Potential role of KRAB-ZFP binding and transcriptional states on DNA methylation of retroelements in human male germ cells
Figures
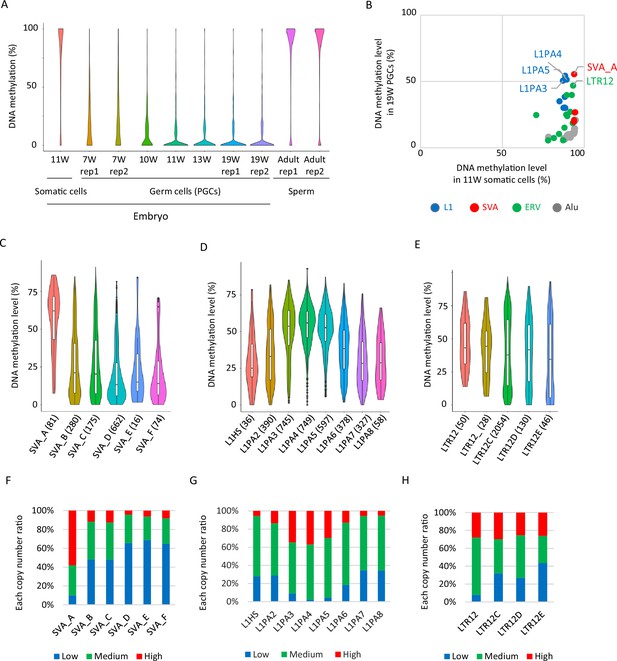
Retroelements showing DNA demethylation resistance.
(A) Violin plots showing DNA methylation levels of each CpG site during human male germ-cell development. DNA demethylation was almost completed at 19 weeks of gestation. (B) Scatter plots showing average DNA methylation level of each retroelement type between somatic cells and male human primordial germ cells (hPGCs) at 19 weeks of gestation. Only full-length copies were used for this analysis, and retroelement types with ≧30 full-length copies were shown. Each plot was colored according to its retroelement family (red: SVA, blue: L1, green: LTR, gray: Alu). (C–E) Violin plots showing DNA methylation level of each retroelement type in hPGCs at 19 weeks of gestation. p-Value was calculated by Tukey’s test and was described in Supplementary file 2. The number in parentheses was analyzed copy number. (F–H) Bar graphs showing the fraction of ‘low’, ‘medium’, and ‘high’ methylated class of each retroelement type in male hPGCs at 19 weeks of gestation. The retroelement copies used in these figures were same as those in (C-E).
-
Figure 1—source data 1
Raw data of graphs in Figure 1.
- https://cdn.elifesciences.org/articles/76822/elife-76822-fig1-data1-v2.xlsx
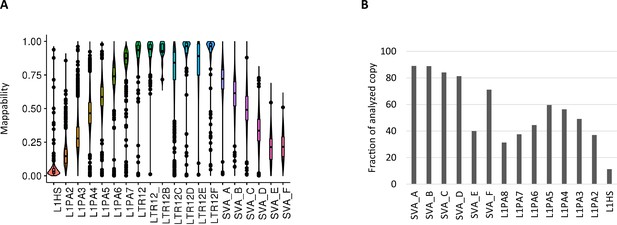
Mappability of whole-genome bisulfite sequencing (WGBS) reads on transposable elements.
(A) Violin plots of WGBS read mappability in each retroelement types. Less than 10% of L1HS-derived reads were uniquely mapped in the majority of L1HS copies. (B) Percentage of copies measured for DNA methylation (≥10 CpG with ≥5 reads).
