VE-cadherin enables trophoblast endovascular invasion and spiral artery remodeling during placental development
Figures
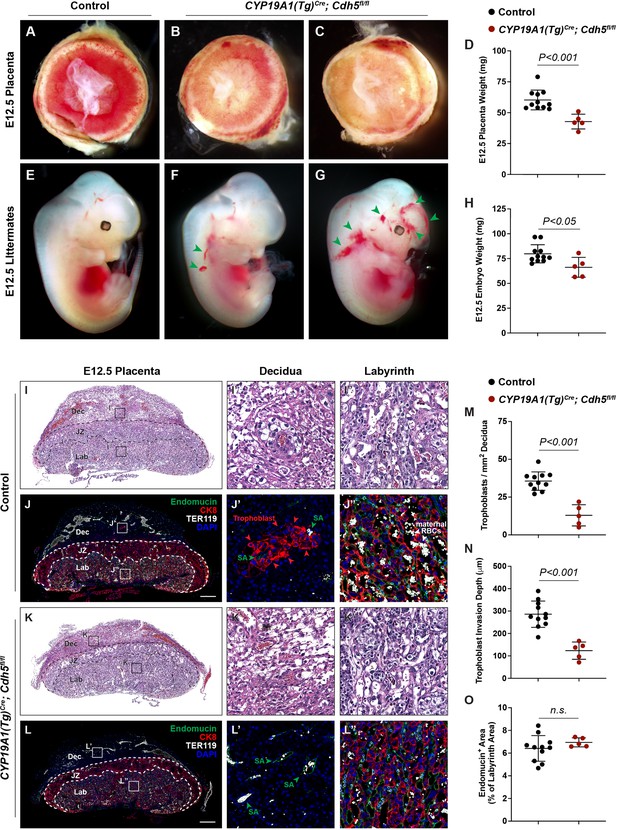
Deletion of VE-cadherin in trophoblasts disrupts cell migration resulting in placental and fetal growth restriction.
(A–D) Gross examination and quantification of E12.5 Control and CYP19A1(Tg)Cre; Cdh5fl/fl placentas and weights. Control n = 11, CYP19A1(Tg)Cre; Cdh5fl/fl n = 5. (E–H) Gross examination and quantification of E12.5 Control and CYP19A1(Tg)Cre; Cdh5fl/fl embryos and weights. Control n = 11, CYP19A1(Tg)Cre; Cdh5fl/fl n = 5. Green arrowheads point to areas of hemorrhage. (I–L) Hematoxylin and eosin (H&E) staining and immunofluorescence staining for Endomucin (green), CK8 (red), and TER119 (gray) of E12.5 Control (I, J) and CYP19A1(Tg)Cre; Cdh5fl/fl (K, L) serial placenta sections. Dotted lines demarcate the different placental regions. Red arrowheads indicate trophoblasts. Green arrowheads indicate spiral arteries (SA). White arrowheads indicate maternal red blood cells (RBCs). Note fewer non-nucleated, maternal TER119+ cells in the labyrinth region of CYP19A1(Tg)Cre; Cdh5fl/fl placentas. Boxes on the left correlate with magnified images on the right, and boxes in H&E and immunofluorescence images are of the same region. Scale bars = 500 μm. Dec (decidua), JZ (junctional zone), Lab (labyrinth). (M–O) Quantification of number of trophoblasts in the decidua (M), trophoblast invasion depth (N), and percent labyrinth Endomucin+ area (O). Control n = 11, CYP19A1(Tg)Cre; Cdh5fl/fl n = 5. Statistical analysis was performed using two-tailed, unpaired Welch’s t-test. Data are shown as means ± SD.
-
Figure 1—source data 1
Excel file containing quantification for embryo weights, placenta weights, trophoblast density, trophoblast migration distance, and fetal labyrinth vasculature in Figure 1.
- https://cdn.elifesciences.org/articles/77241/elife-77241-fig1-data1-v2.xlsx

Deletion of VE-cadherin in CYP19A1(Tg)Cre; Cdh5fl/fl placentas.
