Detecting molecular interactions in live-cell single-molecule imaging with proximity-assisted photoactivation (PAPA)
Figures
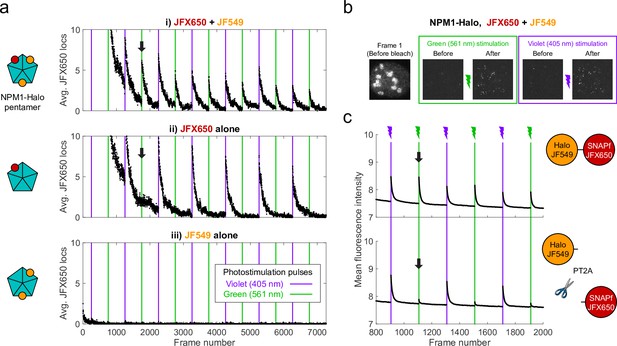
Proximity-assisted photoactivation (PAPA) of JFX650 by JF549.
(a) Green and violet light reactivate JFX650 through distinct JF549-dependent and JF549-independent mechanisms. Left column: schematic of NPM1 pentamers in heterozygously tagged NPM1-Halo U2OS cells labeled with JF549 (orange) and/or JFX650 (red). Right column: average number of localizations in the JFX650 channel as a function of frame number. JFX650 molecules were excited with red (639 nm) light, interspersed with 7 ms pulses of violet (405 nm) and green (561 nm) light (violet and green vertical lines). Reactivation of JFX650 by green light required labeling with JF549 (compare black arrows in i and ii). (b) Sample images of a single cell in the JFX650 channel. Leftmost panel: first movie frame prior to fluorophore bleaching/shelving. Green and violet boxes: maximum-intensity projection of all frames immediately before and after green and violet stimulation pulses, showing reactivation of molecules from the dark state. Image dimensions are 24 µm x 24 µm. (c) Average fluorescence intensity in the JFX650 channel as a function of frame number in cells expressing a Halo-SNAPf fusion with a flexible linker (top panel; N = 40 cells) or a tandem P2A-T2A self-cleaving peptide between Halo and SNAPf (PT2A; bottom panel; N = 20 cells). Halo was labeled with JF549-HTL and SNAPf with JFX650-STL. Reactivation by violet light pulses (violet lines) occurred in both cases, but reactivation by green light pulses (green lines) was mostly eliminated by the self-cleaving peptide (compare black arrows). Raw intensity traces are displayed without background subtraction.
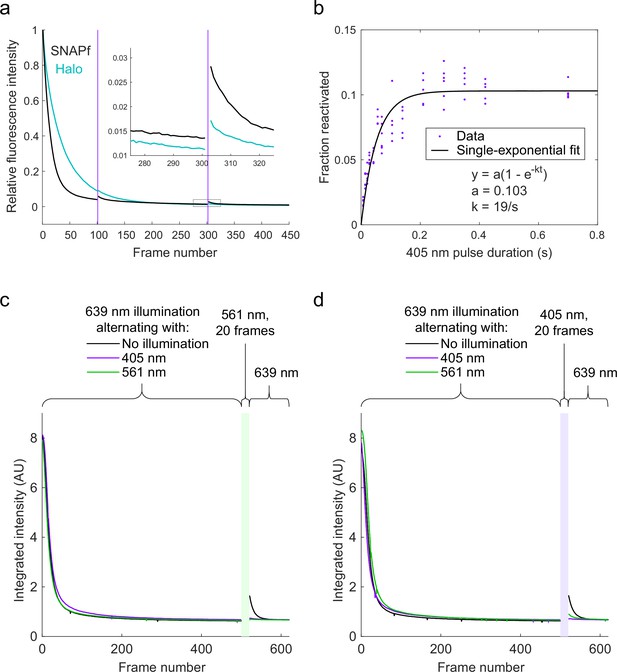
Properties of JFX650 reactivation.
(a) Shelving/bleaching and reactivation of JFX650 bound to Halo (cyan) and SNAPf (black). Relative fluorescence intensity is plotted on the y-axis, averaged over multiple cells (N = 10 for Halo, N = 14 for SNAPf), and frame number is plotted on the x-axis. The frame rate was 7.48 ms/frame. Fluorescence intensity (with red illumination) declined more rapidly for SNAPf-JFX650 than for Halo-JFX650. Violet pulses of 7 ms at frames 101 and 302 induced greater direct reactivation of JFX650-SNAPf than JFX650-Halo. (b) Reactivation of SNAPf-JFX650 as a function of violet pulse duration. SNAPf-JFX650 intensity was measured three times using 20 frames of 1 ms stroboscopic red illumination: (1) before bleaching/shelving, (2) after bleaching/shelving with 400 frames of non-stroboscopic (7 ms/frame) red illumination, and (3) after reactivation by exposure to violet pulses of varying duration. Percent reactivation was calculated by dividing the increase in intensity due to reactivation by the decrease in intensity due to bleaching/shelving. Solid black curve shows a fit to a single-exponential model. (c, d) Mutual occlusion of green and violet reactivation. (c) Halo-SNAPf-expressing U2OS cells labeled with JFX650-STL and JF549-HTL were imaged with red illumination alternating with unrecorded frames with either no illumination (black curve) or violet/green illumination (violet/green curves). Integrated intensity in the JFX650 channel is plotted on the y-axis and frame number on the x-axis. JFX650 intensity initially declined for all conditions due to bleaching and shelving. A 20-frame (140-ms) pulse of green light was applied after frame 500 (green rectangle). This reactivated JFX650 that had been exposed to red light only but failed to reactivate JFX650 that had been exposed to alternating red and green or red and violet light. (d) Same as (c), but with a 20-frame (140-ms) violet pulse after frame 500. Violet reactivation was strongly reduced by preceding green or violet light exposure. Some violet reactivation was still observed for cells exposed to alternating red and green light (green curve). Although this might indicate the presence of additional dark state(s) that can be reactivated by 405 nm light but not PAPA, it might also reflect incomplete labeling of Halo by JF549 or photobleaching of JF549 by green light.