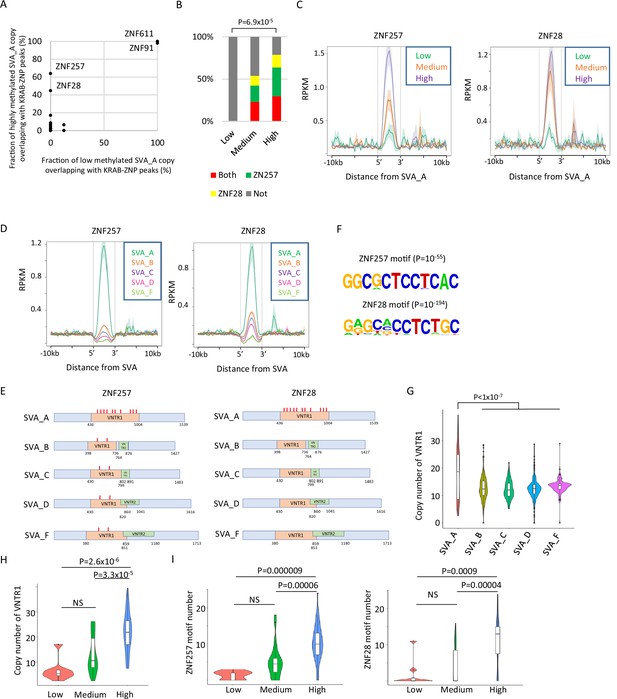
Identification of Krüppel-associated box domain zinc finger proteins (KRAB-ZFPs) associated with DNA demethylation resistance in SINE-VNTR-Alus (SVAs).
(A) Scatter plots showing the fraction of low-methylated or highly methylated SVA_A copies which overlaps of KRAB-ZFP peaks. ZNF257 and ZNF28 peaks were more frequently overlapped with ‘high’ methylated SVA_A than ‘low’ methylated SVA_A. For this analysis, publicly available ChIP-exo data from 250 human KRAB-ZFPs in HEK293T cells were used. (B) Bar graphs showing the fraction of SVA_A copies with ZNF257 and ZNF28 peaks. SVA_A copies were classified by DNA methylation levels in 19 W human primordial germ cells (hPGCs) (N = 8, 26, 47 in low, medium, and high, respectively). p-Value was calculated by chi-square test. (C) Enrichment of ZNF257 and ZNF28 on SVA_A classified by DNA methylation levels in male hPGCs at 19 weeks of gestation. (D) Enrichment of ZNF257 and ZNF28 on each SVA subtype. (E) Position of ZNF257- and ZNF28-binding motifs along SVA consensus sequences. VNTR1 and VNTR2 are composed of multiple copy number of tandem repeats, and the copy number of these number tandem repeats (VNTRs) is highly variable among SVA copies. Both ZNF257- and ZNF28-binding motifs were found within VNTR1 of SVAs. (F) Sequence logo of ZNF257- and ZNF28-binding motifs. (G) Violin plots showing copy number of VNTR1 of each SVA subtype. (H) Violin plots showing VNTR1 copy number of SVA_A classified by its DNA methylation status in male hPGCs at 19 weeks of gestation. (I) Violin plots showing the number of ZNF257 and ZNF28 motifs in SVA_A classified by DNA methylation status in male hPGCs at 19 weeks of gestation. p-Value was calculated by Tukey’s test.
-
Figure 2—source data 1
Raw data of graphs in Figure 2.
- https://cdn.elifesciences.org/articles/76822/elife-76822-fig2-data1-v2.xlsx
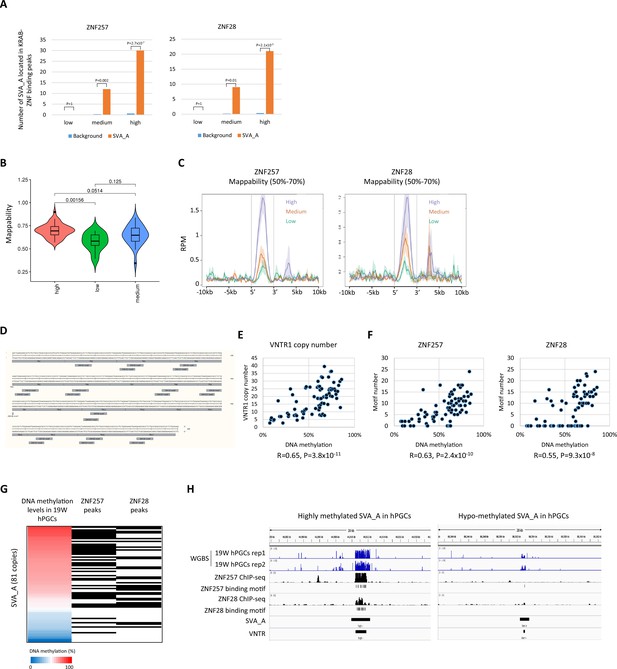
Correlation between ZNF257/ZNF28-binding motifs and DNA methylation levels in SVA_A copies.