(A, B) Immunofluorescence staining and quantification of E12.5 Control and CYP19A1(Tg)Cre; Cdh5fl/fl placentas for VE-cadherin (green) and CK8 (red). Yellow arrowheads indicate VE-cadherin+ trophoblasts, which predominantly line remodeled spiral arteries. White arrowheads indicate VE-cadherin- trophoblasts. Red arrowheads indicate spiral artery endothelial cells (ECs), which are VE-cadherin+ CK8-. VE-cadherin and CK8 colocalized area was divided by total CK8-positive area to determine the extent of VE-cadherin deletion. Note that there are a few VE-cadherin+ trophoblasts in CYP19A1(Tg)Cre; Cdh5fl/fl placentas, however VE-cadherin is deleted from the majority of trophoblasts. The layer of VE-cadherin+ CK8- cells (red arrowheads) in the CYP19A1(Tg)Cre; Cdh5fl/fl placentas are spiral artery endothelial cells that have not been displaced by trophoblasts. Dotted white lines demarcate the decidua and junctional zone (JZ). Positive signal in small, rounded cells in the lumen is the result of erythrocyte autofluorescence. Control n = 4, CYP19A1(Tg)Cre; Cdh5fl/fl n = 3. Scale bars = 100 μm. (C, D) E12.5 placental and embryo weights do not differ between Cre-negative (Cdh5fl/+ or Cdh5fl/fl) and Cre-positive heterozygous (CYP19A1(Tg)Cre; Cdh5fl/fl) controls. Statistical analysis was performed using two-tailed, unpaired Welch’s t-test. Data are shown as means ± SD.
-
Figure 1—figure supplement 1—source data 1
Excel file containing quantification for VE-cadherin expression in trophoblasts, embryo weights, and placenta weights in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/77241/elife-77241-fig1-figsupp1-data1-v2.xlsx
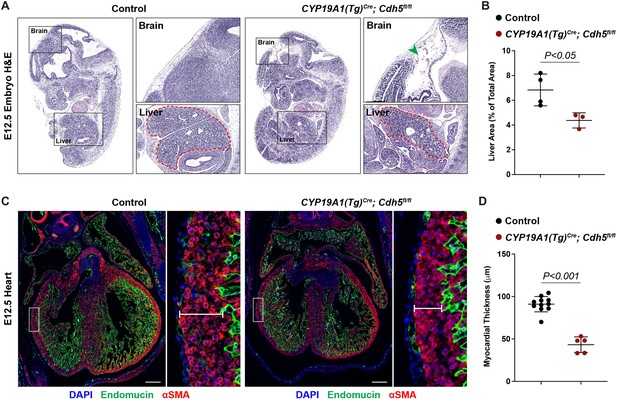
Loss of trophoblast VE-cadherin causes defects in brain, liver, and heart development.
(A) Sagittal hematoxylin and eosin (H&E) sections from E12.5 Control and CYP19A1(Tg)Cre; Cdh5fl/fl embryos. Boxes show magnified regions of the brain, heart, and liver. Note the thinness of the myocardium in CYP19A1(Tg)Cre; Cdh5fl/fl embryos. Dotted red line outlines the liver contour. Green arrowhead points to tissue degradation in the brains of CYP19A1(Tg)Cre; Cdh5fl/fl embryos. (B) Quantification of the liver area as a percent of total embryo area. Control n = 4, CYP19A1(Tg)Cre; Cdh5fl/fl n = 3. (C, D) Immunofluorescence staining of E12.5 Control and CYP19A1(Tg)Cre; Cdh5fl/fl embryonic hearts for Endomucin (green) and alpha-smooth muscle actin (αSMA) (red). Insets demonstrate myocardial thinning and quantified in (D). Control n = 11, CYP19A1(Tg)Cre; Cdh5fl/fl n = 5. Scale bars = 200 μm. Statistical analysis was performed using two-tailed, unpaired Welch’s t-test. Data are shown as means ± SD.