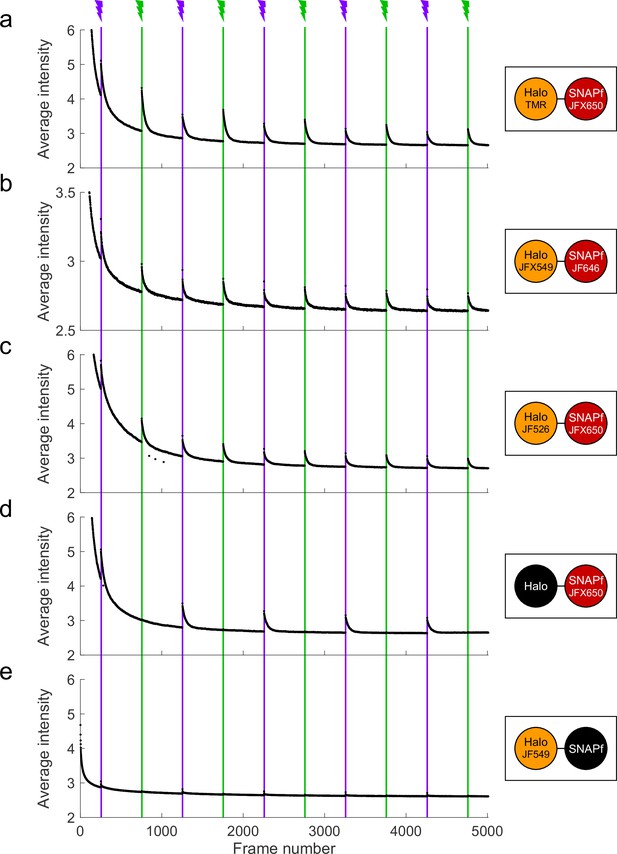
Proximity-assisted photoactivation (PAPA) between other sender–receiver pairs.
U2OS cells expressing Halo-SNAPf-3xNLS were labeled with different SNAP tag ligand (STL) and HaloTag ligand (HTL) fluorophore combinations and imaged with red light alternating with 7-ms pulses of green and violet light. Fluorescence intensity averaged over multiple cells is plotted on the vertical axis and frame number is plotted on the horizontal axis. (a) Tetramethylrhodamine (TMR)-HTL and JFX650-STL. (b) Janelia Fluor X549 (JFX549)-HTL and Janelia Fluor 646 (JF646)-STL. (c) Janelia Fluor 526 (JF526)-HTL and JFX650-STL. (d) JFX650-STL-only control. (e) JF549-only control.
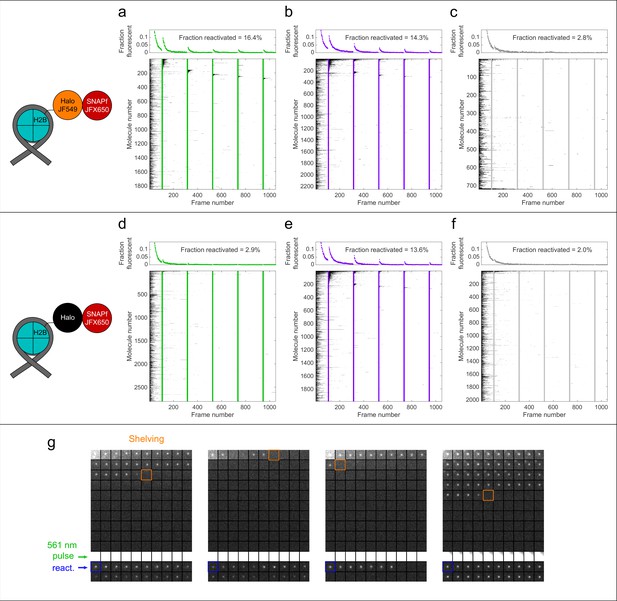
Proximity-assisted photoactivation (PAPA) and direct reactivation (DR) of immobilized single fluorophores.
Cells expressing H2B-Halo-SNAPf were labeled sparsely with JFX650 SNAP ligand, such that individual fluorophores could be resolved without photobleaching. Halo was labeled with JF549 (a–c, g) or left unlabeled as a negative control (d–f). Cells were imaged at 7.48 ms/frame with red illumination alternating with 10 frames of 561 nm (a, d), 405 nm (b, e), or no illumination (c, f), indicated by green, violet, and gray vertical lines, respectively. Top panels in (a–f) show the fraction of fluorophores detected in the first frame that were fluorescent in each subsequent frame. Bottom panels in (a–f) are kymographs of all fluorophores detected in the first frame, in which black squares represent the presence of a localization for that molecule and white squares represent the absence of a localization. For visualization, molecules are sorted based on the first pulse in which they reactivated (if at all). DR by 405 nm light above background was observed independent of the presence of JF549 (b, e), while PAPA by 561 nm light was dependent on JF549 (a, d). (g) Montages of single-molecule images in the JF549-Halo/JFX650-SNAPf double-labeled condition. Each square is a 2.2 × 2.2 µm (14 × 14 pixel) region, and the time interval is 7.48 ms between squares. Shelving of each molecule in a reversible dark state occurred stochastically at different frames, as indicated by the orange outlines. A 561 nm pulse between frames 101 and 110 (green arrowhead) induced fluorophore reactivation via PAPA (blue outlines).
Initial observation of proximity-assisted photoactivation (PAPA). Heterozygous NPM1-Halo knock-in U2OS cells were labeled sparsely with 50 pM JFX650 HTL and densely with 10 nM JF549 HTL.
Cells were alternately imaged at 8ms/frame with 561 nm light to excite JF549 (false-colored green frames) and 639 nm light to excite JFX650 (false-colored red frames). The movie is displayed at 1× normal speed, subsampling every fifth frame. The intensity scale for display is 0–20,000 camera counts for 561 nm illumination and 0–5000 counts for 639 nm illumination. Note that the density of localizations in this movie is higher than we typically use for single-particle tracking. Image dimensions are 24 µm x 24 µm.
