Nr2f1a maintains atrial nkx2.5 expression to repress pacemaker identity within venous atrial cardiomyocytes of zebrafish
Figures
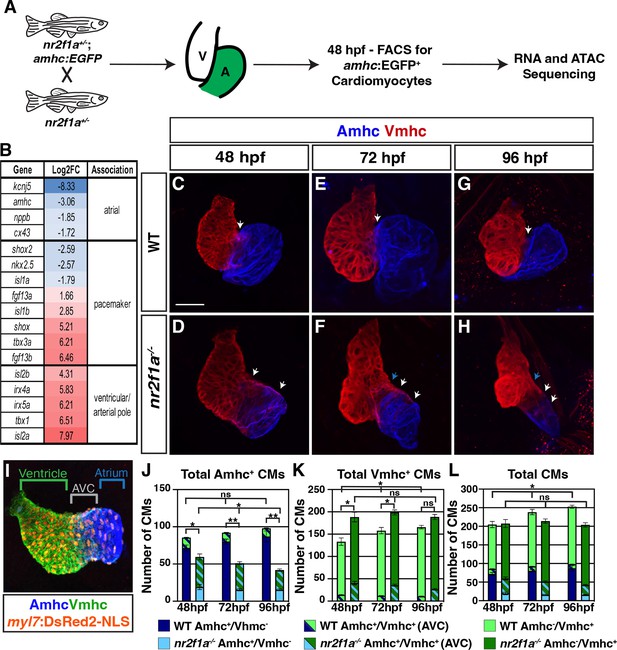
The atrioventricular canal (AVC) resolves to Vmhc-expressing cardiomyocytes in nr2f1a mutant hearts.
(A) Schematic for isolation of atrial cardiomyocytes (ACs) using Tg(amhc:EGFP) transgene for RNA-seq and assay for transpose-accessible chromatin sequencing (ATAC-seq) at 48 hr post-fertilization (hpf). (B) Differential expression of genes associated with ventricular cardiomyocyte (VC)/arterial pole, AC, and pacemaker cardiomyocyte (PC) differentiation in nr2f1a mutants compared to wild-type (WT). (C–H) IHC for Amhc (blue) and Vmhc (red) in nr2f1a and WT hearts at 48, 72, and 96 hpf. White arrows indicate region of overlapping Amhc+ and Vmhc+ cardiomyocytes in the AVC. Blue arrows indicate overt arterial position of the AVC of nr2f1a mutant atria. Number of embryos examined - 48 hpf: WT (n=3), nr2f1a-/- (n=4); 72 hpf: WT (n=4), nr2f1a-/- (n=6); 96 hpf: WT (n=7), nr2f1a-/- (n=10). (I) Schematic for cell quantification: Amhc+/Vmhc- (blue) cardiomyocytes (CMs) mark the atria; Amhc+/Vmhc+ (green/blue) cardiomyocytes mark the AVC; Amhc-/Vmhc+ (green) cardiomyocytes mark the ventricles; DsRed2-NLS (pan-cardiac) marks the cardiomyocyte nuclei. (J–L) Quantification of Amhc+/Vmhc- cardiomyocytes, Amhc+/Vmhc+ (AVC) cardiomyocytes, and Amhc-/Vmhc+ cardiomyocytes. Error bars indicate s.e.m. 48 hpf: WT (n=4), nr2f1a-/- (n=5); 72 hpf: WT (n=5), nr2f1a-/- (n=8); 96 hpf: WT (n=5), nr2f1a-/- (n=12). Scale bar indicates 50 μm. Differences between WT and nr2f1a-/- were analyzed using ANOVA with multiple comparisons. *p=0.05–0.001, **p<0.001.
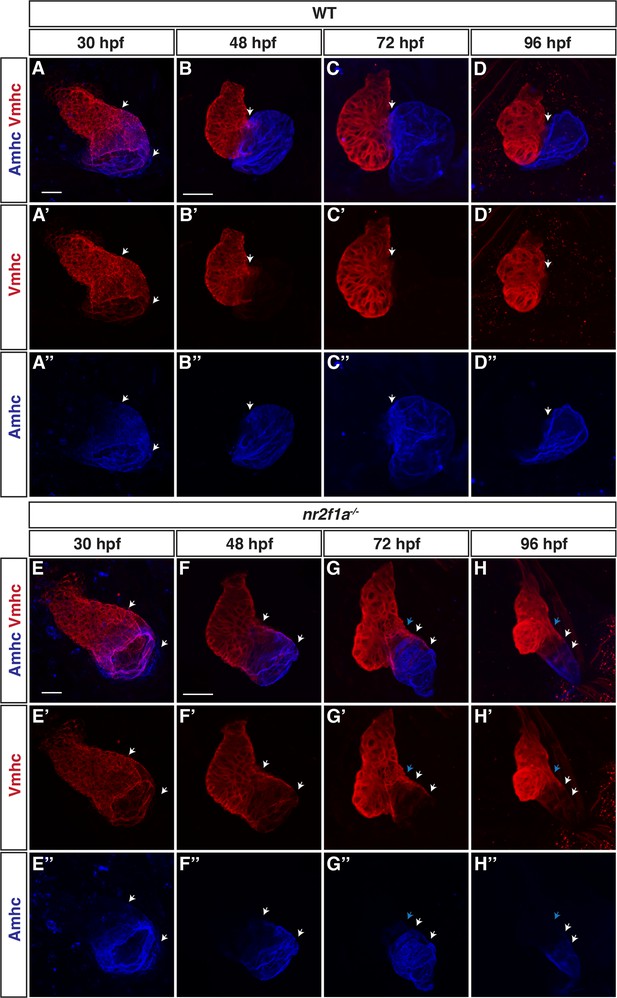
Individual channels showing the resolution of atrioventricular canal (AVC) cardiomyocytes to ventricular cardiomyocyte (VC) identity in nr2f1a mutants.
(A–H’’) IHC for Amhc (blue) and Vmhc (red) at 30, 48, 72, and 96 hr post-fertilization (hpf) in wild-type (WT) and nr2f1a mutant embryos. Forty-eight to 96 hpf images (B–D) and (F–H) are the same as presented in Figure 1C–H. White arrows indicate region of Amhc/Vmhc overlap within the hearts. Blue arrow indicates overt arterial position of AVC in nr2f1a mutant atria. Number of 30 hpf embryos examined: WT (n=20), nr2f1a-/- (n=12). Scale bars indicate 50 µm.
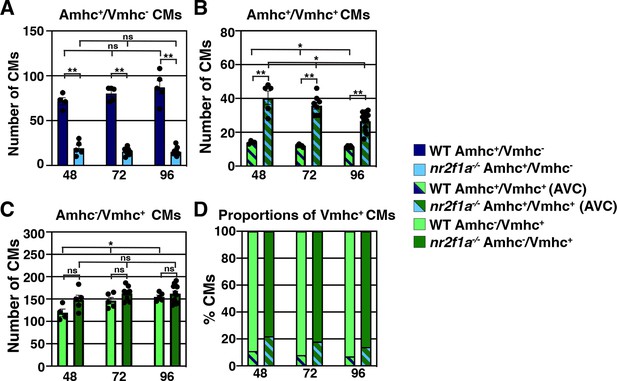
Quantification of cardiomyocytes in nr2f1a mutants.