-
Figure 1—figure supplement 2—source data 1
Excel file containing quantification for liver area and myocardial thickness in Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/77241/elife-77241-fig1-figsupp2-data1-v2.xlsx
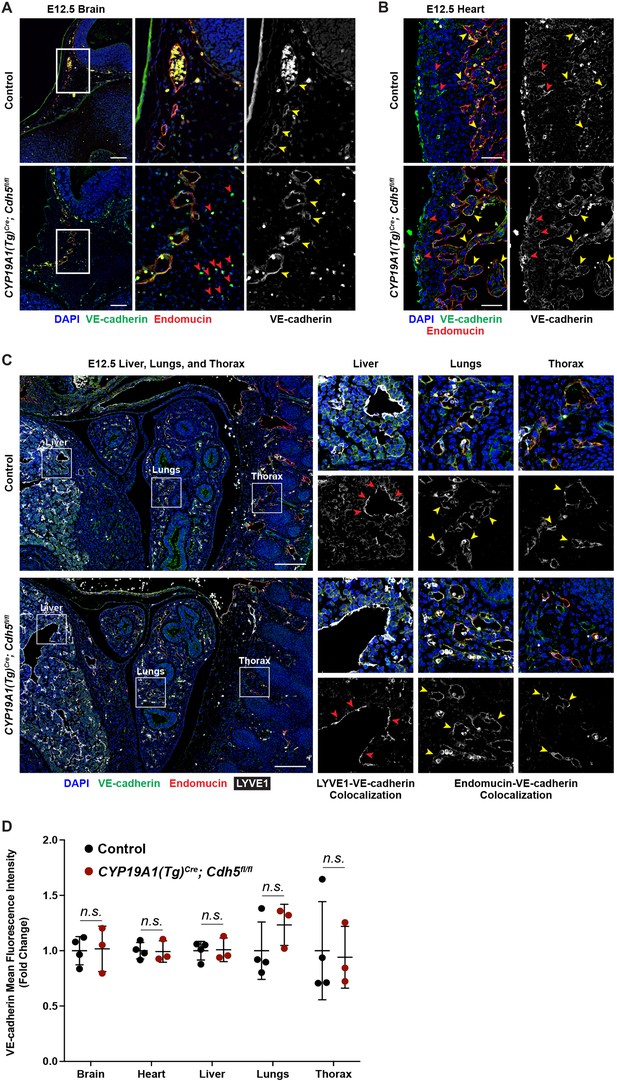
VE-cadherin expression is retained in the vasculature of affected organs in CYP19A1(Tg)Cre; Cdh5fl/fl embryos.
(A) Immunofluorescence staining of E12.5 Control and CYP19A1(Tg)Cre; Cdh5fl/fl brains for VE-cadherin (green) and Endomucin (red) at sites of hemorrhage. Images in gray scale are VE-cadherin alone. Red arrowheads point to extravascular autofluorescent erythrocytes. Yellow arrowheads point to VE-cadherin+ vessels. Scale bars = 50 μm. (B) Immunofluorescence staining of E12.5 Control and CYP19A1(Tg)Cre; Cdh5fl/fl hearts for VE-cadherin (green) and Endomucin (red). Images in gray scale are VE-cadherin alone. Red arrowheads point to VE-cadherin+ developing coronary vessels. Yellow arrowheads point to VE-cadherin+ endocardium. Scale bars = 50 μm. (C) Immunofluorescence staining of E12.5 Control and CYP19A1(Tg)Cre; Cdh5fl/fl hearts for VE-cadherin (green), Endomucin (red), and LYVE1 (gray). Images in gray scale represent VE-cadherin pixels colocalized with either LYVE1 (liver) or Endomucin (lungs and thorax). Red arrowheads point to LYVE1+VE-cadherin+ liver sinusoidal vessels. Yellow arrowheads point to Endomucin+VE-cadherin+ lung and thoracic blood vessels. Scale bars = 200 μm. (D) Quantification of fold change in VE-cadherin mean fluorescence intensity in the brain, heart, liver, lungs, and thorax.
-
Figure 1—figure supplement 3—source data 1
Excel file containing quantification for VE-cadherin expression in various organs in Figure 1—figure supplement 3.
