XAB2 dynamics during DNA damage-dependent transcription inhibition
Figures
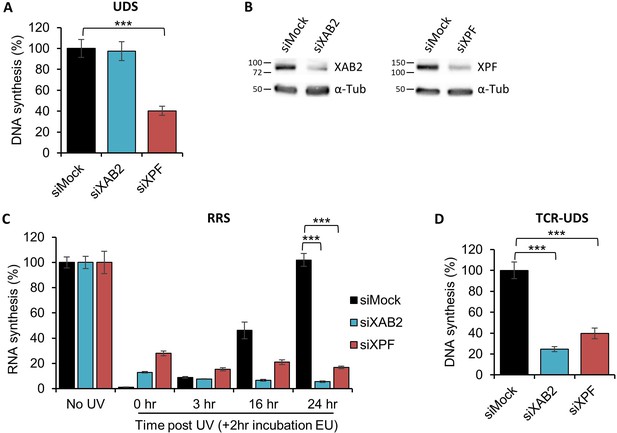
XAB2 is involved in DNA repair.
(A) Quantification of Unscheduled DNA Synthesis (UDS) assay determined by EdU incorporation after local damage (LD) induction with UV-C (100 J/m2) in WT cells (MRC5 cells) treated with siRNAs against indicated factors. Error bars represent the standard error of the mean (SEM) obtained from at least 30 LDs. (B) Western blot on whole-cell extracts of MRC5 cells treated with siRNA against indicated factors. (C) Quantification of RNA Recovery Synthesis (RRS) assay determined by EU incorporation after UV-C (10 J/m2) exposure in WT cells treated with siRNAs against indicated factors. Error bars represent the SEM obtained from at least 50 cells. (D) Quantification of TCR-UDS assay determined by EdU incorporation after LD induction with UV-C (100 J/m2) in GG-NER-deficient cells (XPC−/− cells) treated with siRNAs against indicated factors. Error bars represent the SEM obtained from at least 15 LDs. For all graphs, p-value of Student’s test compared to siMock condition: ***<0.001.
-
Figure 1—source data 1
Source data for Figure 1A: quantification of UDS siXAB2.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig1-data1-v3.xlsx
-
Figure 1—source data 2
Source data for Figure 1C: quantification of RRS siXAB2.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig1-data2-v3.xlsx
-
Figure 1—source data 3
Source data for Figure 1D: quantification of TCR-UDS siXAB2.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig1-data3-v3.xlsx
-
Figure 1—source data 4
Figures with the uncropped blots and relevant bands clearly labeled for Figure 1B: Western blot siXAB2 efficiency.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig1-data4-v3.pdf
-
Figure 1—source data 5
The original files of the full raw unedited gels for Figure 1B: Western blot siXAB2 efficiency.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig1-data5-v3.zip

UDS siXAB2.
Representative images of UDS from Figure 1A for each condition. The position of the local damage is visualized by γH2AX labeling (in red). Scale bar: 3 µm.
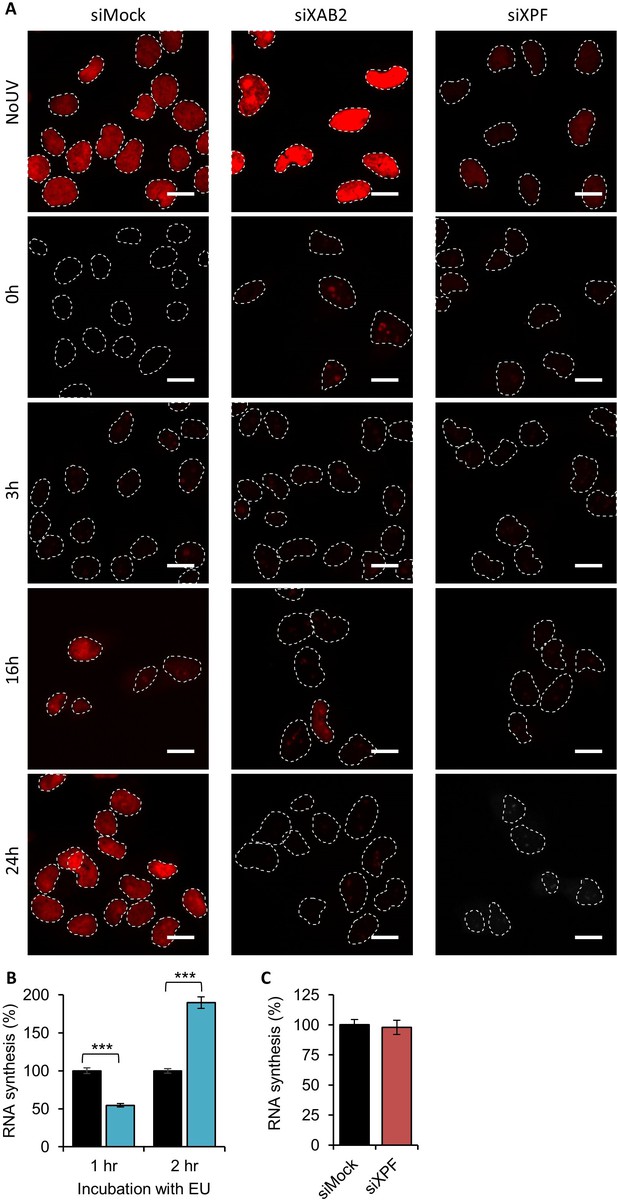
RRS siXAB2.
(A) Representative images of RRS from Figure 1C for each condition. Scale bar: 15 µm. (B) Quantification of RNA synthesis determined by 1 or 2 hr of EU incorporation without UV-C treatment in MRC5 cells transfected or not with siXAB2. (C) Quantification of RNA synthesis determined by 2 hr of EU incorporation without UV-C treatment in MRC5 cells transfected or not with siXPF. Error bars represent the standard error of the mean (SEM) obtained from at least 50 cells. P-value of Student’s test compared to siMock condition: ***<0.001.
-
Figure 1—figure supplement 2—source data 1
Source data for Figure 1—figure supplement 2B, C: quantification of RNA synthesis.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig1-figsupp2-data1-v3.xlsx

TCR-UDS siXAB2.
Representative images of TCR-UDS from Figure 1D for each condition. White arrows show the position of the local damage as visualized by γH2AX labeling. Nuclei are delimited by dashed lines. Scale bar: 3 µm.