(A) Quantification of only Amhc+/Vmhc- cardiomyocytes (CMs) within the wild-type (WT) and nr2f1a mutant hearts. (B) Quantification of only Amhc+/Vmhc+ (AVC) cardiomyocytes within the WT and nr2f1a mutant hearts. (C) Quantification of only Amhc-/Vmhc+ cardiomyocytes within the WT and nr2f1a mutant hearts. (D) Proportions of Amhc+/Vmhc+ and Amhc-/Vmhc+ cardiomyocytes in WT and nr2f1a mutant hearts; 48 hpf: WT (n=4), nr2f1a-/- (n=5); 72 hpf: WT (n=5), nr2f1a-/- (n=8); 96 hpf: WT (n=5), nr2f1a-/- (n=12). Graphs are the same data presented in Figure 1J, K and L. Differences between WT and nr2f1a-/- in A–C were analyzed using ANOVA with multiple comparisons. *p=0.05–0.001, **p<0.001.
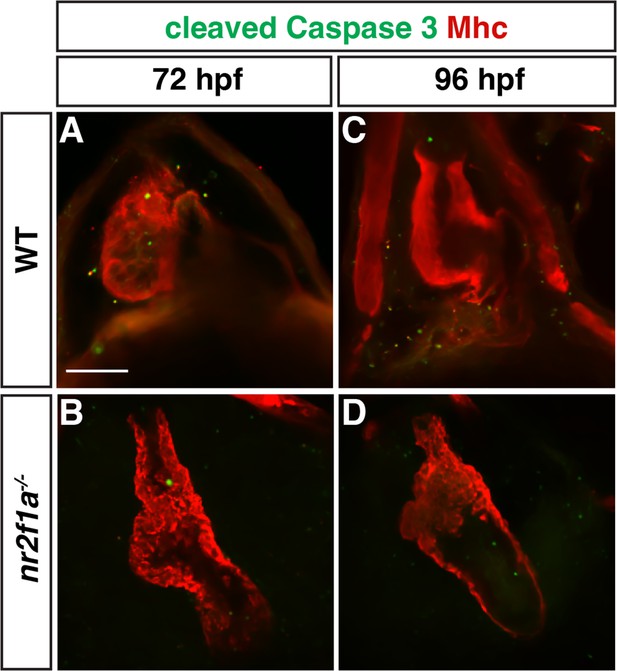
Nr2f1a mutant hearts do not have increased cardiomyocyte death.
(A–D) z-Slices of IHC for cleaved Caspase 3 (green) and Mhc (red) in wild-type (WT) and nr2f1a mutant embryos. The cleaved Caspase 3+ cells shown in the heart were not cardiomyocytes. Number of embryos examined - 72 hr post-fertilization (hpf): WT (n=7), nr2f1a-/- (n=8). 96 hpf: WT (n=9), nr2f1a-/- (n=6). Scale bar indicates 50 µm.
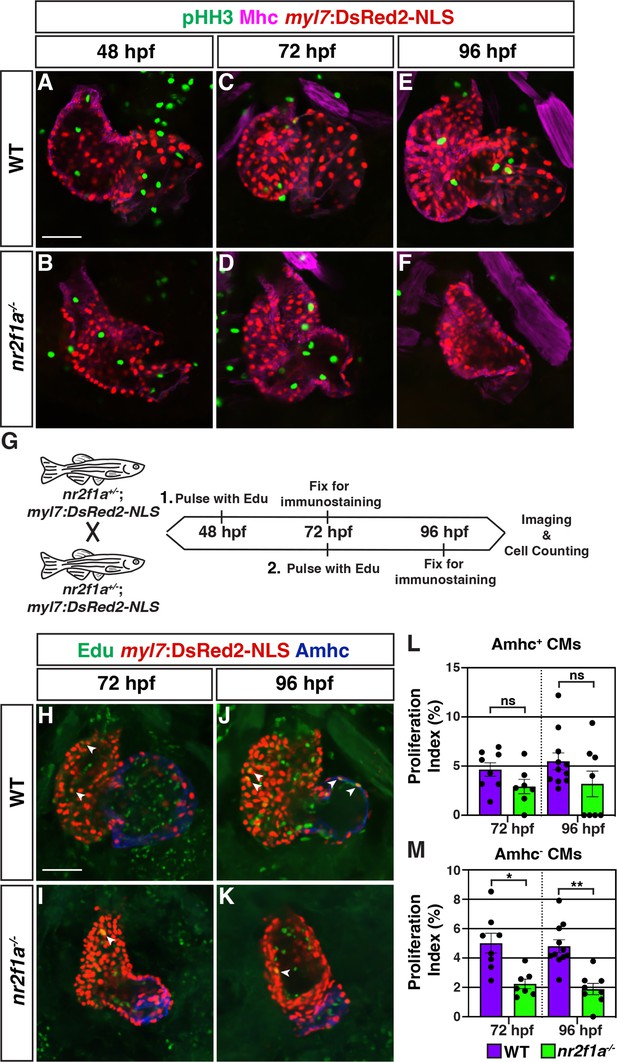
Cardiomyocytes within the nr2f1a mutant atria do not have increased proliferation.
(A–F) z-Slices of IHC for phosphohistone H3 (pHH3) (green), Mhc (pan-cardiac; purple), and myl7:DsRed2-NLS (pan-cardiac; red) in wild-type (WT) and nr2f1a mutant hearts from 48 to 96 hr post-fertilization (hpf). The pHH3+ cells shown in the hearts were not cardiomyocytes. Number of embryos examined - 48 hpf: WT (n=9), nr2f1a-/- (n=7); 72 hpf: WT (n=8), nr2f1a-/- (n=8); 96 hpf: WT (n=9), nr2f1a-/- (n=7). (G) Schematic of the two Edu pulse-chase experiments. (H–K) Representative z-slices of Edu (green) with IHC for myl7:DsRed2-NLS (red) and Amhc (blue). White arrows indicate representative Edu+/myl7:DsRed2-NLS+/Amhc+ and Edu+/myl7:DsRed2-NLS+/Amhc- cardiomyocytes that were proliferating within the hearts. (L,M) Proliferation indices of Amhc+ and Amhc- cardiomyocytes (CMs) in the hearts of WT and nr2f1a mutant embryos; 72 hpf: WT (n=8), nr2f1a-/- (n=7); 96 hpf: WT (n=11), nr2f1a-/- (n=8). Differences between conditions were analyzed using Student’s t-test. *p=0.05–0.001, **p<0.001. Scale bars indicate 50 µm.