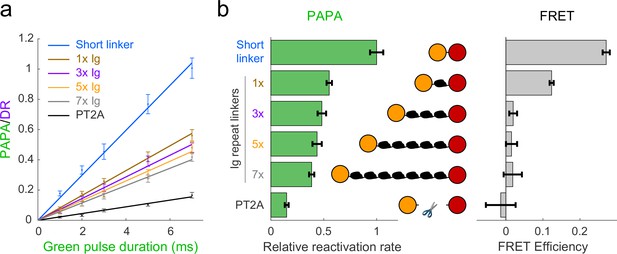
Comparison of distance dependence of proximity-assisted photoactivation (PAPA) and Förster resonance energy transfer (FRET).
(a) PAPA/direct reactivation (DR) ratio vs. green pulse duration for Halo-SNAPf fusions with a short, flexible linker or linkers containing different numbers of tandem Ig domains. Curves are linear fits (y = ax). Error bars, ±2 * SE. PT2A, tandem P2A-T2A self-cleaving peptide. (b) Left panel: relative rates of reactivation by PAPA (slope of fits in a divided by the slope of the short linker construct). Right panel: FRET efficiency measured using fluorescence lifetime imaging (FLIM).
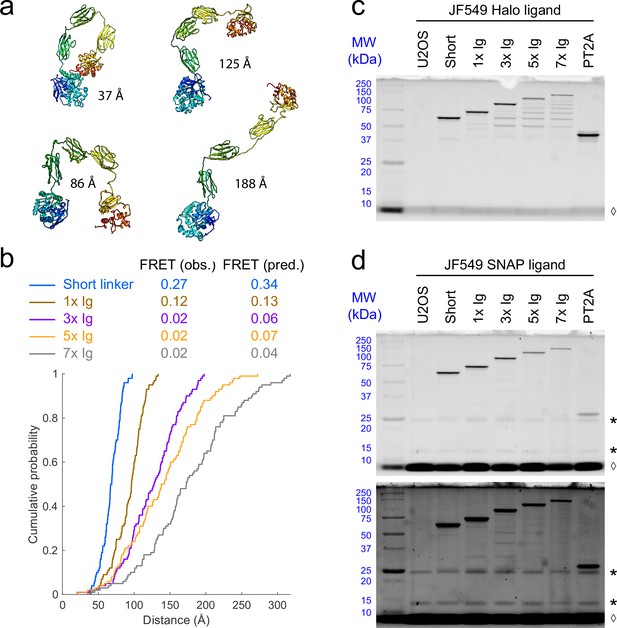
Simulations and SDS-PAGE analysis of linker constructs.
(a) Four example structures of the Halo-3xIg-SNAPf fusion protein obtained from PyRosetta simulations. Inter-dye distances are indicated for each structure. (b) Cumulative distributions of predicted inter-dye distances from Rosetta simulations, with observed (obs.) and predicted (pred.) Förster resonance energy transfer (FRET) values. Predicted FRET efficiency was calculated by averaging EFRET = 1/(1 + (R/R0)6) over all conformations in the ensemble, assuming a theoretical Förster radius of R0 = 58 Å for the JF549-JFX650 pair. (c, d) SDS-PAGE analysis of linker constructs expressed in U2OS cells and labeled with JF549-HTL (c) or JF549-STL (d). The amount of cell lysate loaded in each lane corresponds to 100,000 cells. Lookup table is set between 0 and 3000 counts for (c) and the top image in (d). The bottom image in (d) is the same gel with lookup table set between 0 and 800 counts to highlight faint bands. MW, molecular weight markers in kilodaltons. *, nonspecific bands. ◊, unbound dye. A ladder of smaller fragments is present below the full-length protein for the larger linkers; these are predominantly labeled by Halo ligand but not SNAP ligand (see Supplemental Note 1).
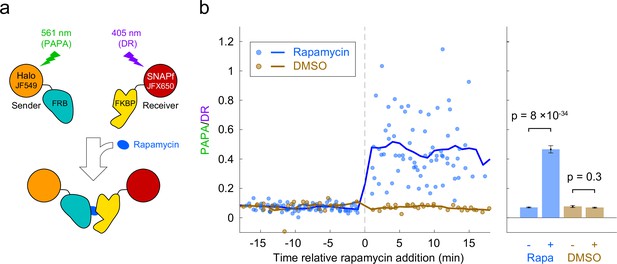
Detection of inducible dimerization using proximity-assisted photoactivation (PAPA).
(a) Halo-FRB was labeled with the sender fluorophore (JF549) and SNAPf-FKBP with the receiver fluorophore (JFX650). After shelving JFX650 with red light, direct reactivation (DR) and PAPA were alternately induced with pulses of violet and green light, respectively. Midway through the experiment, cells were treated with rapamycin (1 µM final concentration) to induce FRB-FKBP dimerization or with dimethylsulfoxide (DMSO) solvent as a negative control. (b) Ratio of fluorescence increase due to PAPA (green reactivation) and DR (violet reactivation) as a function of time after rapamycin addition. Blue, rapamycin. Brown, DMSO solvent-only control. Individual data points represent single cells; solid lines show a 2-min moving average. (c) Average PAPA/DR ratio before (-) and after (+) addition of rapamycin (Rapa) or DMSO. Total number of cells: 75 before and 74 after rapamycin, 30 before and 30 after DMSO. Error bars, ± 2 * SEM. Statistical significance was calculated using a two-tailed t-test.
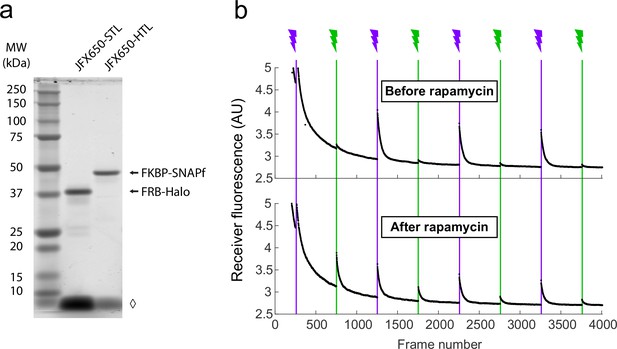
SDS-PAGE and proximity-assisted photoactivation (PAPA) traces of FRB-FKBP.