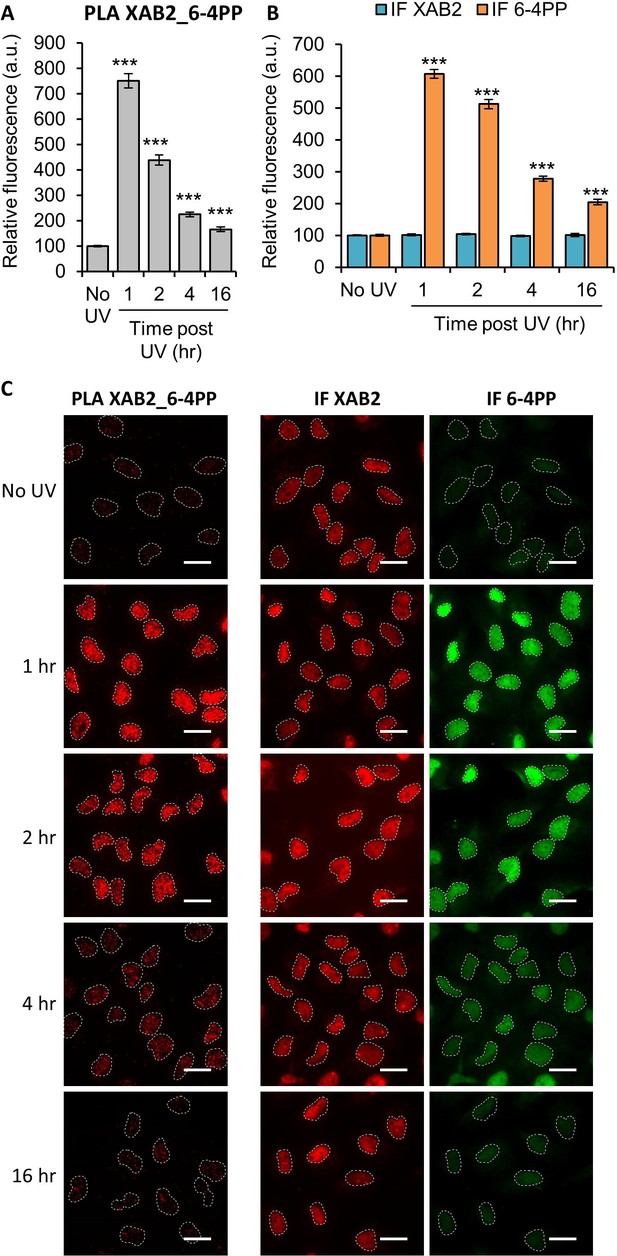
XAB2 interacts with the UV lesion 6-4 Photoproduct (6-4PP).
Quantification of fluorescent signal in the nucleus against the couple XAB2_6-4PP from Proximity Ligation Assay (PLA) experiment (A) or from the immunofluorescence (IF) done in parallel to PLA assay with the same antibodies dilutions (B). Error bars represent the standard error of the mean (SEM) obtained from at least 80 cells. P-value of Student’s test compared to No UV condition: ***<0.001. (C) Representative images of the PLA and IF experiments. Nuclei are delimited by dashed lines. Scale bar: 15 µm.
-
Figure 2—source data 1
Source data for Figure 2A, B: quantification of PLA and IF XAB2_6-4PP.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig2-data1-v3.xlsx
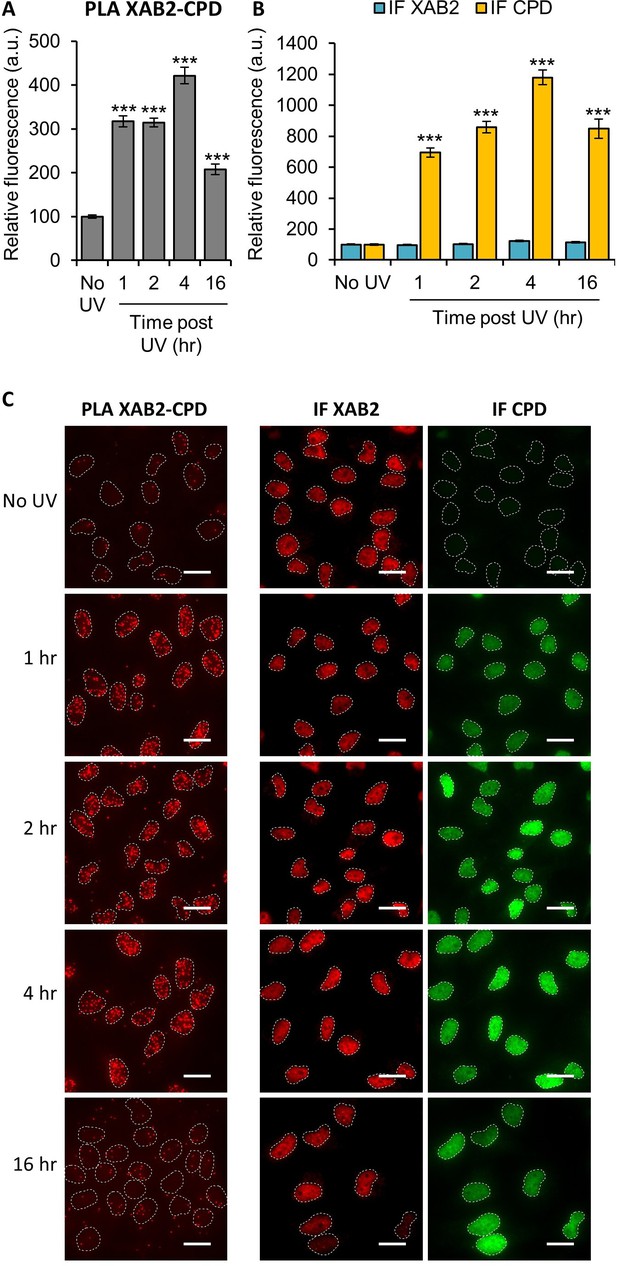
XAB2 interacts with the ultraviolet light (UV) lesions Cyclo-Pyrimidine Dimer (CPD).