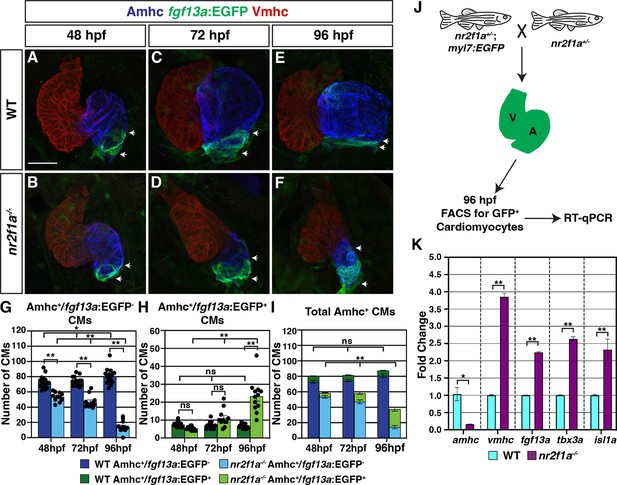
Pacemaker cardiomyocyte (PC) identity expands from the venous pole across the atrium in nr2f1a mutant hearts.
(A–F) IHC for Amhc (blue), Vmhc (red), and fgf13a:EGFP (green). White arrows indicate boundaries of Et(fgf13a:EGFP) expression within the atrium. Number of embryos examined - 48 hr post-fertilization (hpf): wild-type (WT) (n=10), nr2f1a-/- (n=12); 72 hpf: WT (n=10), nr2f1a-/- (n=10); 96 hpf: WT (n=7), nr2f1a-/- (n=10). (G–I) Quantification of Amhc+/fgf13a:EGFP-, Amhc+/fgf13a:EGFP+, and total Amhc+ cardiomyocytes (CMs) using the Tg(myl7:DsRed2-NLS) transgene within the hearts of WT and nr2f1a mutants. Error bars indicate s.e.m.; 48 hpf: WT (n=16), nr2f1a-/- (n=11); 72 hpf: WT (n=14), nr2f1a-/- (n=11); 96 hpf: WT (n=15), nr2f1a-/- (n=12). (J) Schematic for the isolation of cardiomyocytes at 96 hpf using the Tg(myl7:EGFP) transgene. (K) Fold change of marker genes relative to β-actin from real-time quantitative PCR (RT-qPCR) on isolated cardiomyocytes of WT and nr2f1a mutants at 96 hpf. Scale bar indicates 50 μm. Differences between WT and nr2f1a-/- were analyzed using ANOVA with multiple comparisons. *p=0.05–0.001, **p<0.001.
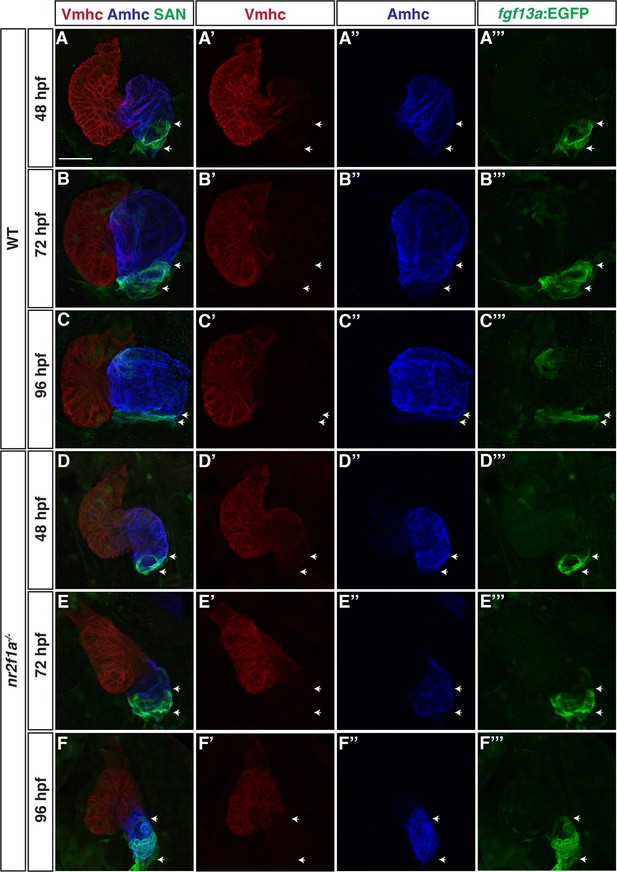
Individual channels showing that pacemaker cardiomyocyte (PC) identity expands from the venous pole across the atrium in nr2f1a mutant hearts.
(A–F’’’) IHC for Vmhc (red), Amhc (blue), and fgf13a:EGFP (green) at 48, 72, and 96 hr post-fertilization (hpf). White arrows indicate boundaries of fgf13a:EGFP expression within the atrium. Images (A–F) are the same as presented in Figure 2A–F. Scale bar indicates 50 µm.
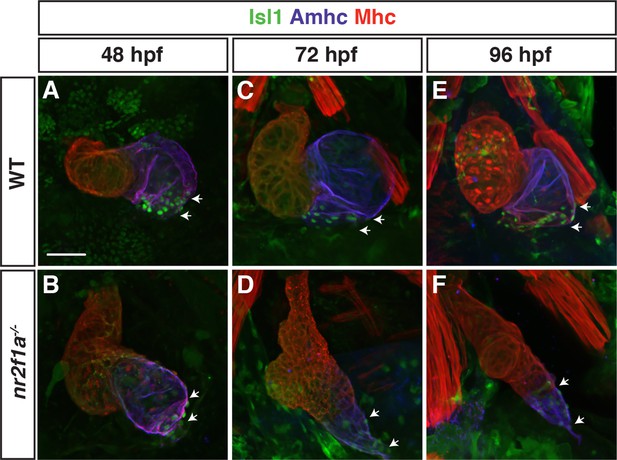
Isl1 expression expands throughout the atrium in nr2f1a mutant embryos.
(A–F) Isl1 (green), Amhc (purple), and Mhc (red) expression in wild-type (WT) and nr2f1a mutant embryos at 48, 72, and 96 hr post-fertilization (hpf). White arrows mark boundaries of Isl1 expression at the venous pole. Number of embryos examined - 48 hpf: WT (n=5), nr2f1a-/- (n=5); 72 hpf: WT (n=5), nr2f1a-/- (n=5); 96 hpf: WT (n=6), nr2f1a-/- (n=10). Scale bar indicates 50 μm.
