The transcription factor Bach2 negatively regulates murine natural killer cell maturation and function
Figures
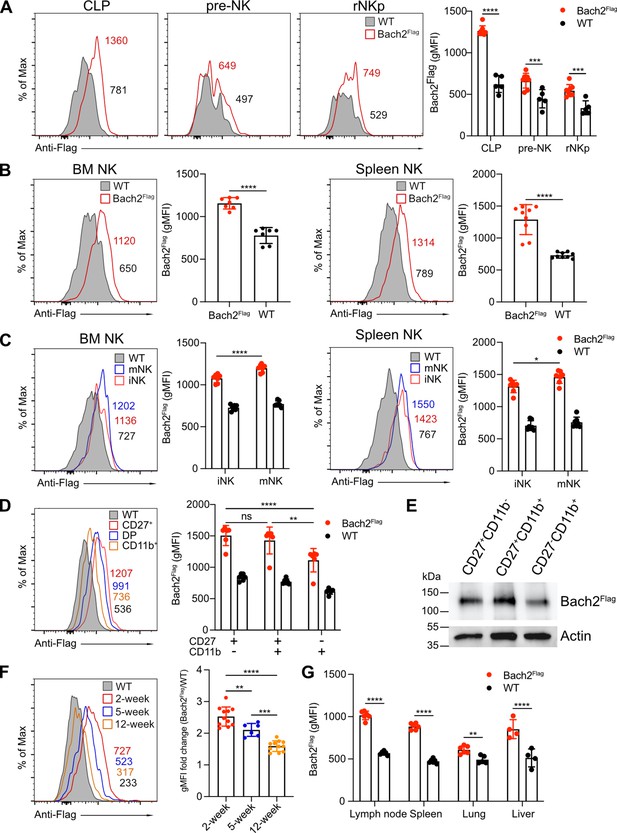
BTB domain And CNC Homolog 2 (Bach2) expression at different natural killer (NK) cell developmental stages by analysis of Bach2Flag knock-in mouse.
(A) Histogram plots of Bach2Flag expression (red) in common lymphoid progenitor (CLP), pre-NK progenitor (pre-NK), and refined NK progenitor (rNKp) as compared to cells from wild-type (WT) mice (gray fill). The geometric MFI (gMFI) number was indicated in the plot. Summary of gMFI was shown (n=5 for each group in three independent experiments). (B) Histogram plots of Bach2Flag expression (red) in NK cell (CD3-CD19-NK1.1+) from bone marrow (BM) and spleen as compared to cells from WT mice (gray fill). Numbers indicate gMFI and summary of gMFI is shown (n=7 or 9 for each group in three to four independent experiments). (C) CD3-CD122+ NK cells from BM and spleen were subdivided into iNK (DX5-NK1.1+) and mNK (DX5+NK1.1+) and analyzed for Bach2Flag expression (red for iNK, blue for mNK) as compared to cells from WT mice (gray fill). Numbers indicate gMFI and summary of gMFI is shown (n=7 for each group in three to four independent experiments). (D) Histogram plot of Bach2Flag expression in CD27+CD11b- (red), CD27+CD11b+ (blue), and CD27+CD11b+ (yellow) NK subsets as compared to cells from WT mice (gray fill). Note that WT plot represents expression of Bach2Flag from CD27-CD11b+ subsets. Numbers indicate gMFI and summary of gMFI is shown (n=6 for each group in three independent experiments). (E) Splenic NK cells were sorted into CD27+CD11b-, CD27+CD11b+ and CD27-CD11b+ subsets. Bach2Flag expression in the subsets was detected using Anti-FLAG M2-Peroxidase (HRP) antibody by western blot. Expression of Actin was used as an internal control. Two individual experiments have been done with one mouse each time. (F) Histogram plot of Bach2Flag expression in splenic NK cell (CD3-CD19-NK1.1+) from mice at 2 weeks’ (red), 5 weeks’ (blue), and 12 weeks’ (orange) age as compared to cells from WT mice (gray fill). Note that WT plot represents expression of Bach2Flag from 12-week age mice. Numbers indicate gMFI and summary of gMFI fold change is shown (n=6 or 11 for each group in three to four independent experiments). (G) Summary of gMFI data for Bach2Flag expression in NK cells from lymph node, spleen, lung, and liver (n=4 or 6 for each group in three independent experiments). NK cells from lymph node, spleen, and lung were gated as CD3-CD19-NK1.1+. NK cells from liver were gated as CD3-CD19-NK1.1+DX5+. Statistical significance was determined by two-way ANOVA (A, C, D, and G), one-way ANOVA, (F) or by Student’s t test (B). Error bars indicate SD. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ns, not significant. See Figure 1—figure supplement 1 for gating strategies.