(A) Enrichment of SVA_A in ZNF257 and ZNF28 peaks. We randomly extracted genomic regions whose length and number matched the SINE-VNTR-Alus (SVA) copy and calculated the overlap with the ZNF257/28 peaks. This procedure was repeated 1000 times, and the average value was used as the background. p-Value was calculated by Prop.test in R. (B) Violin plots of read mappability in SVA_A copies. Read mappability was significantly higher in ‘high’ group of SVA_A copy than other groups. p-Value was calculated by Tukey’s test. (C) Enrichment of ZNF257 and ZNF28 on SVA_A copies classified by DNA methylation levels in male human primordial germ cells (hPGCs) at 19 weeks of gestation. Only SVA_A copies with 50–70% mappability were used in this figure. (D) Position of ZNF28/257-binding motifs along number tandem repeat (VNTR) region of SVA_A consensus sequence. TR represents a unit of VNTR. (E) Scatter plot of DNA methylation levels and VNTR1 copy number of SVA_A copies. Pearson’s r and p-value were calculated by cor.test in R. (F) Scatter plot of DNA methylation levels and the number of ZNF257 (left) or ZNF28 (right) binding motifs in SVA_A copies. (G) Correlation of DNA methylation levels in VNTR of SVA_A and the presence of ZNF257/28 peaks. Black box represents the presence of ZNF257/28 peaks. (H) Representative view of correlation of DNA methylation in SVA_A and ZNF257/28 binding.
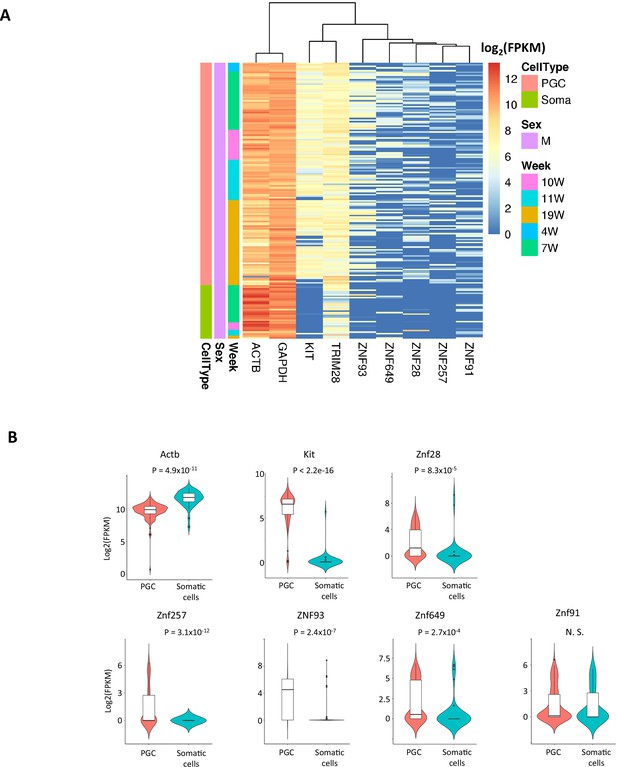
Expression of Krüppel-associated box domain zinc finger proteins (KRAB-ZFP) genes in somatic cells and human primordial germ cells (hPGCs).
(A) Heatmap of expression levels of KRAB-ZFP genes in hPGCs and the neighboring somatic cells. Y-axis represents the data from each cell which categorized by cell type and weeks of gestation. ACTB and GAPDH are house-keeping genes and KIT is germ-cell marker. (B) Violin plot showing expression levels (log2(FPKM)) of KRAB-ZFP genes in hPGCs (N = 233) and the neighboring somatic cells (N = 86). p-Value was calculated by t-test.