- https://cdn.elifesciences.org/articles/77241/elife-77241-fig1-figsupp3-data1-v2.xlsx
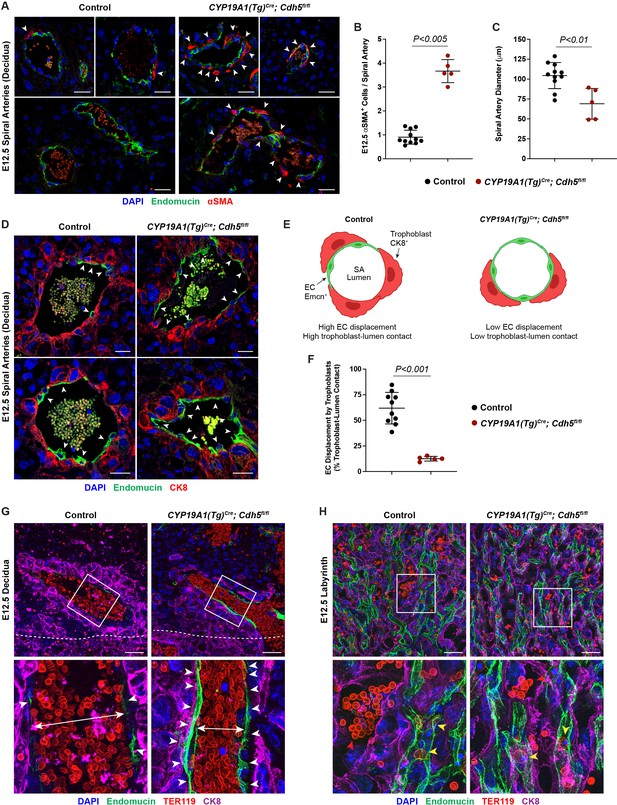
VE-cadherin is required in trophoblasts to remodel spiral arteries (SAs) and to displace SA endothelium.
(A) Immunofluorescence staining of E12.5 Control and CYP19A1(Tg)Cre; Cdh5fl/fl placentas for Endomucin (green) and alpha-smooth muscle actin (αSMA) (red). White arrowheads indicate αSMA+ cells. Scale bars = 25 μm. (B, C) Quantification of αSMA+ cells per SA and SA diameter. Control n = 11, CYP19A1(Tg)Cre; Cdh5fl/fl n = 5. (D) Immunofluorescence staining of E12.5 Control and CYP19A1(Tg)Cre; Cdh5fl/fl placentas for Endomucin (green) and CK8 (red). White arrowheads indicate Endomucin+ SA endothelial cells (ECs). Positive signal in small, rounded cells in the lumen is the result of erythrocyte autofluorescence. (E) Schematic demonstrating differences in trophoblast and SA EC contact with the vessel lumen. Scale bars = 25 μm. (F) Quantification of the percent trophoblast-lumen contact, which was calculated by measuring the circumference of the vessel lumen and then measuring the length of CK8+ trophoblasts in contact with the lumen. Each point represents the average of at least three SAs from an individual placenta. Control n = 10, CYP19A1(Tg)Cre; Cdh5fl/fl n = 5. (G, H) Maximum intensity projections of whole-mount immunofluorescence of the decidua (G) and labyrinth (H) from 200 μm thick placenta sections stained for Endomucin (green), TER119 (red), and CK8 (magenta). Double-headed arrows indicate differences in lumen size. White arrowheads indicate Endomucin+ SA ECs. Red arrowheads indicate maternal red blood cells within the trophoblast-lined vessels. Yellow arrowheads indicate fetal red blood cells within fetal capillaries. Dotted white line demarcates the decidua from the junctional zone. Scale bars = 50 μm. Statistical analysis was performed using two-tailed, unpaired Welch’s t-test. Data are shown as means ± SD.
-
Figure 2—source data 1
Excel file containing quantification for smooth muscle cells per spiral artery, spiral artery diameter, and trophoblast-endothelial cell displacement in Figure 2.