Quantification of fluorescent signal in the nucleus against the couple XAB2_CPD from PLA experiment (A) or from the IF done in parallel to PLA assay with the same antibodies dilutions (B). Error bars represent the standard error of the mean (SEM obtained from at least 70 cells). P-value of Student’s test compared to No UV condition: ***<0.001. (C) Representative images of the PLA and IF experiment. Nuclei are delimited by dashed lines. Scale bar: 15 µm.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1A, B: quantification of PLA and IF XAB2_CPD.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig2-figsupp1-data1-v3.xlsx
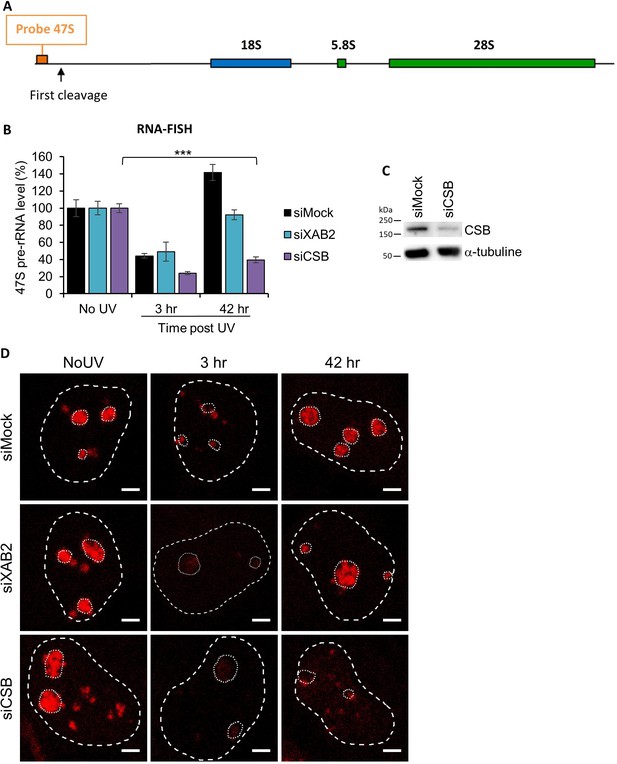
RNA-FISH siXAB2.
(A) Schematic representation of ribosomal DNA (rDNA) unit and localization of the 47S pre-rRNA probe. (B) Quantification of RNA-FISH (Fluorescence in situ hybridization) assay showing the 47S pre-rRNA level after UV-C (16 J/m2) exposure in WT cells treated with siRNAs against indicated factors. Error bars represent the standard error of the mean (SEM) obtained from at least 30 cells. P-value of Student’s test compared to No UV condition: ***<0.001. (C) Western blot on whole-cell extracts of MRC5 cells treated with siRNA against indicated factors. (D) Representative images of RNA-FISH 47S (in red) from the graph B of this figure for each condition. Nuclei and nucleoli are delimited by dashed and dotted lines, respectively. Scale bar: 3 µm.
-
Figure 2—figure supplement 2—source data 1
Source data for Figure 2—figure supplement 2B: quantification of RNA-FISH.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig2-figsupp2-data1-v3.xlsx
-
Figure 2—figure supplement 2—source data 2
Figures with the uncropped blots and the relevant bands clearly labeled for Figure 2—figure supplement 2C: Western blot siCSB efficiency.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig2-figsupp2-data2-v3.pdf
-
Figure 2—figure supplement 2—source data 3
The original files of the full raw unedited gels for Figure 2—figure supplement 2C: Western blot siCSB efficiency.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig2-figsupp2-data3-v3.zip
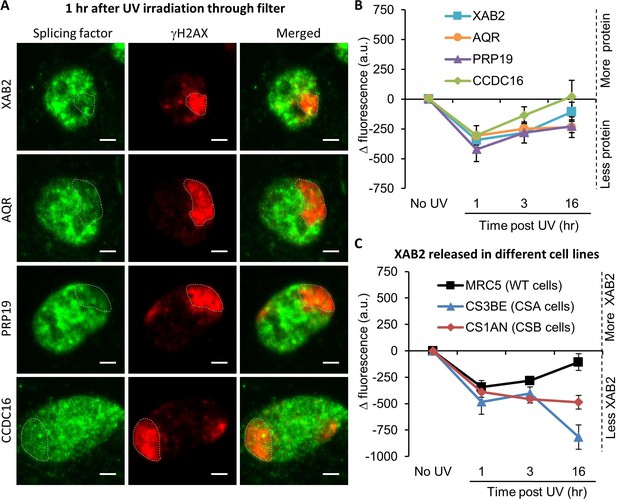
Splicing complex is released from DNA damage.
(A) Representative confocal images of immunofluorescence (IF) against XAB2, AQR, PRP19, or CCDC16 (green) and γH2AX (red) 1 hr after local damage (LD) induction with UV-C (60 J/m2). LDs are indicated by dashed lines. Scale bar: 3 µm. (B) Quantification of the IF signal of the different splicing proteins on the LD after different times of recovery. (C) Quantification of XAB2 signal on LD in different cell lines after different times of recovery. For both graphs, the signal from the local damage has been subtracted from the background of each cell. Error bars represent the standard error of the mean (SEM) obtained from at least 20 cells.
-
Figure 3—source data 1
Source data for Figure 3B: quantification of splicing complex IF.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig3-data1-v3.xlsx
-
Figure 3—source data 2
Source data for Figure 3C: quantification of XAB2 IF.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig3-data2-v3.xlsx

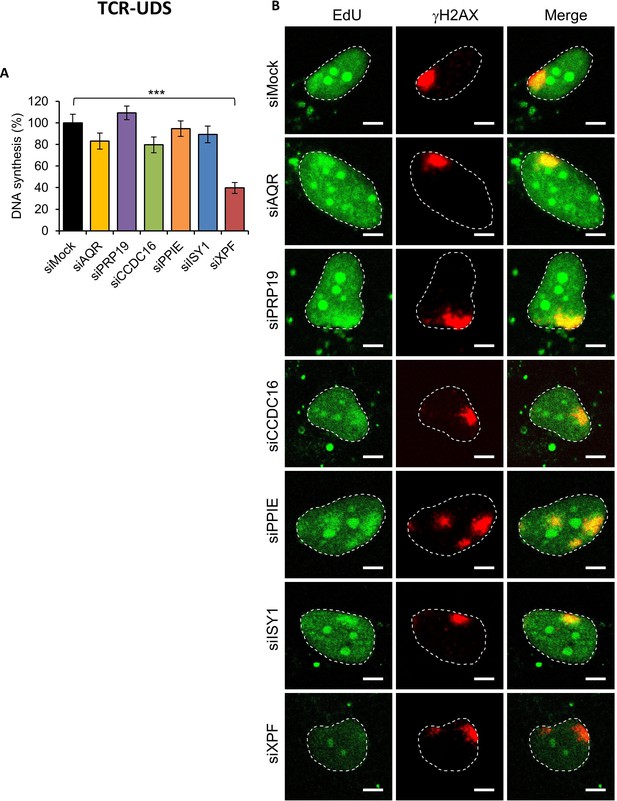
UDS in splicing complex-silenced cell.