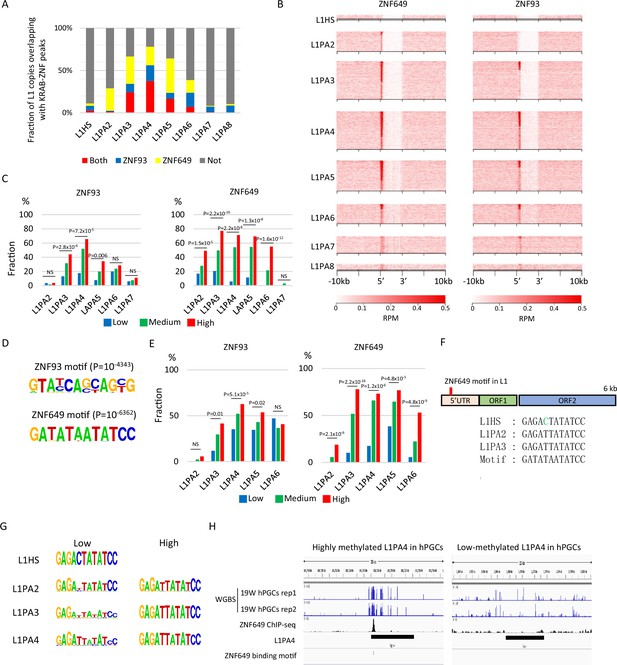
Identification of Krüppel-associated box domain zinc finger proteins (KRAB-ZFPs) associated with DNA demethylation resistance in L1.
(A) Bar graphs showing the fraction of full-length L1 copies with ZNF93 and ZNF649 peaks. (B) Heatmaps showing enrichment of ZNF649 and ZNF93 along full-length L1 copies. ZNF649 binds 5’ regions of L1PA2–PA8, while ZNF93 binds the 5’ regions of L1PA3–PA8. (C) Bar graphs showing the fraction of L1 copies with ZNF93 and ZNF649 peaks. L1 copies were classified by their type and DNA methylation levels in male human primordial germ cells (hPGCs) at 19 weeks of gestation. (D) Sequence logo of ZNF93- and ZNF649-binding motifs. (E) Bar graphs showing the fraction of L1 copies with ZNF93- and ZNF649-binding motifs. The presence of ZNF93- and ZNF649-binding motifs was correlated with higher DNA methylation of L1 in male hPGCs at 19 weeks of gestation (L1PA2 and -PA6 were not significant for ZNF93). p-Value was calculated by Hypothesis Testing for the Difference in the Population Proportions using a function of prop.test by R. (F) Comparison of sequences of ZNF649-binding sites among L1 types. L1HS lost the ZNF649 motif by a base substitution. (G) Comparison of sequences at ZNF649-binding sites between low- and high-methylated L1. (H) Representative view of correlation between DNA methylation of L1PA4 in hPGCs and ZNF649-binding peak.
-
Figure 3—source data 1
Raw data of graphs in Figure 3.
- https://cdn.elifesciences.org/articles/76822/elife-76822-fig3-data1-v2.xlsx
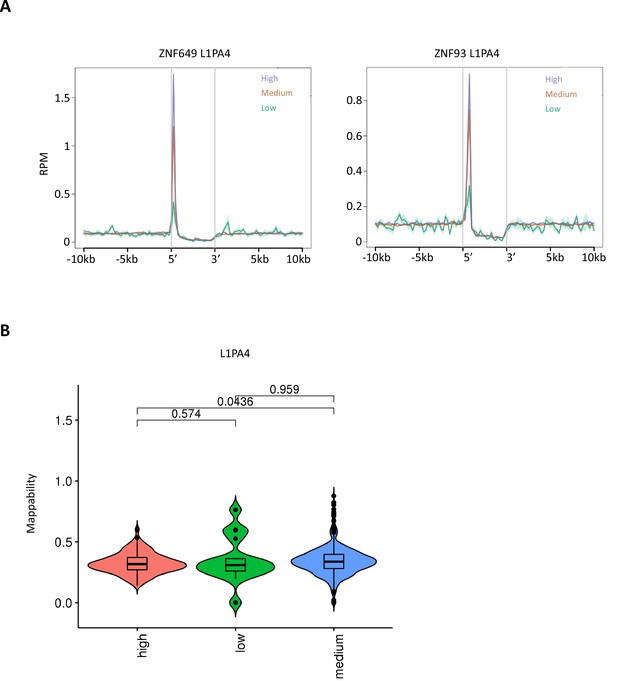
Enrichment of ZNF649/93 on L1PA4.