-
Figure 1—source data 1
Bach2 expression in different subsets by western blot.
Splenic NK cells were enriched from splenocytes of Bach2Flag mice. Enriched NK cells (CD3-NK1.1+) from Bach2Flag mice were further sorted into CD27+CD11b-, CD27+CD11b+, and CD27-CD11b+ subsets. Bach2 expression in the subsets was detected using Anti-FLAG M2-Peroxidase (HRP) antibody by western blot. Expression of Actin was used as an internal control. Splenocytes from wild-type (WT) mice were used as negative control. Splenocytes from Bach2Flag mice were used as positive control. Two individual experiments have been done with one mouse each time.
- https://cdn.elifesciences.org/articles/77294/elife-77294-fig1-data1-v2.zip
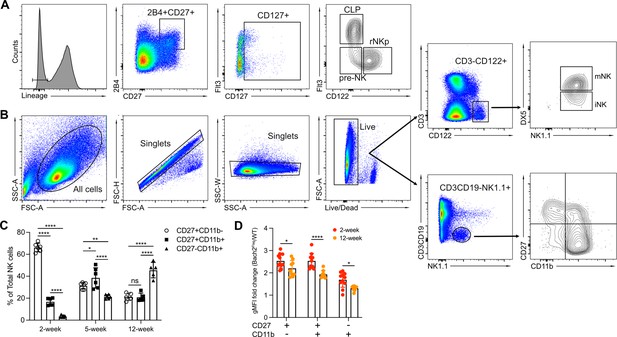
Gating strategy for flow cytometry analysis.
(A) Representative flow cytometry plots show bone marrow (BM) Lin-2B4+CD27+CD127+ cells were subdivided into common lymphoid progenitor (CLP) (Flt3+CD122-), pre-natural killer (NK) progenitor (pre-NK) (Flt3-CD122-), and refined NK progenitor (rNKp) (Flt3-CD122+) subsets. (B) Representative flow cytometry plots show CD3-CD122+ NK cells were subdivided into immature NK cells (DX5-NK1.1+) and mature NK cells (DX5+NK1.1+). Alternatively, CD3-CD19-NK1.1+ NK cells were further subdivided by CD27 and CD11b into maturation subsets. (C) Summary of the percentages of CD27+CD11b-, CD27+CD11b+, CD27-CD11b+ NK subsets among total NK cells in 2 weeks’, 5 weeks’, and 12 weeks’ mice (n=5 or 6 for each group in three independent experiments). (D) Summary of Bach2Flag geometric MFI (gMFI) fold change in NK cell subsets defined by CD27 and CD11b expression from 2- and 12-week mice (n=9 or 11 for each group in four independent experiments). Statistical significance was determined by two-way ANOVA. Error bars indicate SD. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ns, not significant.
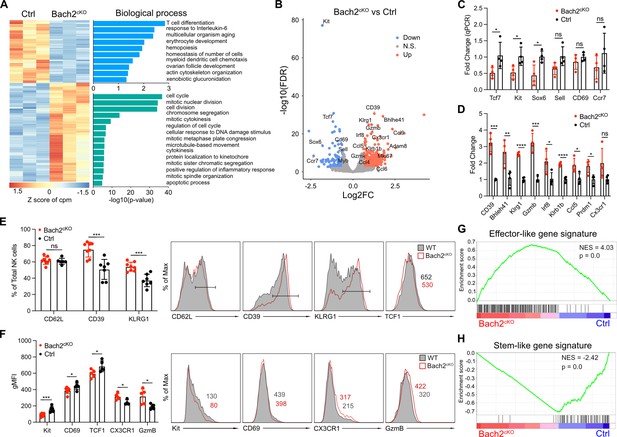
RNA-seq analysis reveals BTB domain And CNC Homolog 2 (Bach2) deficiency in natural killer (NK) cells promotes the terminal maturation of NK cells with elevated effector function.
