Assay | Protein | Biological Replicates | Technical Replicates |
|---|---|---|---|
| Light-chain loading assay (Each biological replicate is from a single unique protein prep diluted across 5 gel lanes, implying 5 technical replicates per biological replicate) | WT 2-hep | 9 | 45 |
| D778V 2-hep | 3 | 15 | |
| L781P 2-hep | 3 | 15 | |
| S782N 2-hep | 3 | 15 | |
| A797T 2-hep | 3 | 15 | |
| F834L 2-hep | 3 | 15 | |
| Actin-activated ATPase assay (Biological replicates are from separate protein preps, technical replicates are from separate wells on the same plate) | WT 2-hep | 15 | 45 |
| WT 25-hep | 5 | 15 | |
| D778V 2-hep | 2 | 6 | |
| D778V 25-hep | 2 | 6 | |
| L781P 2-hep | 2 | 6 | |
| L781P 25-hep | 2 | 6 | |
| S782N 2-hep | 2 | 6 | |
| S782N 25-hep | 2 | 6 | |
| A797T 2-hep | 2 | 6 |
Additional files
-
Supplementary file 1
Full actin-activated ATPase results.
For each mutation, two independent biological experiments were performed with freshly prepared myosins, where each biological replicate was measured with technical triplicates (6 total replicates for each protein). For WT 2- vs 25-hep, five biological replicates were measured with technical triplicates (15 total replicates for each protein). The 2-hep and 25-hep mutant myosins were prepared in tandem with a WT 2-hep control for comparison, given that actin-activated ATPase results show a slight drift from day-to-day. Results in the fourth and fifth columns, respectively, show the fitted values for kcat (s–1) and Kapp (μM) ± SE of the fit for each biological replicate. The kcat ratio of mutant 2-hep/WT 2-hep and mutant 25-hep/mutant 2-hep for each independent biological replicate is shown in the sixth and seventh columns, respectively, where the error is propagated from SE of the fit for each measurement. In the rightmost column (average mutant 25-hep/mutant 2-hep kcat ratio), statistically significant differences for mutant ratios vs WT ratio are shown, where * indicates p≤0.05, ** indicates p≤0.01, and *** indicates p≤0.001.
- https://cdn.elifesciences.org/articles/76805/elife-76805-supp1-v1.pptx
-
Supplementary file 2
Full single ATP turnover results.
Single ATP turnover experiments were performed for both 2-hep and 25-hep myosins. Results show n, the number of independent experiments for each protein, average percent fast and slow phase ± SD, and average fast and slow rates ± SD. Stars denote statistically significant changes as compared to the WT control of the same protein construct, where * indicates p≤0.05 and **** indicates p≤0.0001.
- https://cdn.elifesciences.org/articles/76805/elife-76805-supp2-v1.pptx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76805/elife-76805-transrepform1-v1.docx