(a) Fluorescence image of an SDS-PAGE gel of lysates from a clonal stable U2OS cell line expressing FKBP-SNAPf and FRB-Halo. Amount of lysate loaded in each lane corresponds to approximately 60,000 cells labeled with either JFX650-STL or JFX650-HTL. ◊, unbound dye. (b) Total fluorescence of the JFX650 receiver fluorophore as a function of frame number when illuminated with red light alternating with pulses of violet light to induce direct reactivation (DR) and green light to induce PAPA (vertical lines with lightning bolts). The top panel shows the average of cells imaged prior to rapamycin addition (1 µM final concentration; n = 9), and the bottom panel shows the average of cells imaged between 5 and 15 min after rapamycin addition (n = 15). Fluorescence decreased initially due to bleaching and shelving of JFX650 and recovered upon fluorophore reactivation by green and violet light pulses. PAPA (green reactivation) was low prior to rapamycin addition (top panel) and increased following rapamycin addition (bottom panel). DR (violet reactivation) was observed both before and after rapamycin addition. AU, arbitrary units. Intensity traces are displayed without background subtraction.
Direct reactivation and proximity-assisted photoactivation (PAPA) of an inducible protein–protein interaction.
Reactivation by 561 nm and 405 nm light of FKBP-SNAPf-JFX650 before and after rapamycin-induced dimerization with FRB-Halo-JF549. Violet and green panels are maximum-intensity overlays of three photostimulation cycles by 405 nm and 561 nm light, respectively. The frame with the reactivation pulse occurs halfway through the movie. Numbered green–violet pairs correspond to individual cells that were imaged before (1–15) or after (16–29) addition of rapamycin (1 µM final concentration). Increased reactivation by 561 nm light is seen after rapamycin addition. Display is at 0.22× normal speed, and min–max pixel intensity range is between 0 and 5000 camera counts. Scale bar: 10 µm.
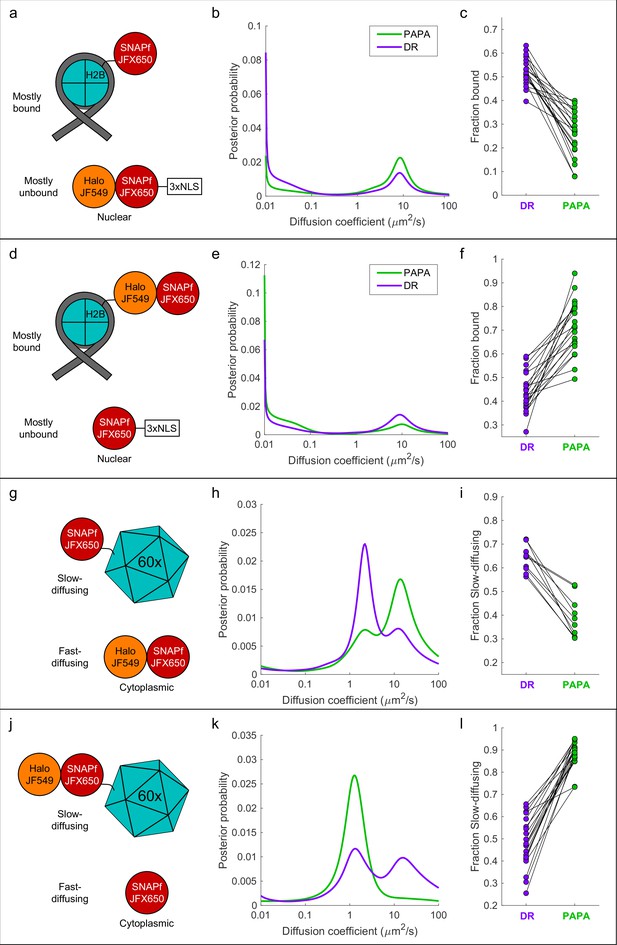
'Unmixing’ of defined two-component mixtures using proximity-assisted photoactivation (PAPA).
.(a) Left column (a, d, g, j): schematic of different defined mixtures of two labeled proteins, in which one protein is labeled with JFX650 only and the other is labeled with both JFX650 and JF549. In (g) and (j), each subunit of the 60-mer is fused to SNAPf or Halo-SNAPf, though only one label is displayed for clarity. (b) Center column (b, e, h, k): inferred diffusion spectra of PAPA (green-reactivated) and direct reactivation (DR) (violet-reactivated) trajectories pooled from 20 cells (b, e, k) or 10 cells (h). (c) Right column: fraction bound (c, f) or fraction slow-diffusing (i, l) of PAPA and DR trajectories from individual cells, obtained from fits to a two-state model (c, f) or three-state model (i, l). Paired, two-tailed t-tests of the comparisons in (c), (f), (i), and (l) showed all differences to be statistically significant with p=9 × 10–9, 8 × 10–8, 1 × 10–5, and 4 × 10–11, respectively.
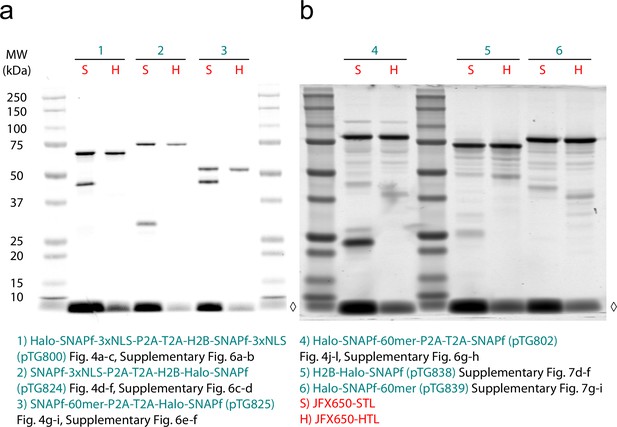
SDS-PAGE gels of defined two-component mixtures and one-component controls.
(a, b) Fluorescent SDS-PAGE gels of lysates from stable U2OS cell lines expressing defined two-component mixtures (1–4) and one-component controls (5 and 6). Cells were labeled with either JFX650-STL (S) or JFX650-HTL (H) prior to lysis, and each lane was loaded with a volume of lysate corresponding to approximately 60,000 cells. Associated figure panels are listed for each construct. ◊, unbound dye.