(A) Heatmap of differentially expressed genes in NK cells compared between control and Bach2cKO mice in RNA-seq analysis (FDR < 0.01 and log2 fold change >1.5). Each column represents total splenic CD3-CD19-NK1.1+NKp46+ cells from an individual mouse. Three biological replicates per group from two individual sorting experiments are shown. The data were analyzed with the Database for Annotation, Visualization and Integrated Discovery (DAVID) Gene Ontology (GO) analysis for the biological process using the genes differentially expressed from NK cells between control and Bach2cKO mice. (B) Volcano plot shows differential gene expression between control and Bach2cKO splenic NK cells. Highlighted are genes discussed in the text. (C and D) RNA expression of indicated genes in Bach2cKO mice or control mice determined by quantitative PCR (qPCR). Data are shown with four mice per group from two independent experiments. (E) Summary of the percentage of NK cells expressing the indicated genes in Bach2cKO mice or control mice (n=7 or 8 for each group in three independent experiments). Representative histogram plots show the expression of indicated protein in NK cells from Bach2cKO mice (red) as compared to NK cells from control mice (gray fill). (F) Summary of the geometric MFI (gMFI) of indicated gene in total NK cells in Bach2cKO mice or control mice (n=5–8 for each group in three independent experiments). Representative histogram plots show the expression of indicated proteins in NK cells in Bach2cKO mice (red) as compared to NK cells from control mice (gray fill). (G and H) Gene set enrichment analysis (GSEA) illustrating enrichment of effector-like (G) and stem-like (H) gene signatures in Bach2cKO and control splenic NK cells. Statistical significance was determined by Student’s t test. Error bars indicate SD. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ns, not significant.
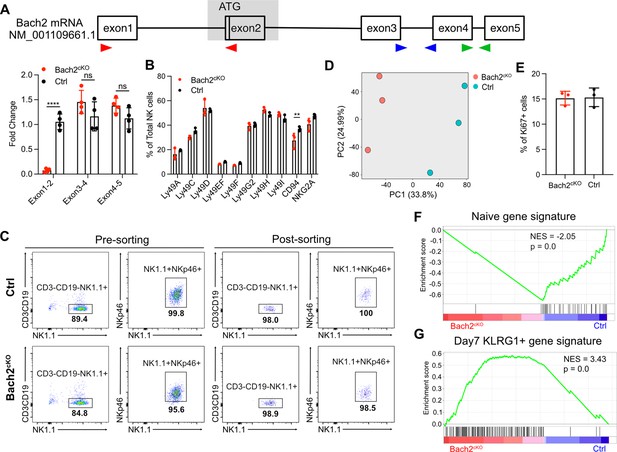
BTB domain And CNC Homolog 2 (Bach2) deficiency in natural killer (NK) cells resemble activated effector CD8+ T cells.
(A) Schematic plot shows quantitative PCR (qPCR) primers targeting regions on Bach2 mRNA. Red arrows represent primers targeting exon 1 to exon 2. Blue arrows represent primers targeting exon 3 to exon 4. Green arrows represent primers targeting exon 4 to exon 5. The gray box denotes the region that has been deleted. RNA expression of Bach2 was determined by qPCR with the indicated exon primer sets. Data are shown with four mice per group from two independent experiments. (B) Summary of the expression profile of various NK receptors (% of total NK cells) in control and Bach2cKO mice (n=3 for each group in three independent experiments). (C) Flow cytometry sorting of splenic NK cells (CD3-CD19-NK1.1+NKp46+) from Bach2cKO or control mice for RNA-seq analysis. Post-sorting flow cytometry shows the purity of the NK cells. (D) Principal component analysis (PCA) of differentially expressed genes (log2 fold change > 1.5 and FDR < 0.01 or log2 fold change < −1.5 and FDR < 0.01) for splenic NK cells isolated from control and Bach2cKO mice. (E) Summary of Ki67-positive cells in Bach2cKO mice or control mice (n=4 for each group in three independent experiments). (F and G) Gene set enrichment analysis (GSEA) plots illustrate the enrichment of naïve (F) and day 7 after infection with rVV-OVA KLRG1-positive (G) CD8+ T cells gene signatures (Roychoudhuri et al., 2016a) in Bach2cKO and control splenic NK cells. Statistical significance was determined by two-way ANOVA. Error bars indicate SD. **p<0.01; ****p<0.0001. ns, not significant.
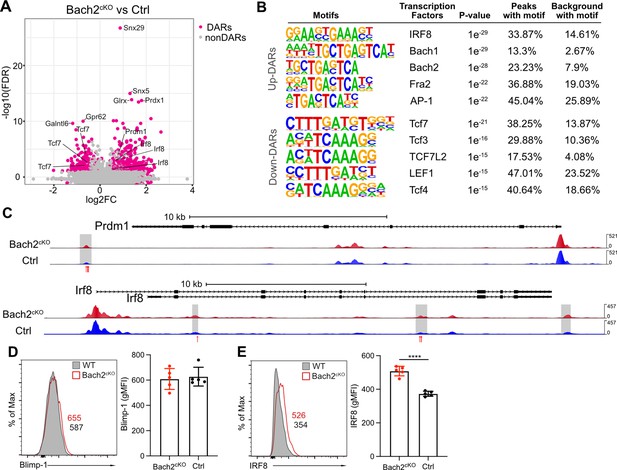
Bach2 deficiency results in increased accessible regions in the genome of natural killer (NK) cells.