(A) Quantification of UDS determined by EdU incorporation after LD induction with UV-C (100 J/m2) in WT cells (MRC5 cells) treated with siRNAs against indicated factors. Error bars represent the standard error of the mean (SEM) obtained from at least 15 LDs. P-value of Student’s test compared to siMock condition: ***<0.001. (B) Representative images of UDS for each condition. The position of the local damage is visualized by γH2AX labeling (in red). Nuclei are delimited by dashed lines. Scale bar: 5 µm. (C) Western blot on whole-cell extracts of cells treated with siRNA against indicated factors.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1A: quantification of UDS in splicing complex-silenced cells.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig3-figsupp1-data1-v3.xlsx
-
Figure 3—figure supplement 1—source data 2
Figures with the uncropped blots and relevant bands clearly labeled for Figure 3—figure supplement 1C: Western blot efficiency of siRNA against splicing complex.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig3-figsupp1-data2-v3.pdf
-
Figure 3—figure supplement 1—source data 3
The orignal files of the full raw unedited gels for for Figure 3—figure supplement 1C: Western blot efficiency of siRNA against splicing complex.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig3-figsupp1-data3-v3.zip
TCR-UDS in splicing complex-silenced cells.
(A) Quantification of TCR-UDS determined by EdU incorporation after local damage induction with UV-C (100 J/m2) in GG-NER-deficient cells (XPC−/− cells) treated with siRNAs against indicated factors. Error bars represent the standard error of the mean (SEM) obtained from at least 15 LDs. P-value of Student’s test compared to siMock condition: ***<0.001. (B) Representative images of TCR-UDS for each condition. The position of the LD is visualized by γH2AX labeling (in red). Nuclei are delimited by dashed lines. Scale bar: 5 µm.
-
Figure 3—figure supplement 2—source data 1
Source data for Figure 3—figure supplement 2A: quantification of TCR-UDS in splicing complex-silenced cells.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig3-figsupp2-data1-v3.xlsx
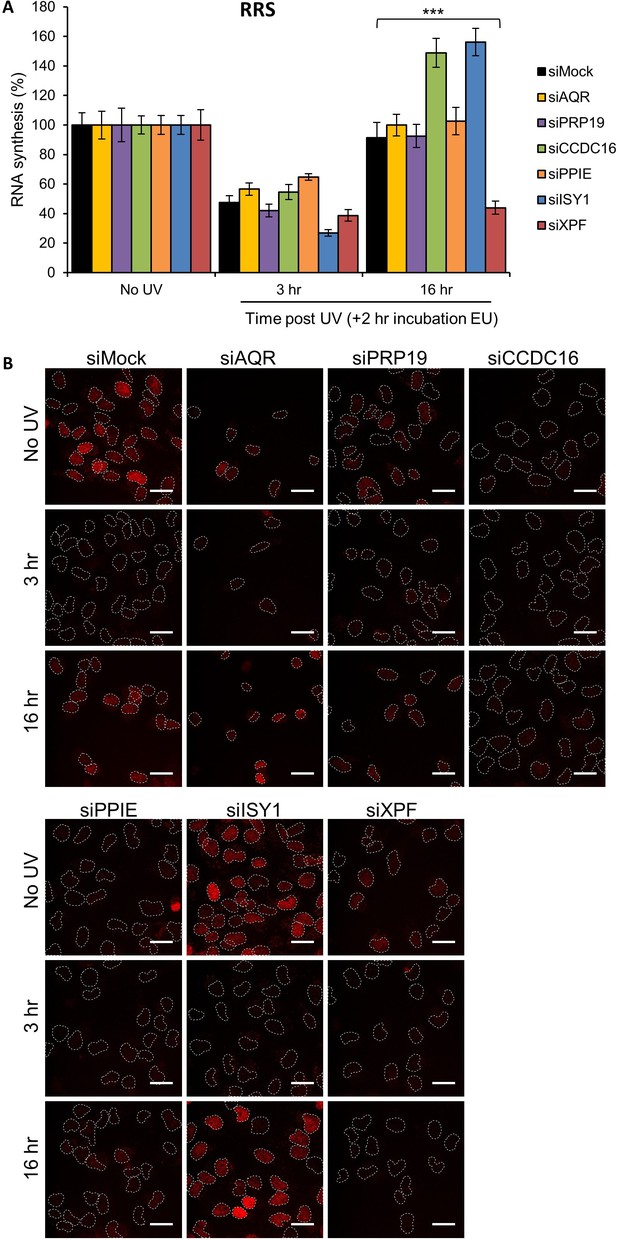
RRS in splicing complex-silenced cells.
(A) RRS determined by EU incorporation after UV-C (10 J/m2) exposure in MRC5 cells treated with siRNAs against indicated factors. Error bars represent the standard error of the mean (SEM) obtained from at least 50 cells. P-value of Student’s test compared to siMock condition: ***<0.001. (B) Representative images of RRS for each condition. Nuclei are delimited by dashed lines. Scale bar: 15 µm.
-
Figure 3—figure supplement 3—source data 1
Source data for Figure 3—figure supplement 3A: quantification of RRS in splicing complex-silenced cells.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig3-figsupp3-data1-v3.xlsx

XAB2 is released from DNA damage also in TC-NER-deficient cells.
Representative confocal images of IF against XAB2 (green) and γH2AX (red) after local damage induction with UV-C (60 J/m2) in different cell lines. LDs are indicated by dotted lines.
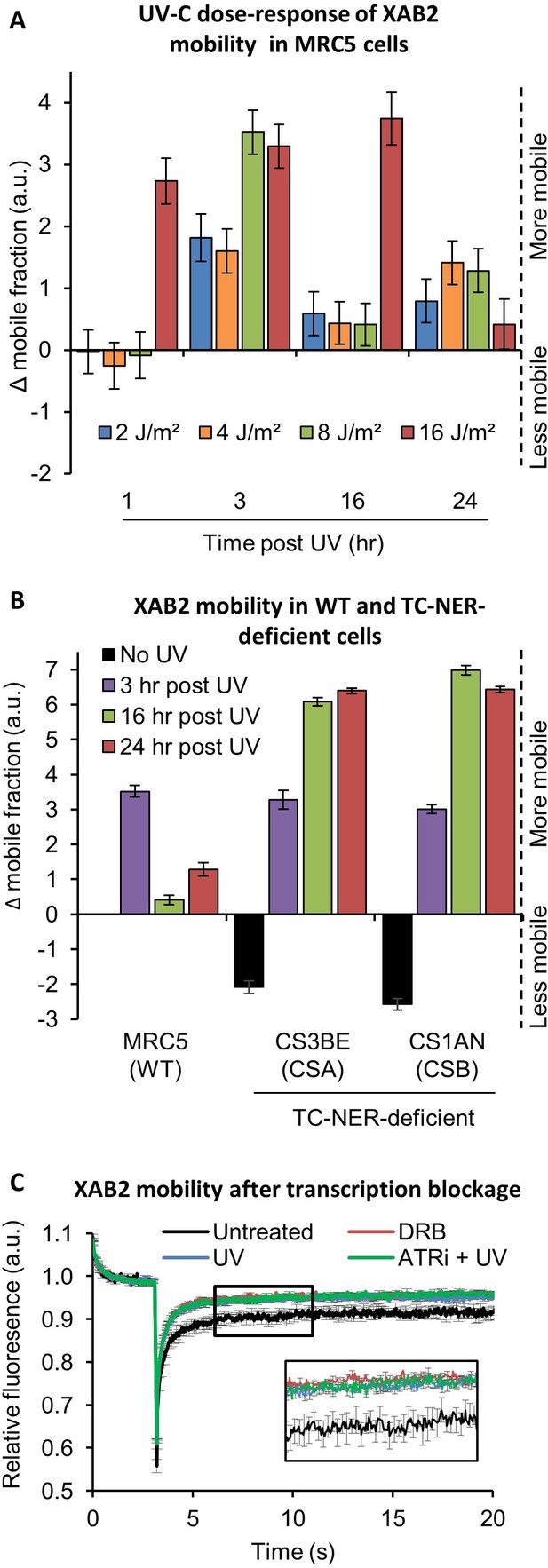
XAB2 is dynamic during TC-NER.