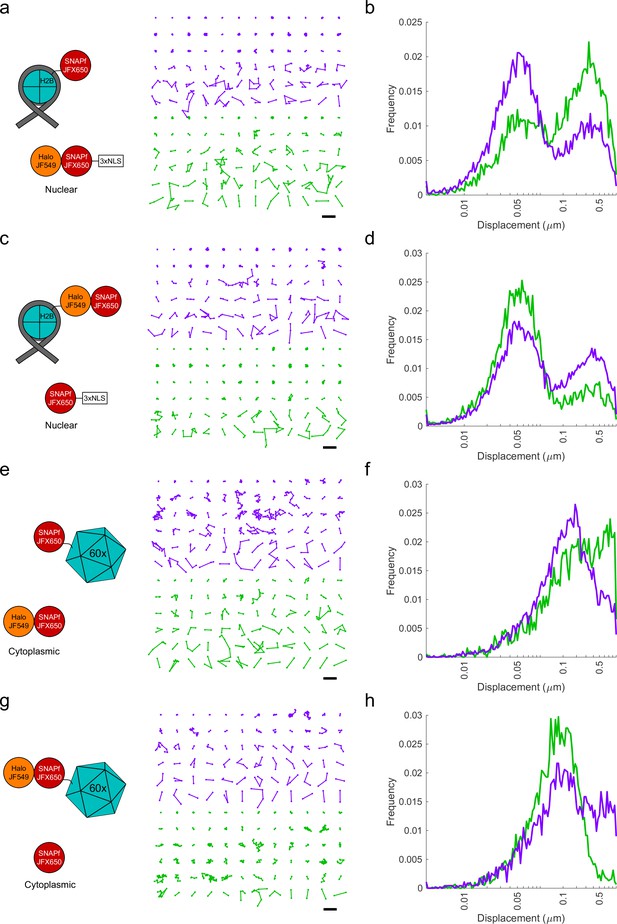
Additional analyses of proximity-assisted photoactivation–single-particle tracking (PAPA-SPT) experiment with two-component controls.
(a, c, e, g) Left panel: schematic of construct tested. Right panel: a random subset of PAPA (green) and direct reactivation (DR) (violet) trajectories from an individual cell. Trajectory centroids are aligned to a grid for visualization, and trajectories are displayed in order of increasing average displacement per step. Black scale bar, 1 µm. (b, d, f, h) Histograms of single-frame displacements for single-particle trajectories from all cells. Green, PAPA trajectories. Violet, DR trajectories. The sharp drop in frequency near 1 µm is due to the maximum displacement cutoff used in the particle tracking algorithm.
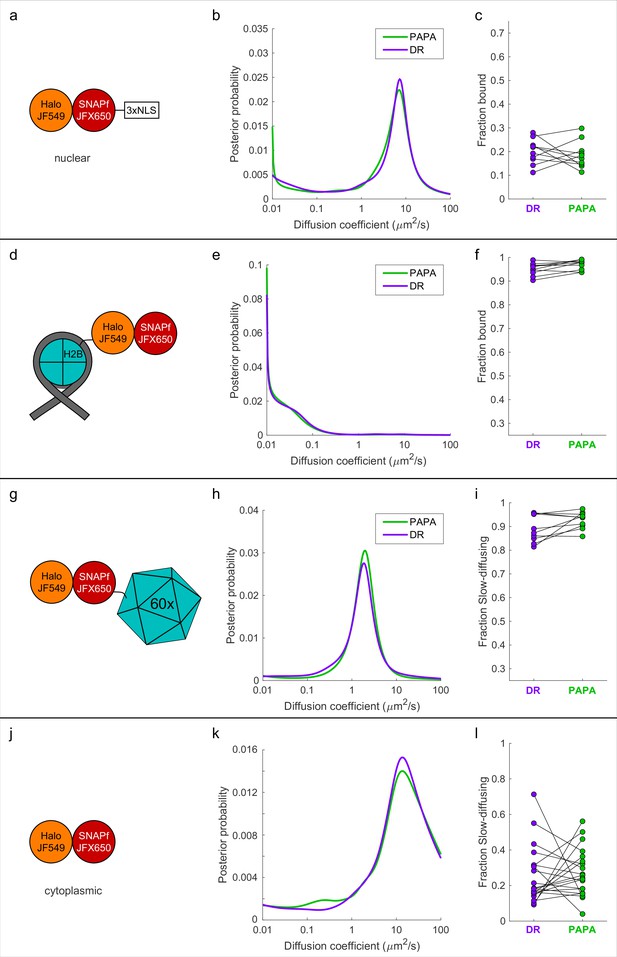
Proximity-assisted photoactivation–single-particle tracking (PAPA-SPT) analysis of single-component controls.
Individual Halo-SNAPf fusion proteins were labeled with a mixture of JF549-HTL and JFX650-STL, imaged with alternating green and violet photostimulation pulses, and analyzed as in Figure 4. (a–c) Halo-SNAPf-3xNLS. (d–f) Halo-SNAPf-H2B. (g–i) Halo-SNAPf-60-mer. Note that all 60 subunits are fused to Halo-SNAPf, but only a single label is shown for clarity. (j–l) cytoplasmic Halo-SNAPf. (b, e, h, k) Inferred diffusion spectra of PAPA (green-reactivated) and direct reactivation (DR) (violet-reactivated) trajectories pooled from 10 cells. (c, f, i, l) Fraction bound (c, f) or slow-diffusing (i, l) among PAPA and DR trajectories from individual cells, obtained from fits to a two-state (c, f) or three-state (i, l) model. p-Values calculated using a two-sided paired t-test were 0.58 (c), 0.017 (f), 0.071 (i), and 0.77 (l).