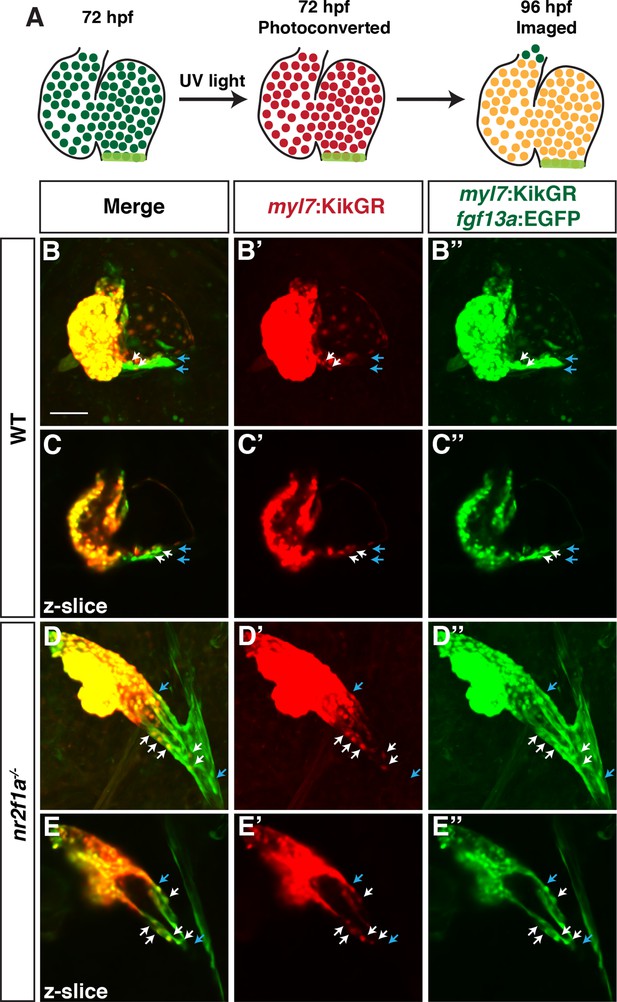
Newly differentiating cardiomyocytes are not added to the atrium in nr2f1a mutant hearts.
(A) Schematic depicting strategy for assessing newly differentiating cardiomyocytes using the Tg(myl7:NLS-KikGR) and Tg(fgf13a:EGFP) transgenes. (B–E”) Wild-type (WT) and nr2f1a mutant embryos photoconverted at 72 hr post-fertilization (hpf) and imaged at 96 hpf. Number of embryos examined - WT (n=6), nr2f1a-/- (n=6). Blue arrows mark boundaries of fgf13a:EGFP expression. White arrows indicate photoconverted (red) cells also expressing fgf13a:EGFP. Scale bar indicates 50 µm.
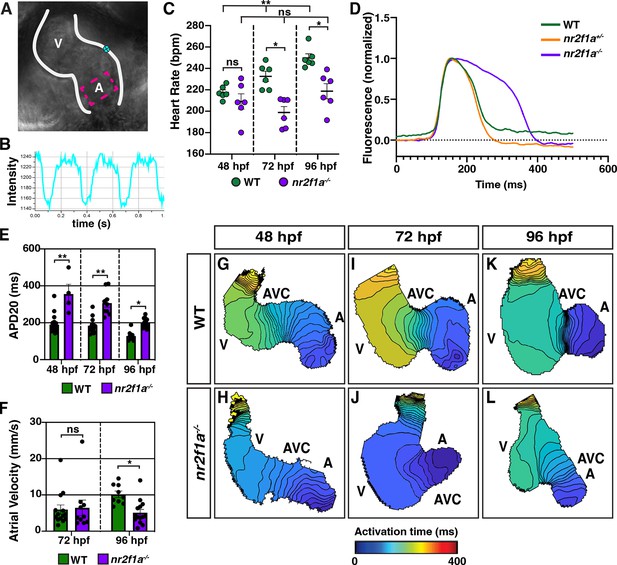
Atrial cardiomyocytes (ACs) in nr2f1a mutants function as pacemaker cardiomyocytes (PCs).
(A) Schematic of point placements to measure heart rate (cyan) and action potential duration at 20% repolarization (APD20) (magenta) in a 48 hr post-fertilization (hpf) heart. (B) Representative kymograph used to analyze heart rate in wild-type (WT) and nr2f1a mutant embryos. (C) Quantification of heart rate in WT and nr2f1a mutant embryos; 48 hpf: WT (n=6), nr2f1a-/- (n=6); 72 hpf: WT (n=6), nr2f1a-/- (n=6); 96 hpf: WT (n=6), nr2f1a-/- (n=6). (D) Representative atrial action potentials of WT, nr2f1a+/-, and nr2f1a-/- embryos at 96 hpf. (E) APD20 in WT and nr2f1a-/- hearts; 48hpf: WT (n=21), nr2f1a-/- (n=4); 72 hpf: WT (n=13), nr2f1a-/- (n=9); 96 hpf: WT (n=9), nr2f1a-/- (n=14). (F) Atrial velocity in WT and nr2f1a-/- hearts; 72 hpf: WT (n=13), nr2f1a-/- (n=9); 96 hpf: WT (n=9), nr2f1a-/- (n=14). (G–L) Representative isochronal maps illustrating the positions of the depolarizing wave front in 5 ms intervals for WT and nr2f1a-/- embryos at 48, 72, and 96 hpf; 48 hpf: WT (n=20), nr2f1a-/- (n=6) 72 hpf: WT (n=12), nr2f1a-/- (n=7) 96 hpf: WT (n=11), nr2f1a-/- (n=14). Differences between WT and nr2f1a-/- analyzed using ANOVA with multiple comparisons. Error bars in all graphs indicate s.e.m. *p=0.05–0.001, **p<0.001.
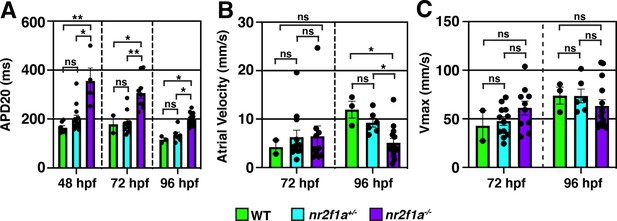
Prolonged repolarization and decreased atrial conduction velocity in nr2f1a mutants compared to wild-type (WT) and heterozygous nr2f1a atria.
(A) Action potential duration at 20% repolarization (APD20) of WT, nr2f1a+/-, and nr2f1a-/- hearts. (B) Atrial velocity of WT, nr2f1a+/-, and nr2f1a-/- hearts. (C) Vmax of WT, nr2f1a+/-, and nr2f1a-/- hearts. Error bars indicate s.e.m.; 48 hr post-fertilization (hpf): WT (n=7), nr2f1a+/- (n=14), nr2f1a-/- (n=4); 72 hpf: WT (n=2), nr2f1a+/- (n=11), nr2f1a-/- (n=9); 96 hpf: WT (n=3), nr2f1a+/- (n=6), nr2f1a-/- (n=14). (A and B) are the same data presented in Figure 3E and F. Differences between WT, nr2f1a+/-, and nr2f1a-/- were analyzed using ANOVA with multiple comparisons. *p=0.05–0.001, **p<0.001.
Heart from 48 hr post-fertilization (hpf) wild-type (WT) embryo.