(A) Fluorescent recovery after photobleaching (FRAP) analysis of XAB2-GFP mobility in WT cells. Cells were treated or not with different doses of UV-C (2–16 J/m2) and XAB2 mobility was measured at different time points after UV-C exposure. The No UV condition was used to calculate the change in bound fraction. (B) FRAP analysis of XAB2-GFP expressed in WT cells (MRC5-SV) and TC-NER-deficient cells (CSA −/− and CSB−/−). Cells were treated or not with 8 J/m2 of UV-C. The No UV condition of the WT cell lines was used to calculate the change in bound fraction. (C) FRAP analysis of XAB2-GFP mobility in WT cells after treatment with 100 µg/ml of DRB for 2 hr (red line) or with 10 J/m2 of UV-C for 3 hr (blue line) or nothing (dark curve). Inhibitor of ATR pathway was added at 10 µM in the medium 1 hr before irradiation (green line). For all graphs, error bars represent the standard error of the mean (SEM) obtained from at least 10 cells.
-
Figure 4—source data 1
Source data for Figure 4A: FRAP XAB2-GFP with different doses of UV-C.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig4-data1-v3.xlsx
-
Figure 4—source data 2
Source data for Figure 4B: FRAP XAB2-GFP in different cell lines.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig4-data2-v3.xlsx
-
Figure 4—source data 3
Source data for Figure 4C: FRAP XAB2-GFP after different treatments.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig4-data3-v3.xlsx

FRAP of XAB2-GFP.
(A) Schematic representation of FRAP experiment and the calculation method used to represent FRAP data in Figure 4A,B and in the graph D of this figure. (B) Schematic representation of the XAB2-GFP construct. (C) Western blot on whole-cell extracts of MRC5 cells transfected with the XAB2-GFP plasmid. (D) FRAP analysis of XAB2-GFP mobility in MRC5 cells. FRAP experiment was performed 3 hr after irradiation with 10 J/m2 of UV-C. Inhibitor of ATM pathway was added at 10 µM in the medium 1 hr before irradiation. The untreated condition was used to calculate the change in bound fraction. Error bars represent the standard error of the mean (SEM) obtained from at least 10 cells.
-
Figure 4—figure supplement 1—source data 1
Source data for Figure 4—figure supplement 1D: FRAP XAB2-GFP after different treatments.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig4-figsupp1-data1-v3.xlsx
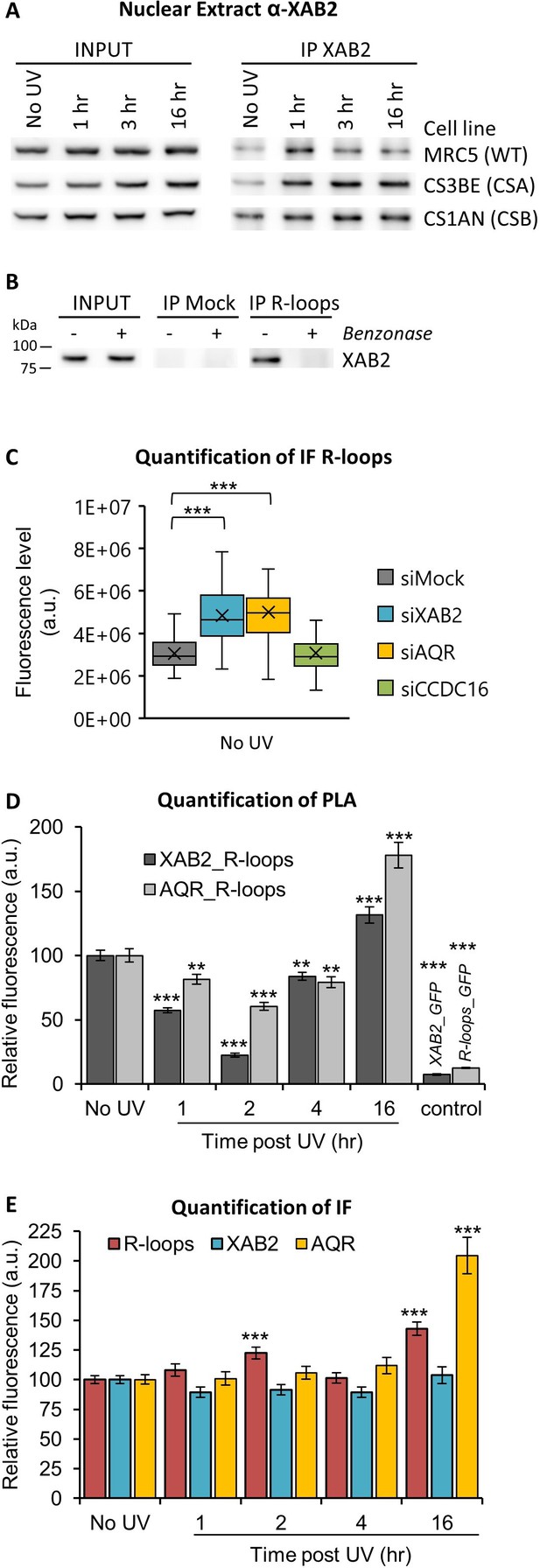
XAB2 and AQR are released from R-loops during DNA repair.
(A) Immunoprecipitation (IF) of XAB2 in nuclear extract from different cell lines treated with 10 J/m2 of UV-C at different times. Bound proteins were revealed by Western blotting with antibodies against XAB2. INPUT, 10% of the lysate used for IP reaction. (B) IP of R-loops in non-crosslinked chromatin extract from WT cells treated or not with Benzonase. XAB2 bounds to R-loops was revealed by Western blotting. INPUT, 10% of the lysate used for IP reaction. (C) Quantification of IF against R-loops in WT cells treated with siRNAs against indicated factors. Quantification of fluorescent signal in the nucleus against the couple XAB2_R-loops or AQR_R-loops from PLA experiments (D) or from the IF done in parallel to PLA assays (E). Error bars represent the standard error of the mean (SEM) obtained from at least 50 cells. P-value of Student’s test compared to No UV or siMock condition: **<0.01; ***<0.001.