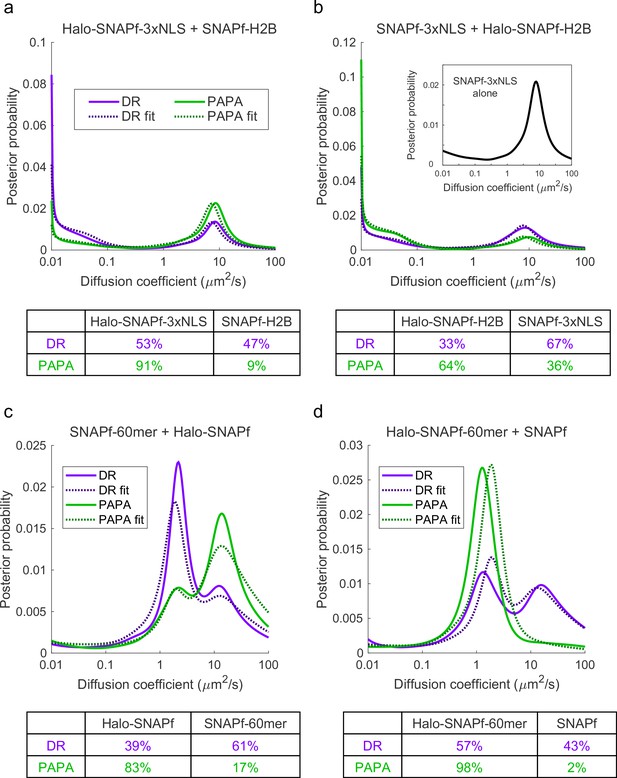
Fitting of two-component diffusion spectra to a mixture of single components.
Diffusion spectra for proximity-assisted photoactivation (PAPA) and direct reactivation (DR) were modeled as a linear combination of diffusion spectra of the corresponding single components, , where p1 is the proportion of component 1 and 1 - p1 is the proportion of component 2.
The MATLAB fminsearch function was used to identify the value of p1 that minimized the Kullback–Leibler divergence between the observed diffusion spectrum, , and the model: (a) Diffusion spectra of Halo-SNAPf-3xNLS + H2B-SNAPf (Figure 4b) modeled as a linear combination of DR diffusion spectra of Halo-SNAPf-3xNLS and H2B-Halo-SNAPf (violet curves in Figure 4—figure supplement 3b and e). (b) SNAPf-3xNLS + H2B-Halo-SNAPf (Figure 4e) modeled as a combination of SNAPf-3xNLS (inset, black curve) and H2B-Halo-SNAPf (violet curve in Figure 4—figure supplement 3b). (c) SNAPf-60mer + cytoplasmic Halo-SNAPf (Figure 4h) modeled as a combination of Halo-SNAPf-60mer and cytoplasmic Halo-SNAPf (violet curves in Figure 4—figure supplement 3h and k). (d) Halo-SNAPf-60mer + cytoplasmic SNAPf (Figure 4k) modeled as a combination of Halo-SNAPf-60mer and cytoplasmic Halo-SNAPf (violet curves in Figure 4—figure supplement 3h and k).
Proximity-assisted photoactivation–single-particle tracking (PAPA-SPT) experiment of a two-component mixture.
Example movies from the two-component mixture experiment in Figure 4j–l. Cells expressing a mixture of cytoplasmic SNAPf and Halo-SNAPf-60mer were double-labeled with JF549 HTL and JFX650 STL. JFX650 was imaged with 639 nm light and stimulated with alternating 405 nm and 561 nm light pulses to elicit direct reactivation (DR) and PAPA, respectively. Short movie segments spanning each stimulation pulse are color-coded violet (405 nm) and green (561 nm). Each numbered violet–green pair corresponds to a single cell. Reactivation occurs after each photostimulation pulse, and it is apparent by eye that 561-nm-reactivated molecules are strongly enriched for the slow-diffusing fraction, while 405-nm-reactivated molecules include a mixture of fast- and slow-diffusing components. Display is at 0.11× normal speed. Scale bar: 10 µm. Displayed min–max pixel intensity range is between 500 and 5000 camera counts.
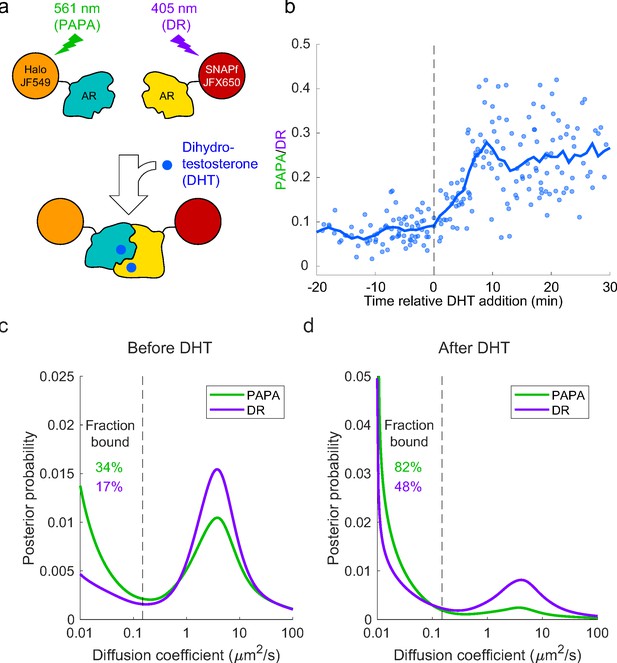
Analysis of mammalian androgen receptor using proximity-assisted photoactivation–single-particle tracking (PAPA-SPT).
(a) Schematic of dihydrotestosterone (DHT)-induced dimerization of JF549-Halo-mAR and JFX650-SNAPf-mAR. (b) PAPA/direct reactivation (DR) ratio as a function of time relative DHT addition. (c) Diffusion spectra of PAPA and DR trajectories. (c) Before addition of DHT; N = 55 cells. (d) After addition of DHT to a final concentration of 10 nM; N = 81 cells. Fraction bound was quantified by summing the portion of each curve below D = 0.15 µm2/s (vertical dashed line).