(A) Volcano plot shows the differentially accessible regions (DARs) and non-DARs between control and Bach2cKO splenic NK cells. (B) De novo motif enrichment at regions of open chromatin as defined by assay for transposase-accessible chromatin coupled with high throughput sequencing (ATAC-seq) in splenic NK cells from Bach2cKO mice compared to cells from control mice. Motifs of interest are listed. (C) Genome browser visualization of ATAC-seq peak near Prdm1 and Irf8 gene loci in NK cells from Bach2cKO mice (red) and control mice (blue). Gray boxes denote differential accessible regions. Red arrows show the location of Bach2-binding motif. (D and E) Histogram plot shows the expression of Blimp-1 (D) and IRF8 (E) in NK cells from Bach2cKO mice (red) as compared to control mice (gray fill). Numbers indicate geometric MFI (gMFI). Summary of gMFI is shown (n=5 for each group in three independent experiments). Statistical significance was determined by Student’s t test. Error bars indicate SD. *p<0.05; ****p<0.0001.
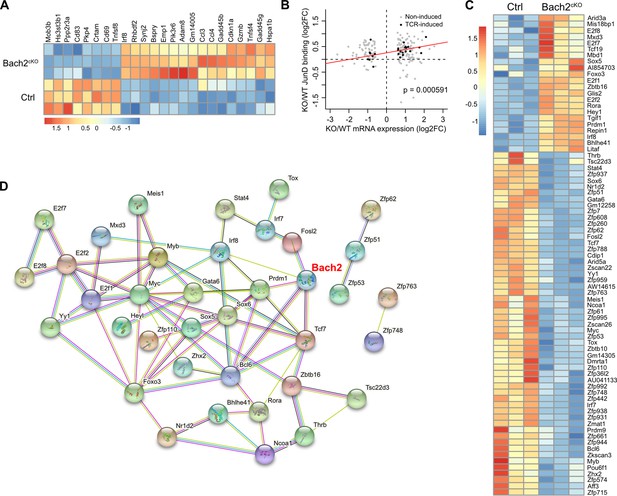
Potential protein interactions between BTB domain And CNC Homolog 2 (Bach2) and differentially regulated transcription factors in natural killer (NK) cells.
(A and B) Differentially expressed genes in NK cells between Bach2cKO mice and control mice were compared for genes that were recognized as Bach2 target genes in CD8+ T cells (Roychoudhuri et al., 2016a). The TCR-induced gene list was used for comparison. Heatmap of the overlapped genes is shown (A). Differences between average JunD binding at Bach2-binding sites (data from published CD8+ T cells) with differences in mRNA expression of associated genes in NK cells (B), using the ratios of Bach2cKO/WT JunD-binding versus expressed genes. All Bach2 target genes in CD8+ T cells including TCR-induced genes and non-TCR-induced genes were used to compare by exact two-sample Kolmogorov-Smirnov test. Im method was used to add the red line. (C) Heatmap of differentially expressed transcription factors in NK cells comparing control and Bach2cKO mice in RNA-seq analysis (FDR < 0.05 and log2 fold change > 0.2). (D) Protein interaction between the differentially expressed transcription factors analyzed at the website STRING (https://string-db.org). Disconnected nodes in the network were hidden.
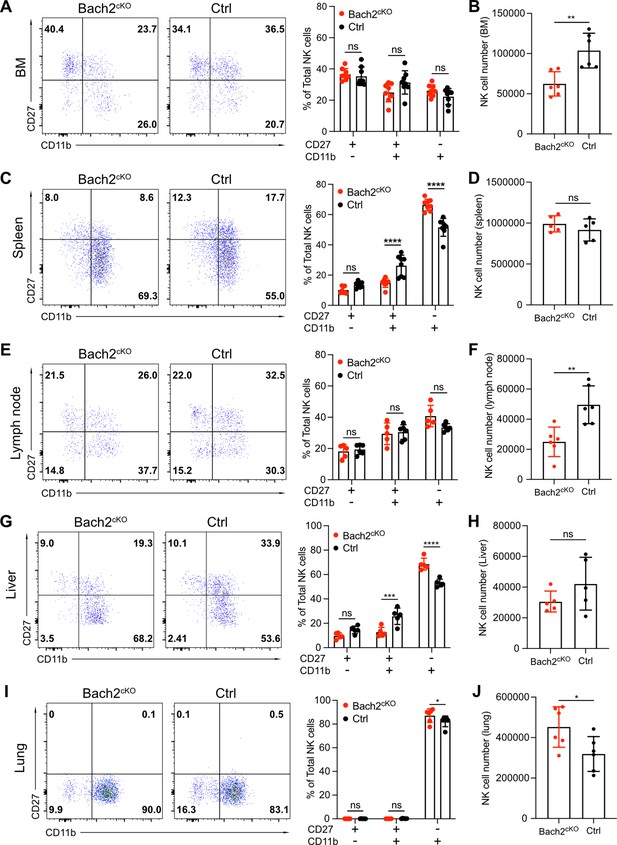
Bach2 deficiency increases natural killer (NK) cells with terminally differentiated phenotype.