-
Figure 5—source data 1
Source data for Figure 5C: quantification of IF R-loops in silenced cells.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-data1-v3.xlsx
-
Figure 5—source data 2
Source data for Figure 5D, E: quantification of PLA and IF XAB2_R-loops and AQR_R-loops.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-data2-v3.xlsx
-
Figure 5—source data 3
Figures with the uncropped blots and relevant bands clearly labeled for Figure 5A: Western blot of IP XAB2.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-data3-v3.pdf
-
Figure 5—source data 4
The original files of the full raw unedited gels for Figure 5A: Western blot of IP XAB2.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-data4-v3.zip
-
Figure 5—source data 5
Figures with the uncropped blots and relevant bands clearly labeled for Figure 5B: Western blot of IP R-loops.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-data5-v3.pdf
-
Figure 5—source data 6
The original files of the full raw unedited gels for Figure 5B: Western blot of IP R-loops.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-data6-v3.zip
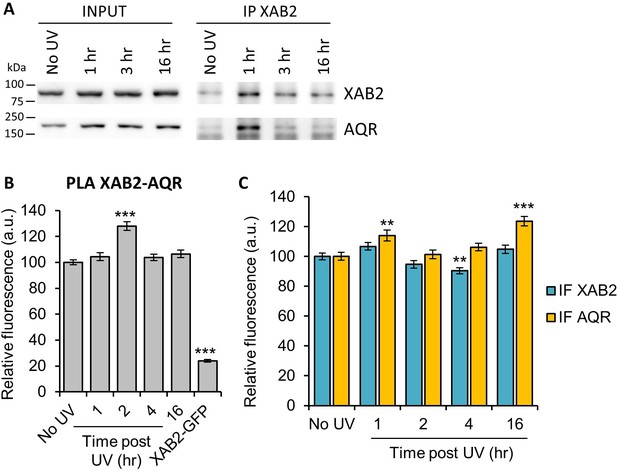
Interaction of XAB2 with the splicing complex AQR after UV damage.
(A) Immunoprecipitation of XAB2 in MRC5 nuclear extract treated with 10 J/m2 of UV-C at different times. Bound proteins were revealed by Western blotting with antibodies against XAB2 and AQR. INPUT, 10% of the lysate used for IP reaction. (B-C) Quantification of fluorescent signal in the nucleus against the couple XAB2_AQR from PLA experiment (B) and from the IF (C) done in parallel to PLA assay with the same antibodies dilution. Error bars represent the standard error of the mean (SEM) obtained from at least 100 cells. P-value of Student’s test compared to No UV condition: **<0.01; ***<0.001.
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1B, C: quantification of PLA and IF XAB2_AQR.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-figsupp1-data1-v3.xlsx
-
Figure 5—figure supplement 1—source data 2
Figures with the uncropped blots and relevant bands clearly labeled for Figure 5—figure supplement 1A: Western blot of IP XAB2 in MRC5.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-figsupp1-data2-v3.pdf
-
Figure 5—figure supplement 1—source data 3
The original files of the full raw unedited gels for Figure 5—figure supplement 1A: Western blot of IP XAB2 in MRC5.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-figsupp1-data3-v3.zip
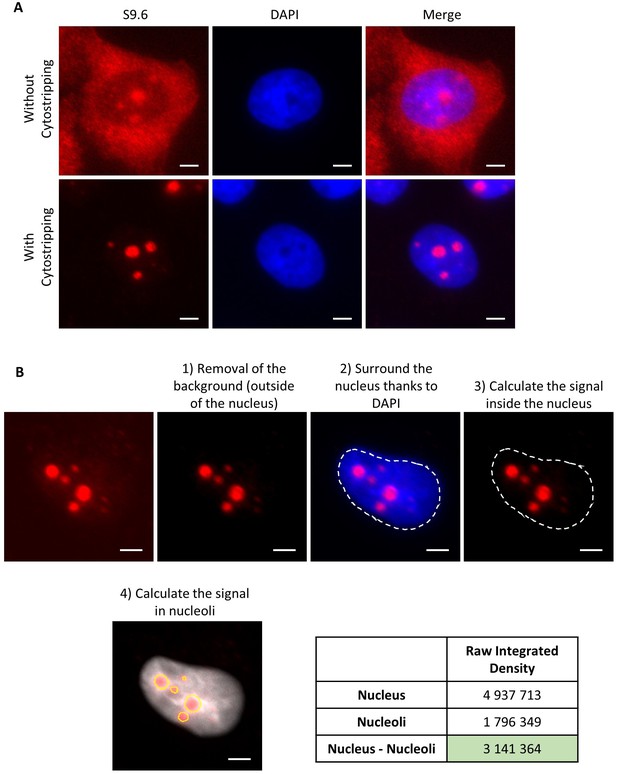
IF R-loops and quantification method.
(A) Immunofluorescence using S9.6 antibody with or without cytostripping before fixation step. (B) Quantification method of DNA:RNA hybrid signal inside the nucleus without the nucleolar signal. First, the background outside the nucleus is removed. Then, the area of the nucleus is determined thanks to the DAPI (4',6-diamidino-2-phenylindole) and the signal inside the entire nucleus is calculated. Next, the signal in nucleoli was also determined. Finally, we subtract the signal obtained in nucleoli from the signal obtained in the entire nucleus. Scale bar: 3 µm.
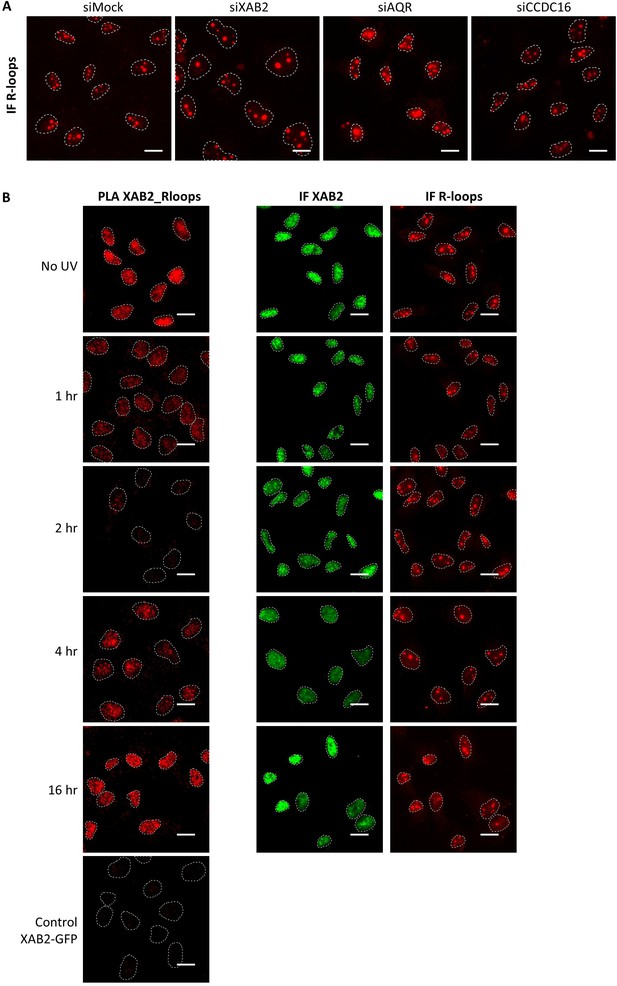
Representatives images of Figure 5C,D,E.