- https://cdn.elifesciences.org/articles/77241/elife-77241-fig2-data1-v2.xlsx
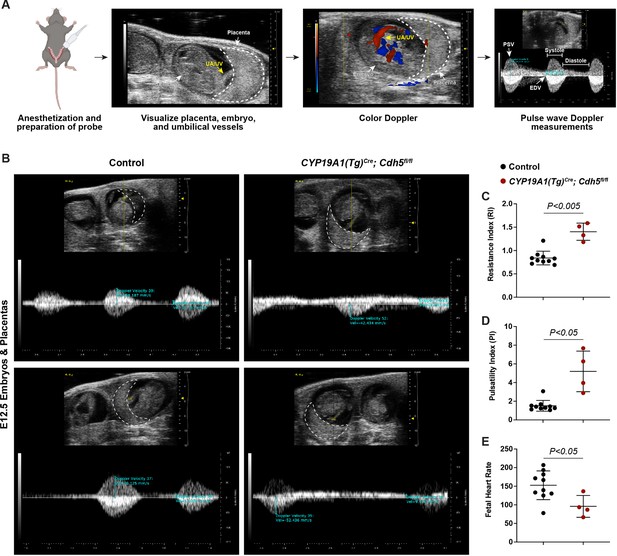
Spiral artery remodeling defects result in placental insufficiency and fetal distress.
(A) Schematic of workflow for Doppler ultrasound of pregnant dams. The embryo, UA/UV (yellow arrow), and placenta (dotted white outline) are labeled. (B) Representative umbilical artery Doppler waveforms from two Control and two CYP19A1(Tg)Cre; Cdh5fl/fl placentas. The placenta is outlined in a dotted white line. Reversal of end-diastolic flow is evident by the change of directional velocity at the end of diastole compared to peak systole (i.e., negative to positive velocity). (C–E) Quantification of resistance index (RI), pulsatility index (PI), and fetal heart rate. PSV (peak systolic velocity), EDV (end diastolic velocity), UA/UV (umbilical artery/umbilical vein). Note that red/blue colors in color Doppler images do not indicate UA/UV, which can only be differentiated based on the Doppler waveform. Control n = 10, CYP19A1(Tg)Cre; Cdh5fl/fl n = 4. Statistical analysis was performed using two-tailed, unpaired Welch’s t-test. Data are shown as means ± SD.
-
Figure 3—source data 1
Excel file containing quantification for ultrasound studies (resistance index, pulsatility index, fetal heart rate) in Figure 3.
- https://cdn.elifesciences.org/articles/77241/elife-77241-fig3-data1-v2.xlsx
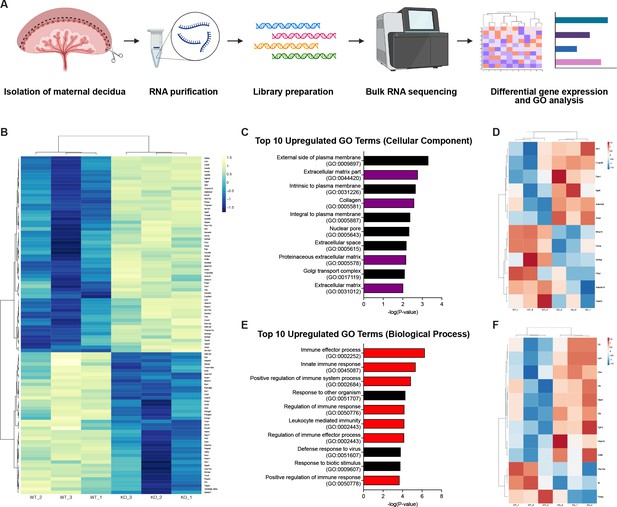
RNA-sequencing reveals defects in the decidual extracellular matrix and immune microenvironment.
(A) Schematic of bulk RNA-sequencing (RNA-seq) workflow on deciduas from Control (WT) and CYP19A1(Tg)Cre; Cdh5fl/fl (KO) E12.5 placentas. (B) Heatmap of the top 100 differentially regulated genes shown by z-score (n = 3 biological replicates). (C) Gene ontology (GO) term analysis of top 10 upregulated cellular components in CYP19A1(Tg)Cre; Cdh5fl/fl placentas. Purple bars indicate GO terms related to the extracellular matrix. (D) Heatmap of significantly differentially expressed genes from GO terms related to the extracellular matrix. (E) GO term analysis of top 10 upregulate biological processes in CYP19A1(Tg)Cre; Cdh5fl/fl placentas. Red bars indicate GO terms related to immune processes. (F) Heatmap of significantly differentially expressed genes from GO terms related to immune processes.