(A) Enrichment of ZNF649 and ZNF93 on L1PA4 classified by DNA methylation levels in male human primordial germ cells (hPGCs) at 19 weeks of gestation. (B) Violin plot showing the mappability of L1PA4 copies classified by DNA methylation levels in male hPGCs at 19 weeks of gestation. p-Value were calculated by Tukey’s test.
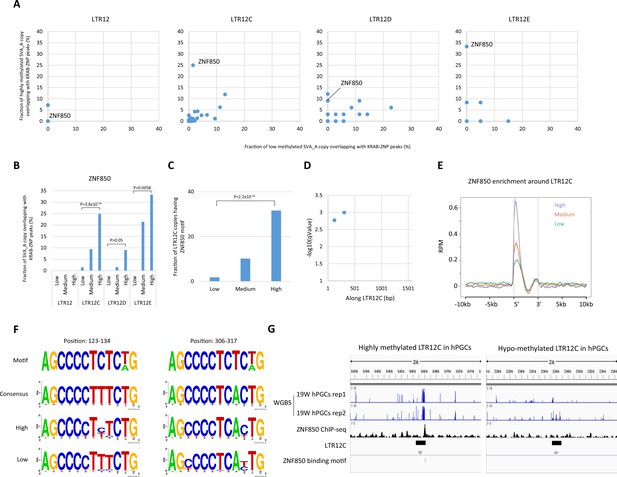
ZNF850 association is correlated with DNA demethylation resistance of LTR12 family in human primordial germ cells (hPGCs).
(A) Scatter plots showing the fraction of low-methylated or highly methylated LTR12 copies which overlaps of Krüppel-associated box domain zinc finger proteins (KRAB-ZFP) peaks. (B) Fraction of LTR12 copies in each group overlapping with ZNF850 peaks. p-Value was calculated by prop.test in R. (C) Fraction of LTR12C copies in each group containing ZNF850-binding motif. p-Value was calculated by prop.test in R. (D) The position of ZNF850-binding motif along LTR12C consensus sequence. (E) Enrichment of ZNF850 ChIP-seq reads within and around LTR12C copies. (F) Comparison of sequences at ZNF850-binding motifs between highly and low-methylated LTR12C copies. (G) Representative view of correlation between ZNF850 peak and DNA methylation of LTR12C in hPGCs.
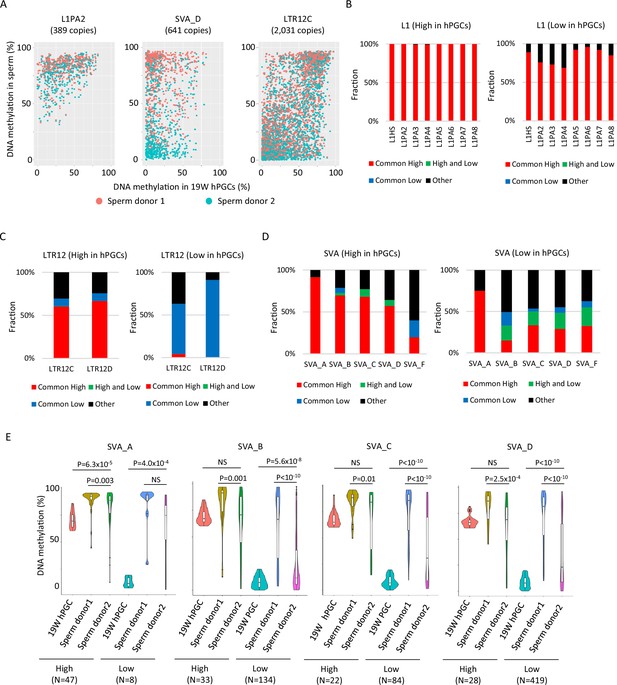
DNA methylation dynamics of retroelements during human spermatogenesis.