(A) Representative images of Figure 5C,IF against R-loops in WT cells treated with siRNAs against indicated factors. Nuclei are delimited by dashed lines. Scale bar: 15 µm. (B) Representative images of Figure 5D, E showing the PLA (red dots) between R-loops (S9.6 rabbit antibodies) and XAB2 (mice antibodies) in MRC5 cells after UV-C treatment (10 J/m2) and the IF done in parallel with the same antibodies dilutions. Nuclei are delimited by dashed lines. Scale bar: 15 µm.

Specificity of R-loops antibody.
(A) Quantification of PLA and IF signal against R-loops and RNA. After fixation, permeabilization and then blocking, cells were treated or not with RNAseH for 2 hr. RNA was detected by incorporating an uridine analog followed by a click-it reaction to add a Biotin tag which can then be recognized by a specific antibody. (B) Representative images of (A). Nuclei are delimited by dashed lines. Scale bar: 15 µm. (C, E) Quantification of PLA and IF signal against R-loops and AQR (C) or R-loops and XAB2 (E). Samples were treated or not with RNAseH. (D, F) Representative images of (C) and (E). Nuclei are delimited by dashed lines. Scale bar: 15 µm. P-value of Student’s test compared to without RNAseH condition: ***<0.001.
-
Figure 5—figure supplement 4—source data 1
Source data for Figure 5—figure supplement 4A: quantification of PLA and IF R-loops_RNA.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-figsupp4-data1-v3.xlsx
-
Figure 5—figure supplement 4—source data 2
Source data for Figure 5—figure supplement 4C, E: quantification of PLA and IF XAB2_R-loops and AQR_R-loops after treatment with RNAseH.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-figsupp4-data2-v3.xlsx
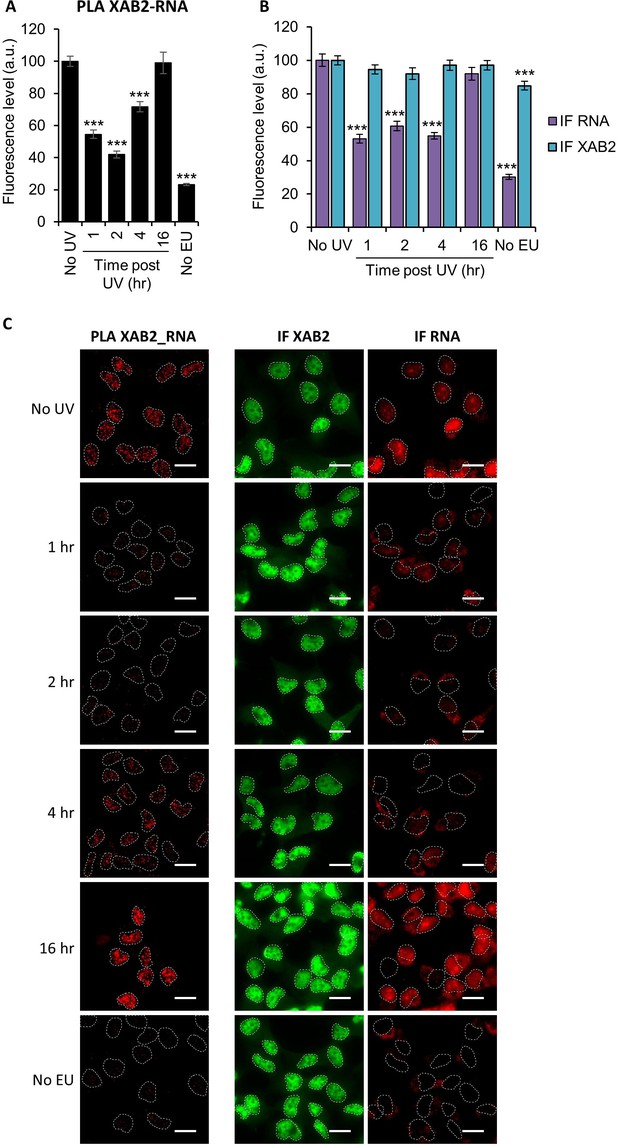
Interaction of XAB2 with RNA.
(A, B) Quantification of fluorescent signal in the nucleus against the couple XAB2-RNA from PLA experiment (A) or from the IF done in parallel to PLA assay. (B) RNA was detected by incorporating an uridine analog followed by a click-it reaction to add a Biotin tag which can then be recognized by a specific antibody. P-value of Student’s test compared to No UV condition: ***<0.001. (C) Representative images of (A) and (B). Nuclei are delimited by dashed lines. Scale bar: 15 µm.
-
Figure 5—figure supplement 5—source data 1
Source data for Figure 5—figure supplement 5A,B: quantification of PLA and IF XAB2_RNA.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig5-figsupp5-data1-v3.xlsx
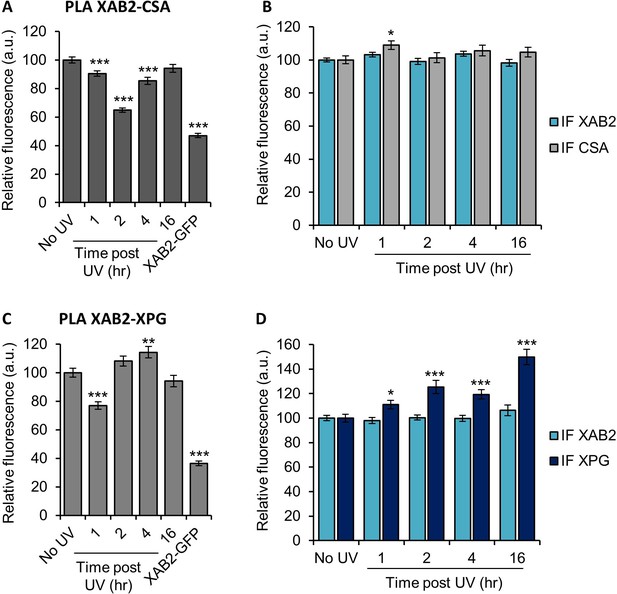
XAB2 is released from CSA and XPG during DNA repair.