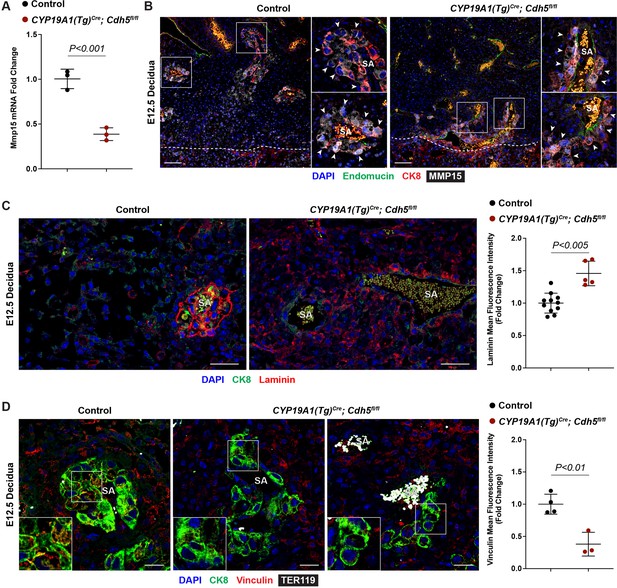
Disrupting trophoblast migration results in decidual extracellular matrix defects.
(A) Mmp15 mRNA gene expression in control and CYP19A1(Tg)Cre; Cdh5fl/fl placentas from RNA-sequencing (RNA-seq). (B) Immunofluorescence staining for Endomucin (green), CK8 (red), and MMP15 (gray) in E12.5 Control and CYP19A1(Tg)Cre; Cdh5fl/fl placentas. White arrowheads point to MMP15+CK8+ trophoblasts which are more abundant in control placentas relative compared to CYP19A1(Tg)Cre; Cdh5fl/fl placentas, particularly around spiral arteries (SA), but there is no difference in MMP15 intensity in trophoblasts. Dotted line demarcates the border of the junctional zone and decidua. Positive yellow signal in small, rounded cells in the SA lumen is the result of erythrocyte autofluorescence. Scale bars = 100 μm. (C) Immunofluorescence staining for laminin (red) and CK8 (green) and quantification of fold change in laminin mean fluorescence intensity in the stroma. In control placentas, laminin is expressed in trophoblasts surrounding SAs but absent in stromal cells compared to CYP19A1(Tg)Cre; Cdh5fl/fl placentas. Scale bars = 50 μm. (D) Immunofluorescence staining for CK8 (green), vinculin (red), and TER119 (gray) and quantification of fold change in vinculin mean fluorescence intensity in trophoblasts. Scale bars = 25 μm. Statistical analysis was performed using two-tailed, unpaired Welch’s t-test. Data are shown as means ± SD.
-
Figure 4—figure supplement 1—source data 1
Excel file containing quantification for Mmp15 gene expression, laminin expression in the decidua, and vinculin expression in trophoblasts in Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/77241/elife-77241-fig4-figsupp1-data1-v2.xlsx
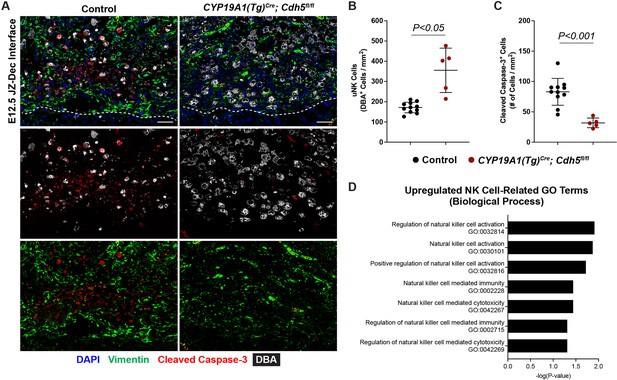
Persistent uterine natural killer (uNK) cells at the junctional zone-decidual interface of Cdh5 knockout placentas.
(A) Immunofluorescence staining for DBA (a lectin that specifically binds uterine natural killer (uNK) cells, gray), vimentin (green), and cleaved caspase-3 (red).