(A) Scatter plots showing DNA methylation levels of each retroelement copy in male human primordial germ cells (hPGCs) at 19 weeks of gestation and sperm. Whole-genome bisulfite sequencing (WGBS) data from two sperm donors (Hammoud et al., 2014) were used for this analysis. Donor 1 and donor 2 were colored by orange and cyan, respectively. (B–D) Bar graphs showing the fraction of groups determined by DNA methylation patterns in two sperm donors in L1 (B), LTR12 (C), and SINE-VNTR-Alus (SVA) (D). ‘Other’ indicates groups except for common high, common low, and high and low, such as low methylated in donor 1 and mediumly methylated in donor 2. Bar graphs were also separated by DNA methylation levels (high or low) in male hPGCs at 19 weeks of gestation. (E) Violin plots showing DNA methylation levels of SVA copies in male hPGCs at 19 weeks of gestation, sperm donor 1, and sperm donor 2. The violin plots were also separated by DNA methylation levels of SVA copies in male hPGCs at 19 weeks of gestation. Although hypomethylated SVA copies in male hPGCs at 19 weeks of gestation acquired DNA methylation during spermatogenesis, the degree of DNA methylation increase was significantly different between sperm donors. p-Value was calculated by Dunnett’s test.
-
Figure 4—source data 1
Raw data of graphs in Figure 4.
- https://cdn.elifesciences.org/articles/76822/elife-76822-fig4-data1-v2.xlsx
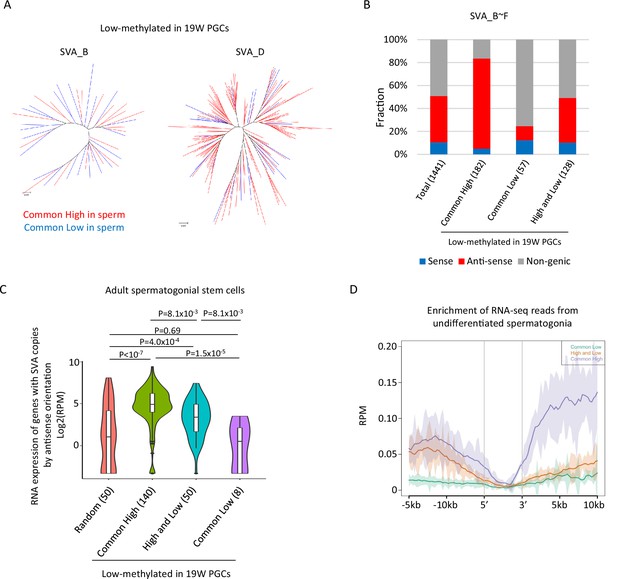
Transcription-associated regulation of DNA methylation of SINE-VNTR-Alus (SVA) during spermatogenesis.
(A) Phylogenetic analysis of SVA_B (left) and SVA_D (right) copies low methylated in male human primordial germ cells (hPGCs) at 19 weeks of gestation. SVA copies highly methylated by both sperm donors were colored by red, while those hypomethylated by both sperm donors were colored by blue. (B) Bar graphs showing the fraction of SVA_B–F copies inserted in a gene body. SVA copies were classified by DNA methylation patterns in two sperm donors. Only low-methylated SVA copies in male hPGCs at 19 weeks of gestation were used for this analysis. The number in parentheses represents analyzed copy number. (C) Violin plots showing the expression of genes in adult spermatogonial stem cells 2 (Sohni et al., 2019). Genes were classified according to the DNA methylation status of SVAs inserted in them in the antisense direction. p-Value was calculated by Tukey’s test. (D) Enrichment of RNA-seq reads from undifferentiated spermatogonia (Tan et al., 2020) around non-genic SVAs. Only low-methylated SVA copies male hPGCs at 19 weeks of gestation were used for the analysis, and SVA copies were classified by DNA methylation patterns in two sperm donors (common low, high, and low and common high).
-
Figure 5—source data 1
Raw data of graphs in Figure 5.