Quantification of fluorescent signal in the nucleus against the couple XAB2-CSA (A, B) and XAB2-XPG (C, D) from PLA experiment (A, C) or from the IF done in parallel to PLA assay (B, D). Error bars represent the standard error of the mean (SEM) obtained from at least 80 cells. P-value of Student’s test compared to No UV condition: *<0.05; **<0.01; ***<0.001.
-
Figure 6—source data 1
Source data for Figure 6A, B: quantification of PLA and IF XAB2-CSA.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig6-data1-v3.xlsx
-
Figure 6—source data 2
Source data for Figure 6C, D: quantification of PLA and IF XAB2-XPG.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig6-data2-v3.xlsx

Interaction of XAB2 with CSB or XPB, repair factors of NER.
Quantification of fluorescent signal in the nucleus against the couple XAB2-CSB (A, B) and XAB2-XPB (C, D) from PLA experiment (A, C) or from the IF done in parallel to PLA assay (B, D). Error bars represent the standard error of the mean (SEM) obtained from at least 80 cells. P-value of Student’s test compared to No UV condition: **<0.01; ***<0.001.
-
Figure 6—figure supplement 1—source data 1
Source data for Figure 6—figure supplement 1A, B: quantification of PLA and IF XAB2-CSB.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig6-figsupp1-data1-v3.xlsx
-
Figure 6—figure supplement 1—source data 2
Source data for Figure 6—figure supplement 1C, D: quantification of PLA and IF XAB2-XPB.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig6-figsupp1-data2-v3.xlsx
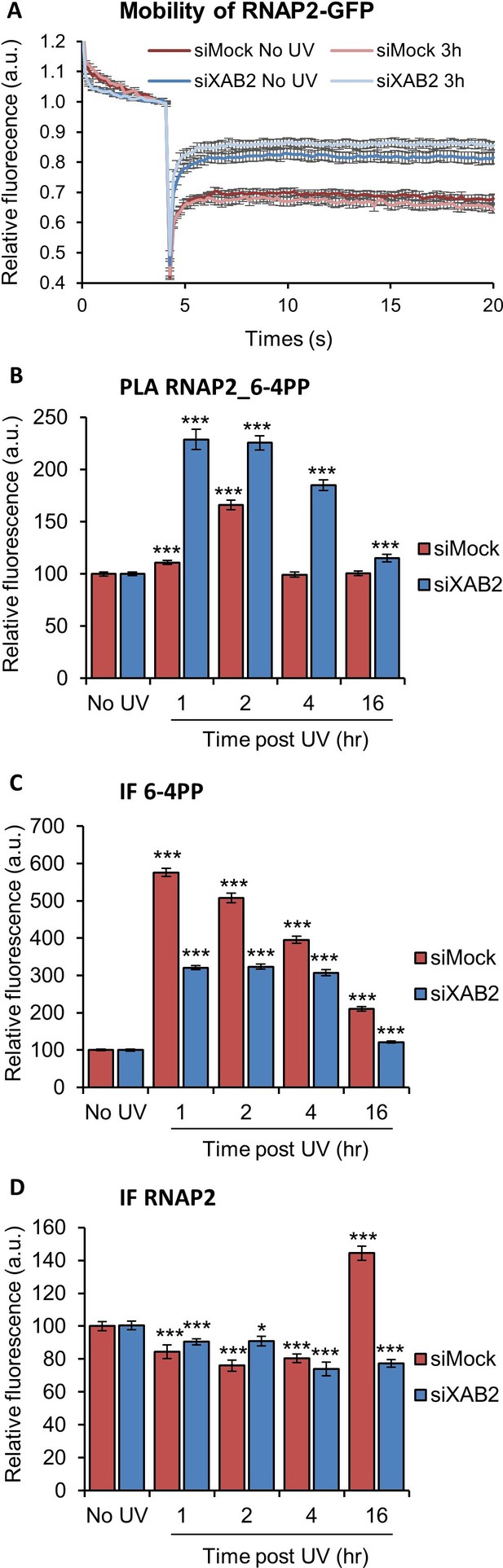
RNAP2 behavior is modified without XAB2.
(A) FRAP analysis of RNAP2-GFP expressing WT cells treated or not with UV-C (10 J/m2) after siRNA-mediated knockdown of the indicated factors. Error bars represent the SEM obtained from at least 10 cells. (B, C, D) Quantification of fluorescent signal in the nucleus against the couple RNAP2_6-4PP from PLA experiment (B) or from the IFndone in parallel to PLA assay (C, D). Error bars represent the standard error of the mean (SEM) obtained from at least 80 cells. P-value of Student’s test compared to No UV condition: *<0.05; ***<0.001.
-
Figure 7—source data 1
Source data for Figure 7A: FRAP RNAP2-GFP.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig7-data1-v3.xlsx
-
Figure 7—source data 2
Source data for Figure 7B–D: quantification of PLA and IF RNAP2_6-4PP.
- https://cdn.elifesciences.org/articles/77094/elife-77094-fig7-data2-v3.xlsx

Representatives images of Figure 7B–D.
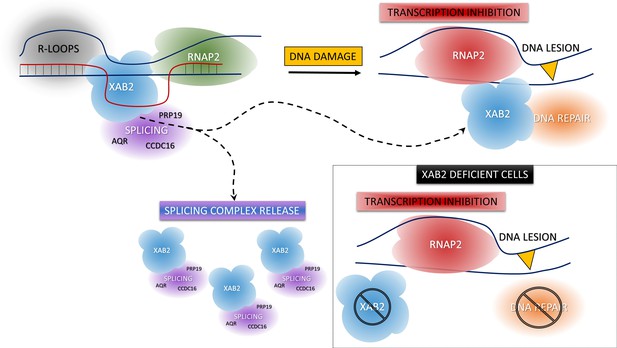
Model of XAB2 dynamics during DNA damage-dependent transcription inhibition.
Considering our results, a hypothetical model of XAB2 roles and dynamics can be sketched. XAB2 is involved in R-loops removal and pre-mRNA splicing, both processes linked to transcription. After DNA damage induction, transcription is blocked and XAB2 (together with some of the proteins involved in the splicing) is massively released from R-loops allowing a subset of XAB2 molecules to interact with UV-stalled RNA polymerase 2 (RNAP2) and participate in the TC-NER process. In the absence of XAB2, TC-NER is defective and as a consequence, RNAP2 remains longer in the proximity of DNA lesions.