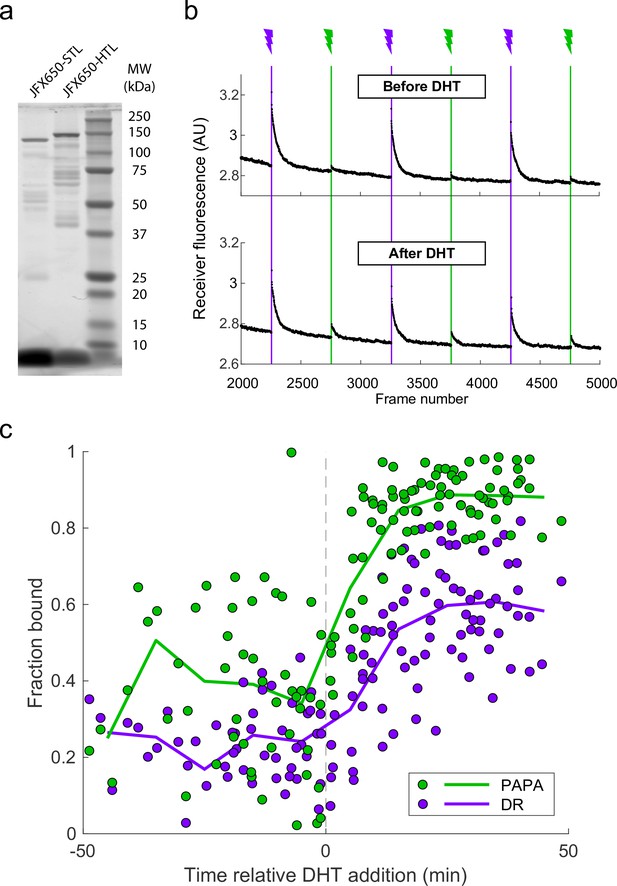
Proximity-assisted photoactivation (PAPA) analysis of androgen receptor.
(a) Fluorescent SDS-PAGE gel of lysates from a clonal stable U2OS cell line expressing SNAPf-mAR and Halo-mAR. Cells were stained with JFX650-STL or JFX650-HTL, and a volume of lysate corresponding to 60,000 cells was loaded per lane. MW, molecular weight in kilodaltons. (b) Ensemble PAPA analysis (see Figure 3—figure supplement 1b legend) of interaction between SNAPf-mAR and Halo-mAR. PAPA signal (green reactivation) increased after addition of dihydrotestosterone (DHT) to a final concentration of 10 nM. Intensity traces are displayed without background subtraction. (c) Fraction bound as a function of time relative DHT addition (vertical dashed line) for PAPA trajectories (green) and direct reactivation (DR) trajectories (violet), based on fits to a two-state model. Each data point corresponds to PAPA or DR trajectories from a single cell. Solid lines show moving averages over 10-min intervals.
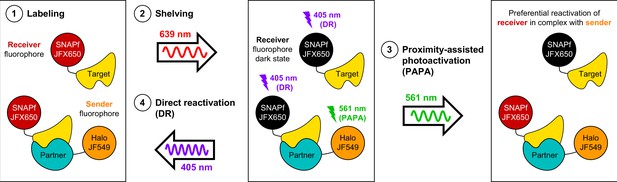
Using proximity-assisted photoactivation (PAPA) to spotlight protein–protein interactions.
(1) Label a SNAPf-tagged Target protein with a receiver fluorophore (e.g., JFX650) and a Halo-tagged Partner protein with a sender fluorophore (e.g., JF549). (2) Shelve the receiver fluorophore in the dark state using intense 639 nm illumination. Image receiver molecules with 639 nm light while alternately illuminating with (3) pulses of 561 nm light to induce PAPA of receiver-labeled Target molecules in complex with sender-labeled Partner molecules, and (4) pulses of 405 nm light to induce direct reactivation (DR) of receiver fluorophores, independent of proximity to the sender.
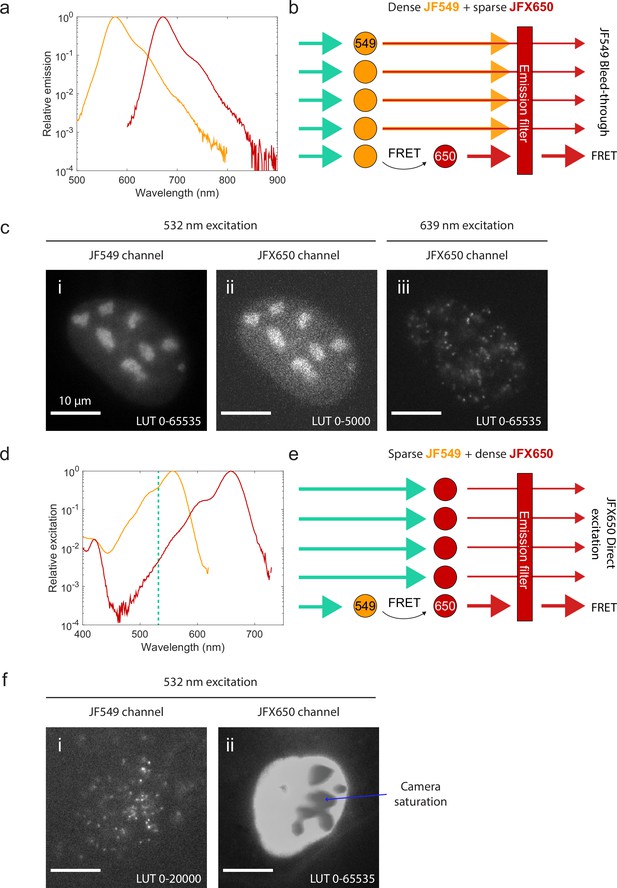
Spectral crosstalk impedes the use of single-molecule Förster resonance energy transfer (smFRET) as an interaction sensor in live cells.