Heart from 48 hr post-fertilization (hpf) nr2f1a mutant embryo.
Heart from 72 hr post-fertilization (hpf) wild-type (WT) embryo.
Heart from 72 hr post-fertilization (hpf) nr2f1a mutant embryo.
Heart from 96 hr post-fertilization (hpf) wild-type (WT) embryo.
Heart from 96 hr post-fertilization (hpf) nr2f1a mutant embryo.
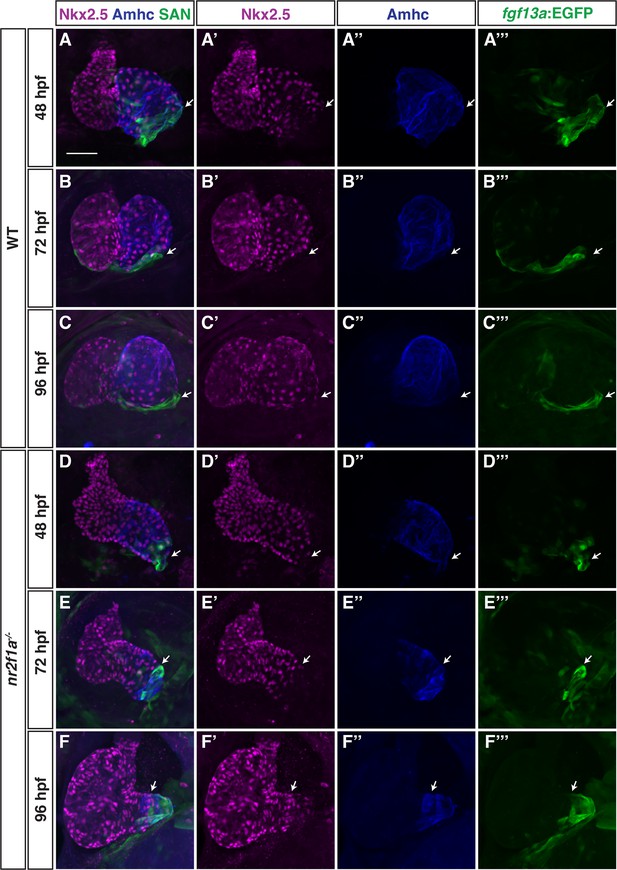
Nkx2.5 expression recedes from venous pole in nr2f1a mutant atria.
(A–F’’’) IHC for Nkx2.5 (purple), Amhc (blue), and fgf13a:EGFP (SAN - green) in wild-type (WT) and nr2f1a mutant embryos from 48 to 96 hr post-fertilization (hpf). White arrows indicate border of Nkx2.5+ and fgf13a:EGFP+ cardiomyocytes. Number of embryos examined - 48 hpf: WT (n=7), nr2f1a-/- (n=6); 72 hpf: WT (n=7), nr2f1a-/- (n=10); 96 hpf: WT (n=14), nr2f1a-/- (n=23). Scale bar indicates 50 μm.
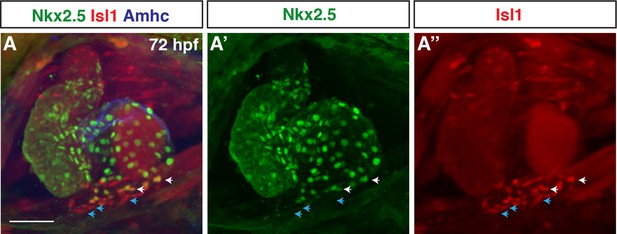
Nkx2.5 is predominantly excluded from pacemaker cardiomyocytes (PCs) at the venous pole of the atrium.
(A–A”) IHC for Nkx2.5 (green), Isl1 (red), and Amhc (blue) in wild-type (WT) hearts at 72 hr post-fertilization (hpf) (n=4). White arrows indicate cardiomyocytes expressing both Nkx2.5 and Isl1. Blue arrows mark cardiomyocytes expressing Isl1 only. Scale bar indicates 50 µm.
Nkx2.5:ZsYellow expression recedes toward the arterial pole of the atrium in nr2f1a mutants.
(A–D’’’) IHC for nkx2.5:ZsYellow (nkx:ZY - purple), Amhc (blue), and fgf13a:EGFP (green) in the atria of wild-type (WT) and nr2f1a mutant embryos. White arrows indicate border of nkx2.5:ZsYellow+ and fgf13a:EGFP+ cardiomyocytes within the atria. Number of embryos examined - 48 hr post-fertilization (hpf): WT (n=8), nr2f1a-/- (n=10); 96 hpf: WT (n=9), nr2f1a-/- (n=12). Scale bar indicates 50 μm.
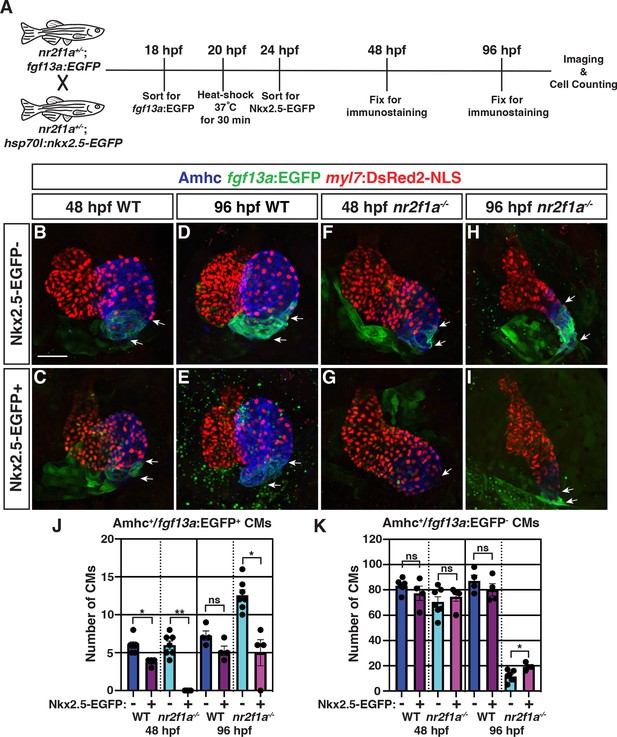
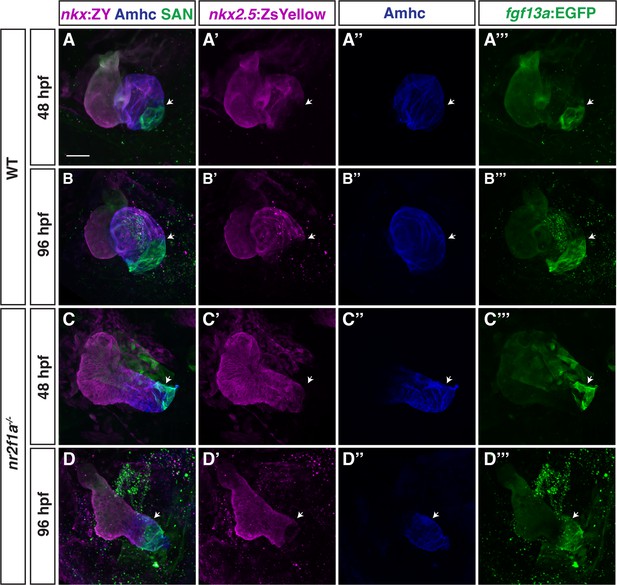
Nkx2.5 induction at 20 hr post-fertilization (hpf) represses pacemaker cardiomyocyte (PC) marker expansion in nr2f1a mutant atria.