- https://cdn.elifesciences.org/articles/76822/elife-76822-fig5-data1-v2.xlsx
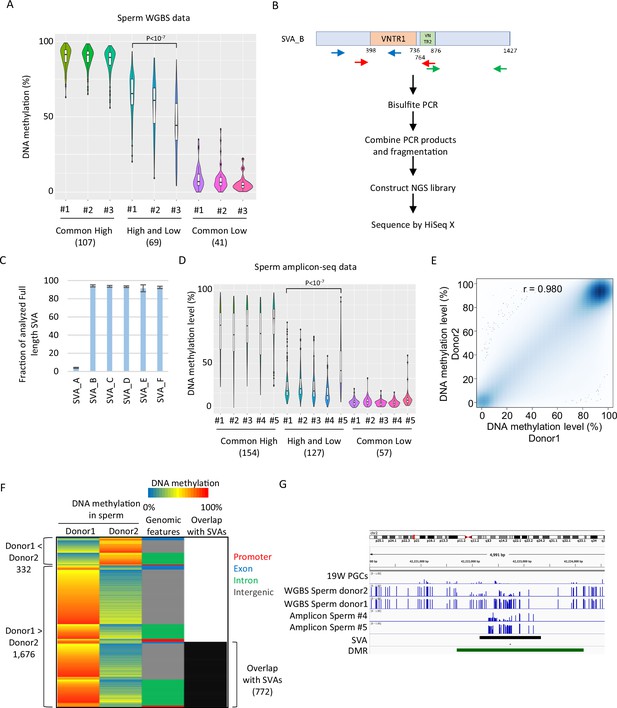
SINE-VNTR-Alus (SVAs) constitute a major source of inter-individual epigenetic variations in sperm.
(A) Violin plots showing DNA methylation of SVA copies in previously reported three sperm donors (#1–#3) (Okae et al., 2014). Only low-methylated SVA copies in male human primordial germ cells (hPGCs) at 19 weeks of gestation were used for the analysis. SVA copies were classified by DNA methylation levels of two sperm donors from Hammoud et al., 2014. Donor #1 showed significantly higher DNA methylation levels in ‘high and low’ SVA copies than other sperm donors. p-Value was calculated by Tukey’s test. (B) Scheme of amplicon sequencing (amplicon-seq) for analyzing SVA methylation. (C) Bar plots showing the fraction of analyzed full-length SVA copies by amplicon-seq. (D) Violin plots showing DNA methylation levels of SVA copies in five sperm donors from amplicon-seq. Only low-methylated SVA copies in male hPGCs at 19 weeks of gestation were used for the analysis. SVA copies were classified by DNA methylation levels of two sperm donors from Hammoud et al. Donor #5 showed significantly higher DNA methylation levels in ‘high and low’ SVA copies than other sperm donors. p-Value was calculated by Tukey’s test. (E) Scatter plot showing the DNA methylation between sperm donor 1 and sperm donor 2 from Hammoud et al. DNA methylation levels between these two donors were highly correlated. (F) Heatmap showing DNA methylation levels, genomic distribution, and overlap with SVAs of differentially methylated regions (DMRs). (G) Representative view of DMRs overlapping with SVA. Black and green boxes represent SVA and DMR, respectively.
-
Figure 6—source data 1
Raw data of graphs in Figure 6.
- https://cdn.elifesciences.org/articles/76822/elife-76822-fig6-data1-v2.xlsx
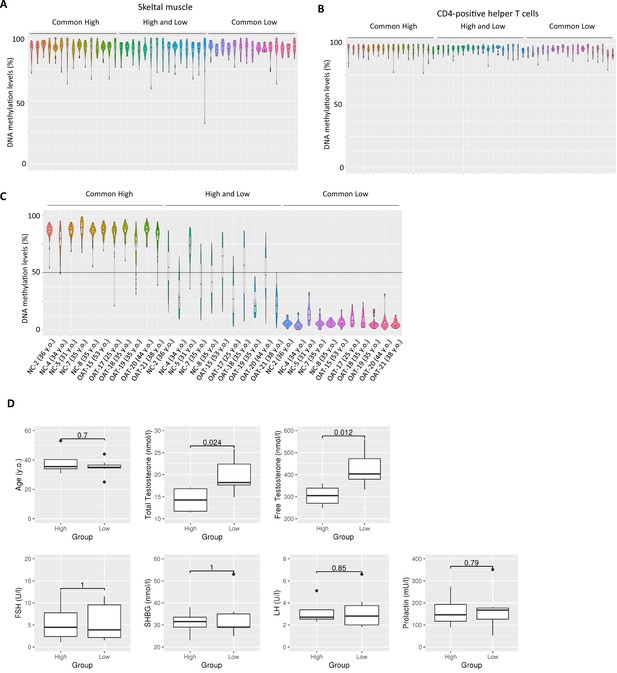
Inter-individual epigenetic variation in SINE-VNTR-Alus (SVAs) was correlated with testosterone levels in bloods.