Representative flow cytometry plots show NK cells (CD3-CD19-NK1.1+NKp46+) separated into maturation stages by CD27 and CD11b expression. The percentage of CD27+CD11b-, CD27+CD11b+, CD27-CD11b+ NK cells among all NK cells (left, representative flow plots; right, bar graph summary) from bone marrow (BM) (A), spleen (C), lymph node (E), liver, (G) and lung (I) in Bach2cKO mice and control mice were plotted (n=5–8 for each group from three independent experiments). Note that NK cells from liver were gated on CD3-CD19-DX5+NK1.1+NKp46+. Summary of the total number of NK cells from BM (B), spleen (D), lymph node (F), liver (H), and lung (J) in Bach2cKO mice and control mice were plotted (n=5–8 for each group from three independent experiments). Statistical significance was determined by two-way ANOVA (A, C, E, G, and I) or by Student’s t test (B, D, F, H, and J). Error bars indicate SD. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ns, not significant.
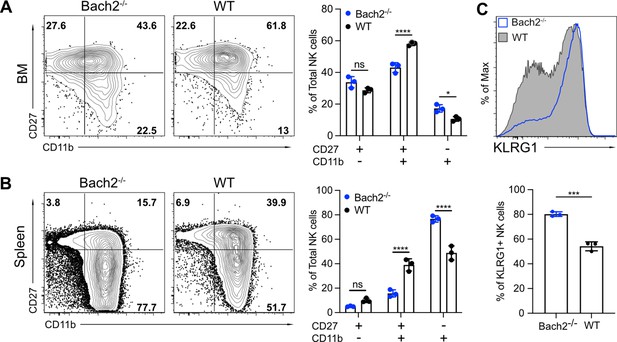
BTB domain And CNC Homolog 2 (Bach2) deficiency results in a more mature phenotype.
(A and B) Representative flow cytometry plots and summary of total natural killer (NK) cells (CD3-CD19-NK1.1+NKp46+) maturation stages determined by CD27 and CD11b expression from bone marrow (BM) (A) or spleen (B) in Rag1-/-Bach2-/- (Bach2-/-) and Rag1-/- (wild-type [WT]) mice. The percentage of different subsets among total NK cells was plotted (n=3 for each group in one experiment). (C) Representative histogram of KLRG1 expression on splenic NK cells (CD3-CD19-NK1.1+NKp46+) from Rag1-/-Bach2-/- (Bach2-/-) and Rag1-/- (WT) mice. The percentages of NK cells that express KLRG1 are shown in the bar graph (n=3 for each group in one experiment). Statistical significance was determined by two-way ANOVA with Bonferroni correction (A and B) or by Student’s t test (C). Error bars indicate SD. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; ns, not significant.
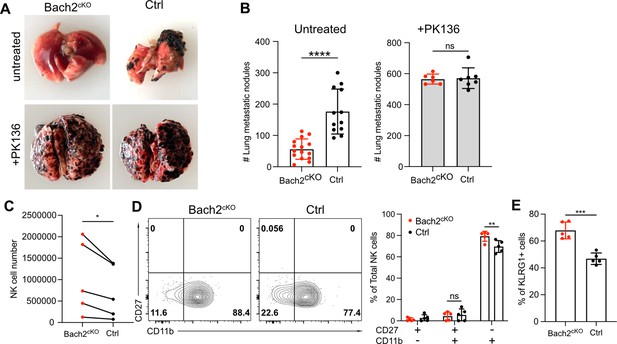
Lack of BTB domain And CNC Homolog 2 (Bach2) expression in natural killer (NK) cells suppresses B16F10 tumor metastasis.