(A) Schematic of timeline for heat-shock experiments. (B–I) IHC of representative hearts for Amhc (blue), fgf13a:EGFP (green), and myl7:DsRed2-NLS (red) used to quantify Amhc+/fgf13a:EGFP+ cardiomyocytes (CMs) marking PCs from wild-type (WT) and nr2f1a mutant embryos with and without Nkx2.5-EGFP. White arrows indicate boundaries of Tg(fgf13a:EGFP) expression. Scale bar indicates 50 μm. (J–K) Quantification of Amhc+/fgf13a:EGFP+/myl7:DsRed2-NLS+ (PCs) and Amhc+/fgf13a:EGFP-/myl7:DsRed2-NLS+ cardiomyocytes in the atria with and without Nkx2.5-EGFP induction; 48 hpf WT: Nkx2.5-EGFP- (n=6), Nkx2.5-EGFP+ (n=4); 48 hpf nr2f1a-/-: Nkx2.5-EGFP- (n=7), Nkx2.5-EGFP+ (n=5); 96 hpf WT: Nkx2.5-EGFP- (n=4), Nkx2.5-EGFP+ (n=4); 96 hpf nr2f1a-/-: Nkx2.5-EGFP- (n=7), Nkx2.5-EGFP+ (n=4). Differences between WT and nr2f1a-/- at the different time points were analyzed using Student’s t-test. Error bars indicate s.e.m. *p=0.05–0.001, **p<0.001.
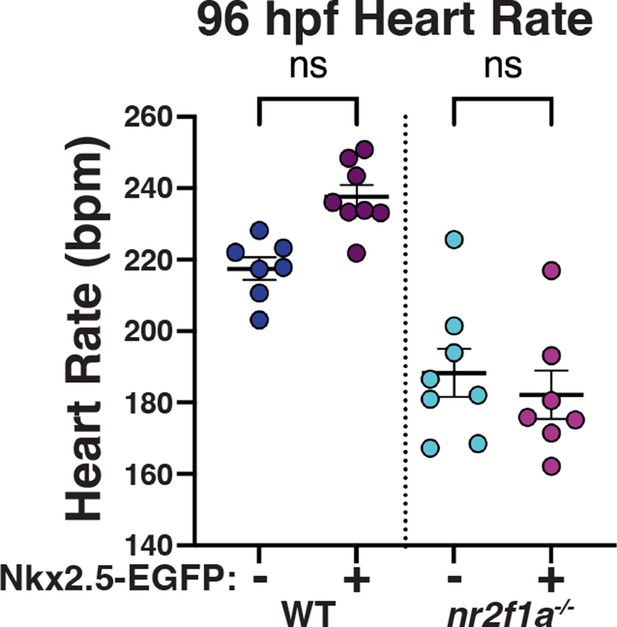
Induction of Nkx2.5 at 20 hr post-fertilization (hpf) does not rescue heart rate.
Quantification of heart rate in wild-type (WT) and nr2f1a mutant embryos at 96 hpf following induction of Nkx2.5 at the 20 somite stage; 96 hpf WT: Nkx2.5-EGFP- (n=7), Nkx2.5-EGFP+ (n=8); 96 hpf nr2f1a-/-: Nkx2.5-EGFP- (n=8), Nkx2.5-EGFP+ (n=7). Analyzed using Student’s t-test.
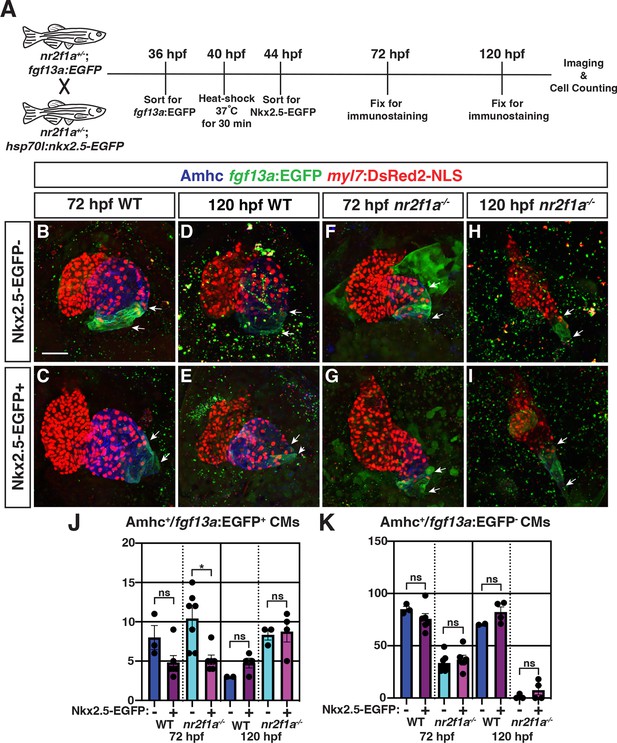
Nkx2.5-EGFP induction at 40 hr post-fertilization (hpf) partially represses the expansion of pacemaker cardiomyocyte (PC) identity.
(A) Schematic of heat-shock experiments. (B–I) IHC for Amhc (blue), fgf13a:EGFP (green), and myl7:DsRed2-NLS (red) of representative hearts from wild-type (WT) and nr2f1a mutant embryos with and without Nkx2.5-EGFP. White arrows indicate boundaries of fgf13a:EGFP expression. Scale bar indicates 50 μm. (J–K) Quantification of Amhc+/fgf13a:EGFP+ cardiomyocytes (CMs) and Amhc+/fgf13a:EGFP- cardiomyocytes using the myl7:DsRed2-NLS transgene with and without Nkx2.5-EGFP induction at 72 and 120 hpf; 72 hpf WT: Nkx2.5-EGFP- (n=3), Nkx2.5-EGFP+ (n=6); 72 hpf nr2f1a-/-: Nkx2.5-EGFP- (n=7), Nkx2.5-EGFP+ (n=6); 120 hpf WT: Nkx2.5-EGFP- (n=2), Nkx2.5-EGFP+ (n=4); 120 hpf nr2f1a-/-: Nkx2.5-EGFP- (n=3), Nkx2.5-EGFP+ (n=4). Differences between WT and nr2f1a-/- were analyzed using Student’s t-test. Error bars indicate s.e.m. *p=0.05–0.001, **p<0.001.