(a) Emission spectra of JF549 (orange) and JFX650 (red). Data from fpbase.org are replotted with a logarithmic y-axis scale to show the long tail of emission. (b) Schematic of donor bleed-through. Weak far-red emission from the donor fluorophore (JF549) passes through the acceptor emission filter (JFX650), overwhelming the much weaker FRET signal if the donor is densely labeled and the acceptor is sparsely labeled. (c) Experimental example of high background due to donor bleed-through. U2OS cells with endogenously tagged NPM1-Halo were double-labeled with a high concentration of JF549 HTL and a lower concentration of JFX650 HTL. Excitation of JF549 with 532 nm light yielded diffuse bleed-through signal in the JFX650 channel (ii), in contrast to the discrete single-molecule spots seen with direct excitation of JFX650 with 639 nm light (iii). (d) Absorption spectra of JF549 (orange) and JFX650 (red). Data from fpbase.org are replotted with a logarithmic y-axis scale to show the long tail of absorption. The wavelength used for donor excitation in this figure, 532 nm, is shown as a vertical dashed line. (e) Schematic of acceptor direct excitation. While the acceptor fluorophore (JFX650) is excited by 532 nm light more weakly than the donor fluorophore (JF549), this direct excitation can overwhelm the FRET signal if the acceptor is in large excess of the donor. (f) Experimental example of high background due to acceptor direct excitation. U2OS cells with endogenously tagged NPM1-Halo were double-labeled with a high concentration of JFX650 HTL and a lower concentration of JF549 HTL. Excitation of JF549 with 532 nm light yielded sparse single-molecule localizations in the JF549 channel but extremely strong, diffuse signal in the JFX650 channel due to JFX650 direct excitation. This background fluorescence was so intense that it saturated the EMCCD camera, leading to artifactual gray splotches in the image (blue arrow).
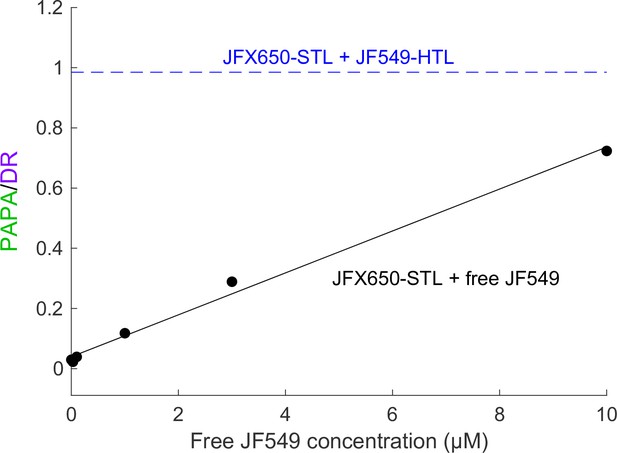
Background reactivation by free JF549 dye.
U2OS cells expressing Halo-SNAPf-3xNLS were labeled with JFX650-STL only and incubated with various concentrations of free JF549 dye. PAPA/DR ratio (black points) was measured as in the main text figures by calculating the ratio of reactivation due to 7 ms pulses of 561 nm and 405 nm light. Solid black line shows a linear fit. For comparison, the dashed blue line shows the PAPA/DR ratio measured on the same day for Halo-SNAPf-3xNLS double-labeled with JFX650-STL and JF549-HTL.
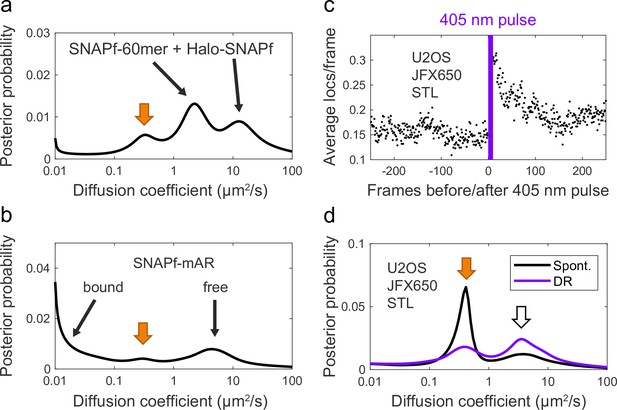
SNAP ligand background staining.
(a, b) Two example diffusion spectra of spontaneously reactivated trajectories in which an additional unidentified peak was observed at ~0.3–0.4 µm2/s (orange arrow). (a) SNAPf-60mer + Halo-SNAPf 2-component mixture. (b) SNAPf-mAR + Halo-mAR with 10 nM dihydrotestosterone (DHT). (c) 405 nm reactivation of JFX650 STL background staining in U2OS cells not expressing a SNAPf-tagged protein, averaged over 10 cycles of photostimulation. (d) Diffusion spectrum of JFX650 STL background staining in U2OS cells not expressing a SNAPf-tagged protein. Background staining consists of distinct slow-diffusing (orange arrow, ~0.3–0.4 µm2/s) and fast-diffusing (white arrow, ~3–4 µm2/s) components.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/76870/elife-76870-transrepform1-v3.docx
-
Source data 1
Raw data of the plots in Figure 2, Figure 3, Figure 4, and Figure 5.
fig22_FLIM.mat: MATLAB data file containing calculated fluorescence lifetimes (t1) and Förster resonance energy transfer (FRET) efficiencies (FRETe) for each condition in Figure 2. fig22_PAPA.csv: mean and standard error of proximity-assisted photoactivation/direct reactivation (PAPA/DR) ratio as a function of 561 nm pulse duration for each condition in Figure 2. fig33_rawdata.mat: single-cell PAPA/DR ratio measurements for all cells in the rapamycin-treated (allrapa) and dimethylsulfoxide (DMSO)-treated control (alldmso) conditions in Figure 3. The top row gives the time in minutes before/after treatment, while the lower row gives the PAPA/DR ratio. fig44.zip: subfolders contain data plotted in the corresponding figure panels. Subfolders b, e, h, and k contain two files called 0_rbme_marginal_posterior.csv and 1_rbme_marginal_posterior.csv, which correspond to diffusion spectra of PAPA and DR trajectories, respectively. The first column contains diffusion coefficients, while the second column contains probabilities. Subfolders c, f, i, and l contain the fraction bound (c, f) or fraction slow-diffusing (i, l) for PAPA and DR trajectories from single cells, obtained from analysis with a reduced model. fig5b.mat: single-cell PAPA/DR ratio measurements for all cells in the experiment in Figure 5b. The top row gives the time in minutes before/after DHT treatment, while the lower row gives the PAPA/DR ratio. fig5c.mat: diffusion spectra for PAPA and DR trajectories in Figure 5d. fig5d.mat: diffusion spectra for PAPA and DR trajectories in Figure 5d.
- https://cdn.elifesciences.org/articles/76870/elife-76870-data1-v3.zip