(A) Representative picture of lung metastatic nodules in Bach2cKO mice and control mice under steady-state or anti-NK1.1 (PK136) depletion. (B) Summary of the number of B16F10 metastatic nodules in lung from Bach2cKO and control mice with or without anti-NK1.1 (PK136) depletion. Data were pooled from three to four independent experiments with a total of six to fifteen mice per group. (C–E) 2×105 B16F10 cells were intravenously injected to Bach2cKO mice or control mice. After 24 hr, NK cells from lung were harvested and analyzed for total number (C), maturation by CD27 and CD11b (D), and the percentage of KLRG1+ NK cells (E). Average NK cell numbers were calculated for each experiment with one to two mice per group, and data were pooled together from five independent experiments (C). Data were pooled from three independent experiments with five mice per group (D and E). Statistical significance was determined by Student’s t test (B and E), Student’s t test (paired t test) (C), or by two-way ANOVA (D). Error bars indicate SD. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ns, not significant.
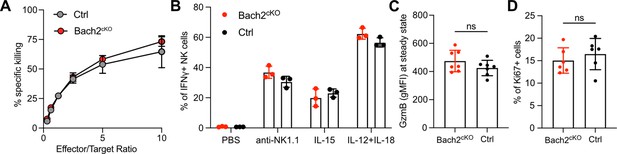
Natural killer (NK) cell effector function was unchanged with BTB domain And CNC Homolog 2 (Bach2) deficiency.
(A) Killing of B16F10 tumor cells by lymphokine-activated killer (LAK) cells from spleen of Bach2cKO mice or control mice. Data are shown as of one representative experiment with three mice per group from three independent experiments. (B) Splenocytes from Bach2cKO mice or control mice were treated with anti-NK1.1, IL-15, or IL-12 plus IL-18 and analyzed for interferon-γ (IFNγ) expression. Summary of IFNγ-positive NK cells is shown. Data are shown as of one representative experiment with three mice per group from three independent experiments. (C) Summary of GzmB expression in lung NK cells from Bach2cKO mice or control mice at steady state is shown (n=7 for each group in three independent experiments). (D) 2×105 B16F10 cells were intravenously injected into Bach2cKO mice or control mice. After 24 hr, cells from the lung were harvested and analyzed for Ki67-positive cells. Summary of Ki67-positive NK cells in the lung from Bach2cKO mice or control mice after tumor inoculation (n=6 or 7 for each group in three independent experiments). Statistical significance was determined by Student’s t test. Error bars indicate SD. ns, not significant.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C56BL/6J, wild type | The Jackson Laboratories | RRID:IMSR_JAX:000664 | |
| Strain, strain background (Mus musculus) | RAG1-/-, wild type | The Jackson Laboratories | RRID:IMSR_JAX:034159 | |
| Strain, strain background (Mus musculus) | Bach2Flag | Tomohiro Kurosaki | ||
| Strain, strain background (Mus musculus) | Bach2fox/flox, flox-Bach2 | Tomohiro Kurosaki | ||
| Strain, strain background (Mus musculus) | NKp46iCre; Ncr1tm1.1(icre)Viv | Eric Vivier | MGI:5308410 | |
| Strain, strain background (Mus musculus) | Bach2-/-, Bach2tm1e | EuComm | Bach2tm1e(EUCOMM)Wtsi | ES clone EPD0689_1_H03, ES line JM8A3.N |
| Cell line (Mus musculus) | B16F10 | ATCC | CRL-6475 | |
| Antibody | Anti-Mouse CD127 PE-Cyanine7, rat monoclonal | eBioscience | Catalog# 25-1271-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD3e FITC, armenian hamster monoclonal | eBioscience | Catalog# 11-0031-85 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD19 FITC, rat monoclonal | eBioscience | Catalog# 11-0193-86 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD49b (Integrin α2) FITC, rat monoclonal | eBioscience | Catalog# 11-5971-85 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse NK1.1 PE-Cyanine7, mouse monoclonal | eBioscience | Catalog# 25-5941-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD27 APC, armenian hamster monoclonal | eBioscience | Catalog# 17-0271-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD11b eFluor 450, rat monoclonal | eBioscience | Catalog# 48-0112-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse NKp46 PerCP-eFluor 710, rat monoclonal | eBioscience | Catalog# 