Dotted line demarcates the border of the junctional zone and decidua. Scale bars = 50 μm. (B, C) Quantification of DBA+ cell density and cleaved caspase-3+ cell density in the decidua. (D) Gene ontology (GO) analysis of upregulated biological process terms related to NK cells. Statistical analysis was performed using two-tailed, unpaired Welch’s t-test. Data are shown as means ± SD.
-
Figure 4—figure supplement 2—source data 1
Excel file containing quantification for uterine natural killer cell density and apoptotic cell density in Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/77241/elife-77241-fig4-figsupp2-data1-v2.xlsx
Tables
Decreased survival of CYP19A1(Tg)Cre; Cdh5fl/fl mutants in late gestation.
Cdh5fl/fl male mice were crossed with CYP19A1(Tg)Cre; Cdh5fl/+ females to generate litters with mixed genotypes. The expected percentage is listed under the genotype label. The observed number of each genotype is shown with the corresponding percentage given in parentheses. P-values were calculated using Fisher’s exact test at stages E10.5-12.5 (pooled), E14.5-16.5 (pooled), and P21. E designates embryonic day and P designates postnatal day.
| CYP19A1(Tg)Cre; Cdh5fl/fl (25%) | CYP19A1(Tg)Cre; Cdh5fl/+ (25%) | Cdh5fl/fl (25%) | Cdh5fl/+ (25%) | Total (100%) | Fisher Exact Test | |
|---|---|---|---|---|---|---|
| E10.5-E12.5 | 13* (26%) | 13 (26%) | 12 (24%) | 12 (24%) | 50 (100%) | P=1.0000 |
| E14.5-E16.5 | 1 (2.9%) | 13 (38.2%) | 12 (35.3%) | 8 (23.5%) | 34 (100%) | P=0.0033 |
| P21 | 1 (2.2%) | 11 (24.4%) | 15 (33.3%) | 18 (40.0%) | 45 (100%) | P=0.0333 |
-
*
One embryo at E12.5 was dead.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculcus) | CYP19A1(Tg)-Cre | Wenzel and Leone, 2007 | ||
| Genetic reagent (Mus musculcus) | Cdh5 flox | Yang et al., 2019 | ||
| Antibody | Anti-Endomucin (goat polyclonal) | R&D | AF4666 | IF(1:400) |
| Antibody | Anti-Endomucin (rat monoclonal) | Abcam | ab106100 | IF(1:300) |
| Antibody | Anti-TER119 (rat monoclonal) | Abcam | ab91113 | IF(1:300) |
| Antibody | Anti-VE-cadherin (goat polycloncal) | R&D | AF1002 | IF(1:200) |
| Antibody | Anti-CK8 (rabbit monoclonal) | Abcam | ab53280 | IF(1:300) |
| Antibody | Anti-CK8 (rat monoclonal) | DSHB | TROMA-1 | IF(1:400) |
| Antibody | Anti-αSMA-Cy3 (mouse monoclonal) | Sigma | C6198 | IF(1:300) |
| Antibody | Anti-Cleaved Caspase-3 (rabbit polyclonal) | Millipore Sigma | AB3623 | IF(1:100) |
| Antibody | Anti-MMP15 (rabbit polyclonal) | Thermo Fisher Scientific | PA5-13184 | IF(1:200) |
| Antibody | Anti-Laminin (rabbit polyclonal) | Sigma | L9393 | IF(1:200) |
| Antibody | Anti-Vimentin (goat polyclonal) | R&D | AF2105 | IF(1:300) |
| Antibody | Anti-Vinculin (mouse monoclonal) | Sigma | V9131 | IF(1:200) |
| Commercial assay or kit | Direct-zol RNA Miniprep Kits | Zymo Research | R2053 | |
| Software, algorithm | ImageJ | NIH, Bethesda, MD, USA | RRID:SCR_003070 | |
| Software, algorithm | GraphPad Prism | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | Picard v2.17.11 | Picard | RRID:SCR_006525 | |
| Other | DBA-Biotin | Vector Labs | B-1035 | (1:500) |