(A–C) Violin plots showing DNA methylation levels of SVAs in adult skeletal muscle (A), adult CD-4-positive helper T-cell (B) and sperm from healthy and oligozoospermic men (C). In (C), NC and OAT represent normal control and oligozoospermic men, respectively. (D) Boxplots showing various physiological conditions between donors with high methylated ‘high and low’ SVAs (N = 4) and donors with low-methylated ones (N = 7) in (C). p-Value was calculated by Wilcoxon signed-rank test.
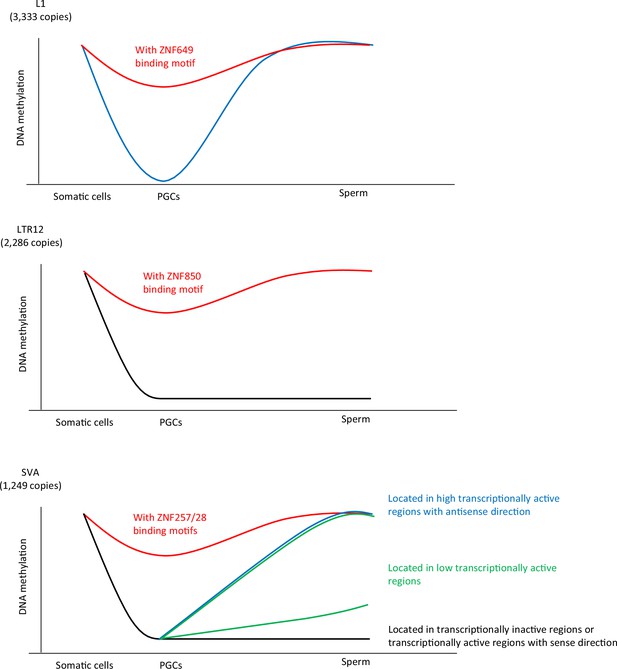
Summary of this study.
Our data demonstrated an association between KRAB-ZFP-binding motifs and the DNA demethylation resistance of L1, SVA, and LTR12. ZNF649, ZNF257/28, and ZNF850 were associated with DNA demethylation resistance of L1, SVA, and LTR12C, respectively. The dynamics of DNA methylation during spermatogenesis are largely different among retroelement types. The majority of L1 copies acquired DNA methylation during spermatogenesis, whereas the DNA methylation status of LTR12 in human primordial germ cells (hPGCs) tended to be maintained during spermatogenesis. The mode of DNA methylation changes in SINE-VNTR-Alus (SVAs) during spermatogenesis largely differs between copies and individuals. SVA copies located in highly transcriptionally active regions acquire DNA methylation during spermatogenesis, while those located in transcriptionally inactive regions maintain a hypomethylated state during spermatogenesis. In contrast, the degree of DNA methylation in sperm in SVA copies located in low transcriptionally active regions was highly variable among the individuals. These results suggest that SVAs may be methylated by transcription-directed DNA methylation mechanisms during spermatogenesis, and their activity varies among individuals.
Additional files
-
Supplementary file 1
List of full-length L1/SVA/LTR12.
- https://cdn.elifesciences.org/articles/76822/elife-76822-supp1-v2.xlsx
-
Supplementary file 2
Statistical analysis of DNA methylation levels of repeat elements in human primordial germ cells (PGCs) (related to Figure 1C–E).
- https://cdn.elifesciences.org/articles/76822/elife-76822-supp2-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76822/elife-76822-transrepform1-v2.docx