46-3351-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD39 PE, rat monoclonal | eBioscience | Catalog# 12-0391-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse Granzyme B PE, rat monoclonal | eBioscience | Catalog# 12-8898-80 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse IFNγ gamma eFluo450, rat monoclonal | eBioscience | Catalog# 48-7311-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse Ly49A/D PE, rat monoclonal | eBioscience | Catalog# 12-5783-81 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse Ly49D APC, rat monoclonal | eBioscience | Catalog# 17-5782-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse Ly49E/F APC, rat monoclonal | eBioscience | Catalog# 17-5848-80 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse Ly49I FITC, mouse monoclonal | eBioscience | Catalog# 11-5895-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD94 FITC, rat monoclonal | eBioscience | Catalog# 11-0941-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse NKG2A PerCP eFluor 710, mouse monoclonal | eBioscience | Catalog# 46-5897-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse TER-119 Biotin, rat monoclonal | eBioscience | Catalog# 13-5921-85 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD117 APC, rat monoclonal | eBioscience | Catalog# 17-1171-81 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD62L eFluor450, rat monoclonal | eBioscience | Catalog# 48-0621-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD122 eFluor 450, rat monoclonal | eBioscience | Catalog# 48-1222-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD244.2 FITC, mouse monoclonal | BD Pharmingen | Catalog# 553305 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse TCF-7/TCF-1 PE, mouse monoclonal | BD Pharmingen | Catalog# 564217 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse Ly49F PE, mouse monoclonal | BD Pharmingen | Catalog# 550987 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD122 FITC, rat monoclonal | BD Pharmingen | Catalog# 553361 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD69 PE, armenian hamster monoclonal | BD Pharmingen | Catalog# 553237 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse Ly49-G2 APC, rat monoclonal | BD Pharmingen | Catalog# 555316 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD135 Brilliant Violet 421, rat monoclonal | Biolegend | Catalog# 135313 | Flow cytometry (1:100) |
| Antibody | Anti-DYKDDDDK tag APC, rat monoclonal | Biolegend | Catalog# 637307 | Flow cytometry (1:100) |
| Antibody | Anti- DYKDDDDK tag PE, rat monoclonal | Biolegend | Catalog# 637310 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse KLRG1 (MAFA) Biotin, syrian hamster monoclonal | Biolegend | Catalog# 138406 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CX3CR1 PE, mouse monoclonal | Biolegend | Catalog# 149005 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse Ly49H Biotin, mouse monoclonal | In-house | Flow cytometry (1:100) | |
| Antibody | Anti-Mouse Ly49C Biotin, mouse monoclonal | In-house | Flow cytometry (1:100) | |
| Antibody | Anti-Mouse CD3 Biotin, armenian hamster monoclonal | eBioscience | Catalog# 145–2C11 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD19 Biotin, rat monoclonal | eBioscience | Catalog# 13-0193-85 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse NK1.1 Biotin, mouse monoclonal | Biolegend | Catalog# 108704 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD11b Biotin, rat monoclonal | Biolegend | Catalog# 101204 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse TER-119 Biotin, rat monoclonal | eBioscience | Catalog# 13-5921-85 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse Ly-6G and Ly-6C Biotin, rat monoclonal | BD Pharmingen | Catalog# 553125 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse CD135 Biotin, rat monoclonal | BD Horizon | Catalog# 562537 | Flow cytometry (1:100) |
| Antibody | Anti-Human/Mouse IRF8 APC, mouse monoclonal | eBioscience | Catalog# 17-9852-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse Blimp-1 PE, rat monoclonal | eBioscience | Catalog# 12-9850-82 | Flow cytometry (1:100) |