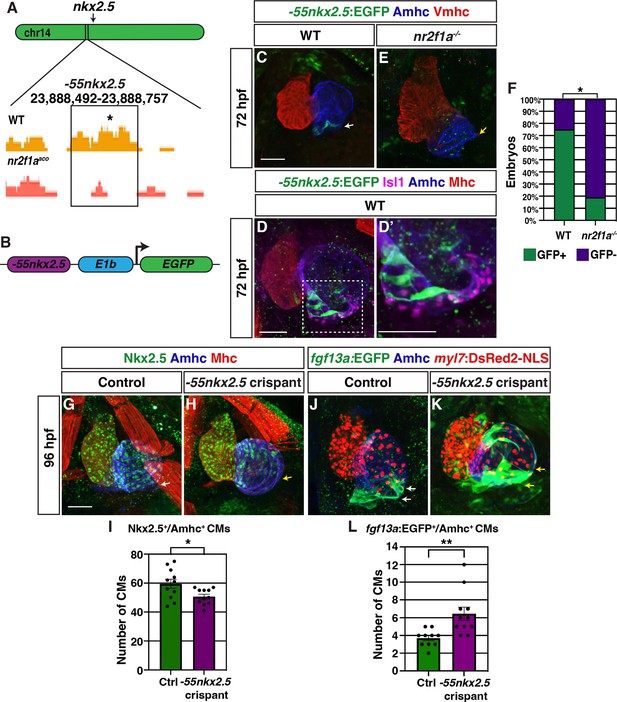
A putative nkx2.5 enhancer is expressed in atrial cardiomyocytes (ACs) adjacent to the sinoatrial node (SAN).
(A) Comparison of assay for transpose-accessible chromatin sequencing (ATAC-seq) peaks from wild-type (WT) and nr2f1a mutant amhc:EGFP+ cardiomyocytes at 48 hr post-fertilization (hpf), ~55 kb upstream of nkx2.5. Asterisk denotes canonical Nr2f binding site. (B) Schematic of –55 kb nkx2.5 enhancer reporter construct. (C) IHC for Tg(–55nkx2.5:EGFP) enhancer (green), Amhc (blue), and Vmhc (red) in hearts at 72 hpf. White arrow indicates –55nkx2.5:EGFP expression. (D–D’) IHC for –55nkx2.5:EGFP (green), Isl1 (purple), Amhc (blue), and sarcomeric myosin heavy chain (Mhc: pan-cardiac - red). The transgenic enhancer reporter is expressed in the ACs directly adjacent to the Isl1+ pacemaker cardiomyocytes (PCs). (E) IHC for Tg(–55nkx2.5:EGFP) enhancer (green), Amhc (blue), and Vmhc (red) in an nr2f1a mutant heart at 72 hpf. Expression of transgenic enhancer reporter is lost in nr2f1a mutants. Yellow arrow indicates venous pole devoid of –55nkx2.5:EGFP expression. (F) Comparison of percentage of WT and nr2f1a mutant embryos with –55nkx2.5:EGFP expression in the heart at 72 hpf WT (n=16); nr2f1a-/- (n=16), using Fisher’s exact test. (G,H) IHC for Nkx2.5 (green), Amhc (purple), and Mhc (red) in uninjected control and –55nkx2.5 crispant embryo hearts at 96 hpf. White arrow in (G) indicates the border of Nkx2.5+ cardiomyocytes extends close to the venous pole. Yellow arrow in (H) indicates that the Nkx2.5+ cardiomyocyte border is located farther from the venous pole of the atrium in crispant embryos. (I) Quantification of Nkx2.5+/Amhc+ cardiomyocytes in uninjected control (n=11) and –55nkx2.5 crispant (n=11) embryos at 96 hpf. (J,K) IHC for fgf13a:EGFP (green), Amhc (blue), and myl7:DsRed2-NLS (red) in uninjected control and –55nkx2.5 crispant Tg(fgf13a:EGFP)+ embryo hearts at 96 hpf. White arrows in (J) indicate the region of fgf13a:EGFP+ cardiomyocytes at the venous pole. Yellow arrows in (K) indicates that the region of fgf13a:EGFP+ cardiomyocytes is expanded from the venous pole in the atrium of crispant embryos. (L) Quantification of fgf13a:EGFP+/Amhc+ cardiomyocytes in uninjected control (n=10) and –55nkx2.5 crispant (n=11) embryo hearts at 96 hpf. Differences between uninjected control and –55nkx2.5 crispants were analyzed using Student’s t-test. Error bars indicate s.e.m. *p=0.05–0.001, **p<0.001. Scale bars in all images indicate 50 μm.
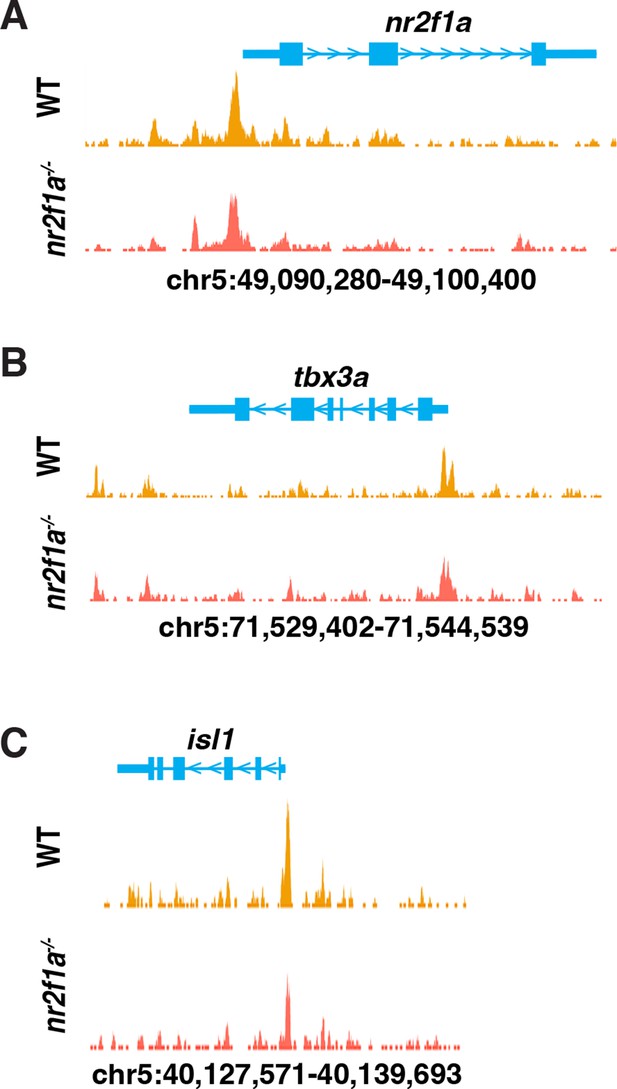
Peaks showing open chromatin surrounding representative loci from the assay for transpose-accessible chromatin sequencing (ATAC-seq) data.