| Antibody | Anti-Mouse Ki-67 eFluor, rat monoclonal | eBioscience | Catalog# 48-5698-82 | Flow cytometry (1:100) |
| Peptide, recombinant protein | Streptavidin PerCP | BD Pharmingen | Catalog# 554064 | Flow cytometry (1:200) |
| Peptide, recombinant protein | Streptavidin PE | BD Pharmingen | Catalog# 554061 | Flow cytometry (1:200) |
| Other | Fixable Viability Dye eFluor 506 | eBioscience | Catalog# 65-0866-14 | Flow cytometry (1:1000) |
| Antibody | Anti-FLAG M2-Peroxidase (HRP) mouse monoclonal | Sigma | Catalog# A8592 | Western blot (1:1000) |
| Antibody | Beta-Actin antibody rabbit polyclonal | Cell Signaling | Catalog# 4967S | Western blot (1:1000) |
| Antibody | Anti-rabbit IgG HRP-linked antibody affinity purified from goat | Cell Signaling | Catalog# 7074S | Western blot (1:1000) |
| Commercial assay or kit | EasySep Mouse NK cell isolation kit | STEMCELL Technologies | Catalog# 19855 | NK cell enrichment |
| Commercial assay or kit | PureLink RNA Mini Kit | Ambion | Catalog# 12183018A | RNA isolation |
| Sequence-based reagent | Tcf7 | IDT DNA | Mm.PT.58.13528979 | Fwd: CTTCAATCTGCTCATGCCCTA Rev: TGTTCGTAGAGTGGAGAAAGC |
| Sequence-based reagent | Gzmb | IDT DNA | Mm.PT.58.42155916 | Fwd: AAGAGAGCAAGGACAACACTC Rev: CATGTCCCCCGATGATCTC |
| Sequence-based reagent | Klrg1 | IDT DNA | Mm.PT.58.30803964 | Fwd: GCTCACATCTCCTTACATTTCCG Rev: TCCTCAAGCCGATCCAGTA |
| Sequence-based reagent | Kit | IDT DNA | Mm.PT.58.33701407 | Fwd: CGGCTAACAAAGGGAAGGAT Rev: GTATAAGTGCCTCCTTCTGTGC |
| Sequence-based reagent | Sell | IDT DNA | Mm.PT.58.13849728 | Fwd: CTTCATTCCTGTAGCCGTCAT Rev: CCATCCTTTCTTGAGATTTCTTGC |
| Sequence-based reagent | CD69 | IDT DNA | Mm.PT.58.32284621 | Fwd: ACGGAAAATAGCTCTTCACATCT Rev: ACCACTATTAACACAGCCCAAG |
| Sequence-based reagent | Klrb1b | IDT DNA | Mm.PT.58.41786742 | Fwd: CTAGCCAGGATCCAAGAACC Rev: CAATCACGACCAGCACAAGA |
| Sequence-based reagent | Bhlhe41 | IDT DNA | Mm.PT.31539347 | Fwd: GGAACATAGGGATTTTATAGGACTGG Rev: GCATTCATTAATTCGGTCTCGTC |
| Sequence-based reagent | Sox6 | IDT DNA | Mm.PT.58.30547031 | Fwd: CCGTACAGTTCATTCCGTCAA Rev: GTCACTTATGCCCTTTAGCCT |
| Sequence-based reagent | Ccr7 | IDT DNA | Mm.PT.58.312575202 | Fwd: GAGACAAGAACCAAAAGCACAG Rev: GGAAAATGACAAGGAGAGCCA |
| Sequence-based reagent | CD39 | IDT DNA | Mm.PT.58.8557349 | Fwd: CGAGAAGGAGAATGACACAGG Rev: GTATCAGTTCGGTGGACAGTTC |
| Sequence-based reagent | Irf8 | IDT DNA | Mm.PT.58.30819027 | Fwd: TGTCTCCCTCTTTAAACTTCCC Rev: GAAGACCATGTTCCGTATCCC |
| Sequence-based reagent | Ccl5 | IDT DNA | Mm.PT.58.43548565 | Fwd: GCTCCAATCTTGCAGTCGT Rev: CCTCTATCCTAGCTCATCTCCA |
| Sequence-based reagent | Cx3cr1 | IDT DNA | Mm.PT.58.17555544 | Fwd: TCCCTTCCCATCTGCTCA Rev: CACAATGTCGCCCAAATAACAG |
| Sequence-based reagent | Prdm1 | IDT DNA | Mm.PT.58.10253822 | Fwd: GAACCTGCTTTTCAAGTATGCTG Rev: TTCCCTTCGGTATGTACTCCT |
| Sequence-based reagent | Actb | IDT DNA | Mm.PT. 39a.22214843.g | Fwd: GATTACTGCTCTGGCTCCTAG Rev: GACTCATCGTACTCCTGCTTG |
| Sequence-based reagent | Bach2 Exon1-2 | IDT DNA | Mm.PT.58.12332872 | Fwd: TGTAGCCTTCTCATCTCTTCCT Rev: ATCCACAGACATGCCGTTC |
| Sequence-based reagent | Bach2 Exon3-4 | IDT DNA | Mm.PT.58.11332004 | Fwd: GAAGCAGACAGTGAGTCGTG Rev: ACTGTTCTGAGGTTAGCTTGTG |
| Sequence-based reagent | Bach2 Exon4-5 | IDT DNA | Mm.PT.58.11289601 | Fwd: GAGTTCATCCACGACATCCG Rev: CAGTTTTTCCTTCTCGCACAC |
| Chemical compound, drug | DNase | Sigma | Catalog# DN25 | |
| Chemical compound, drug | Hyaluronidase | Sigma | Catalog# H2126 | |
| Chemical compound, drug | Liberase | Roche | Catalog# 05401119001 | |
| Peptide, recombinant protein | Recombinant murine IL-15 | PeproTech | Catalog# 210–15 | |
| Peptide, recombinant protein | Recombinant murine IL-12 p70 | PeproTech | Catalog# 200–12 | |
| Peptide, recombinant protein | Recombinant mouse IL-18 | MBL | Catalog# B002-5 | |
| Chemical compound, drug | Calcein-AM | Biolegend | Catalog# 425201 | |
| Software, algorithm | Prism 9 | GraphPad | https://www.graphpad.com/ | |
| Software, algorithm | FlowJo 10 | Treestar | https://www.flowjo.com/ |