(A–C) Regions of open chromatin using images from the UCSC genome browser were found at the promoters and adjacent to nr2f1a, tbx3a, and isl1 in the sorted amhc:EGFP+ cardiomyocytes from wild-type (WT) and nr2f1a mutant embryos.
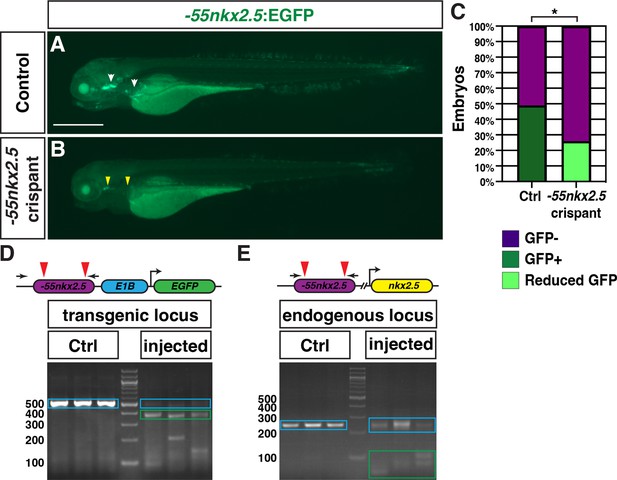
Deletion of the putative nkx2.5 enhancer in crispants.
(A,B) Transgenic Tg(–55nkx2.5:EGFP) control and crispant embryos at 96 hr post-fertilization (hpf). Images are lateral views with anterior to the left and dorsal up. White arrows in (A) indicate expression in dorso-lateral jaw and fin. Yellow arrowheads in (B) indicate reduced expression in the dorso-lateral jaw and loss of expression in the fin in Tg(–55nkx2.5:EGFP) crispants. Scale bar is 500 µm. (C) Comparison of the percentage of Tg(–55nkx2.5:EGFP) uninjected control and crispant embryos with EGFP expression at 72 hpf. Number of Tg(–55nkx2.5:EGFP) embryos examined - uninjected control (n=184); –55nkx2.5 crispant (n=78). *p<0.001 using Fisher’s exact test. Experiments were performed by outcrossing hemizygous Tg(–55nkx2.5:EGFP) to wild-type (WT) embryos. Hence, 50% of the control embryos did not carry the transgene. (D) PCR for the –55nkx2.5:EGFP transgenic locus in individual –55nkx2.5:EGFP uninjected control and crispant embryos. The expected product size for the transgenic –55nkx2.5 enhancer is 512 bp (blue box in controls). The –55nkx2.5 transgenic element is almost completely eliminated (blue box in crispants). Green box indicates deletions. (E) PCR for the endogenous –55nkx2.5 locus in individual uninjected control and crispant embryos. The expected product for the endogenous –55nkx2.5 enhancer from uninjected control (WT) embryos is 246 bp (blue box in controls). The endogenous locus is significantly affected in crispants (blue box in crispants). Green box indicates products showing deletion of the locus. Red arrowheads in D and E indicated location of dgRNA pairs. Black arrows indicate location of the primers. Schema are not drawn to scale.
-
Figure 6—figure supplement 2—source data 1
Uncropped gel pictures of PCR analysis for efficacy of the dgRNA pairs in generating deletions of the transgenic and endogenous –55nkx2.5 loci.
Numbers in the labeled gels indicate individual control uninjected and CRISPR/Cas9-injected embryos. Control uninjected embryos 6–8 and CRISPR/Cas9-injected embryos 1–3 are shown in Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/77408/elife-77408-fig6-figsupp2-data1-v1.zip
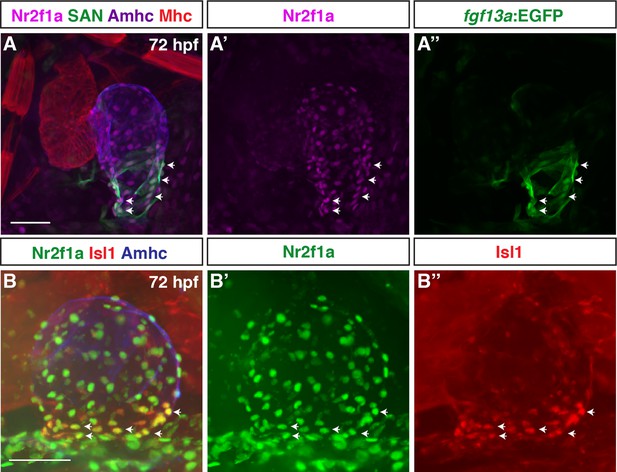
Nr2f1a is expressed throughout cardiomyocytes in the atrium.
(A–A”) IHC for Nr2f1a (purple), fgf13a:EGFP (green), Amhc (blue), and Mhc (red) in wild-type (WT) hearts at 72 hr post-fertilization (hpf) (n=7). White arrows indicate Nr2f1a+ and fgf13a:EGFP+ cardiomyocytes. (B–B”) IHC for Nr2f1a (green), Isl1 (red), and Amhc (blue) in WT hearts at 72 hpf (n=4). White arrows mark Nr2f1a+ and Isl1+. Scale bars indicate 50 µm.
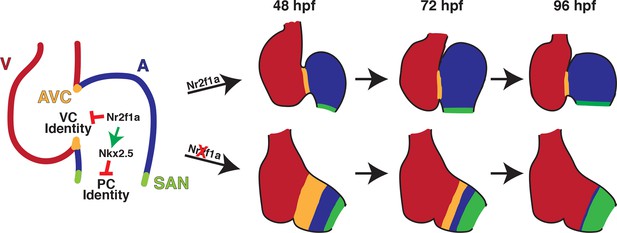
Model summarizing the consequences of Nr2f1a loss on atrial development.
Following its initial requirement promoting atrial cardiomyocyte (AC) differentiation (Duong et al., 2018), Nr2f1a is required to maintain AC identity within the arterial (outflow) and venous (inflow) regions of the zebrafish atrium. In the absence of Nr2f1a, cardiomyocytes within the enlarged atrioventricular canal (AVC), where there is expression of both AC and ventricular cardiomyocyte (VC) differentiation markers, progressively lose expression of AC differentiation markers and only express VC differentiation markers. Concurrently, more venous ACs progressively gain PC differentiation marker expression.
Additional files
-
Supplementary file 1
(xlsx) Sheets with integrated and primary RNA-seq and assay for transpose-accessible chromatin sequencing (ATAC-seq) data with differential gene expression, changes in called peaks, and putative Nr2f binding sites.
- https://cdn.elifesciences.org/articles/77408/elife-77408-supp1-v1.xlsx
-
Supplementary file 2
Antibody information.
- https://cdn.elifesciences.org/articles/77408/elife-77408-supp2-v1.xlsx
-
Supplementary file 3
Primer information.
- https://cdn.elifesciences.org/articles/77408/elife-77408-supp3-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/77408/elife-77408-mdarchecklist1-v1.pdf





