Decoding the IGF1 signaling gene regulatory network behind alveologenesis from a mouse model of bronchopulmonary dysplasia
Figures
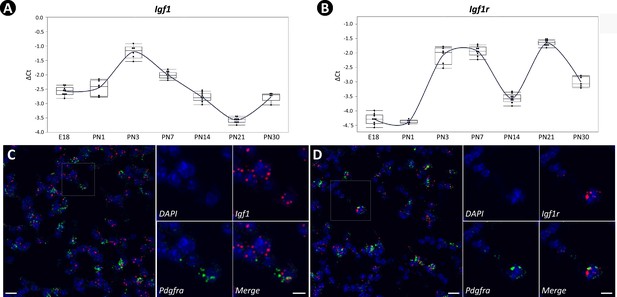
Temporal and spatial expression of Igf1 and Igf1r during neonatal lung development in mice.
(A, B) Temporal expression of Igf1 (A) and Igf1r (B) from embryonic day 18 (E18) to postnatal day 30 (PN30), quantified by RT-PCR and normalized to Gapdh. RNA used was collected from the whole lung, and data was presented as box plots with each stage represented by at least five lungs. (C, D) Spatial localization of mRNA for Igf1 (C) and Igf1r (D) in PN7 lungs as detected by RNAscope and their overlapped expression with Pdgfra. Outlined area on the left is magnified on the right. Scale bars: 20 um under the whole view and 10 um under the magnified view.
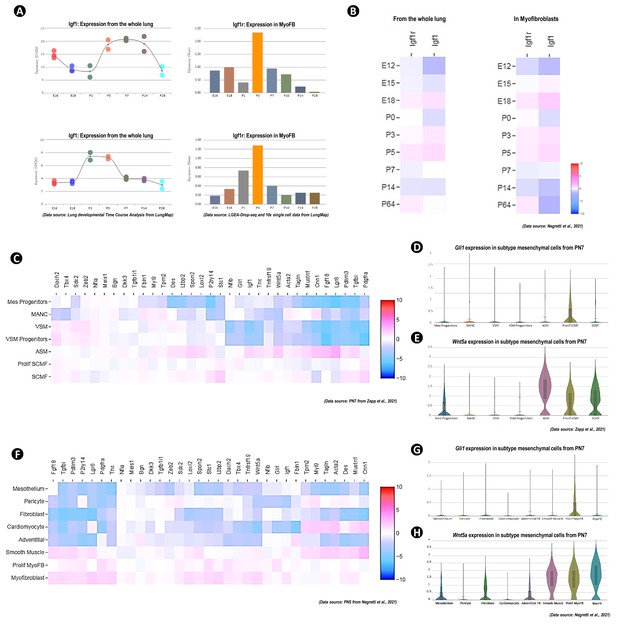
Igf1, Igf1r, and secondary crest myofibroblasts (SCMF)/myofibroblast marker gene’s expression in mouse lung from published public data sources.
(A, B) Expression profiling of Igf1 and Igf1r from whole lung and in SCMF/myofibroblast cells during lung development as calculated on data from LungMap (A) and from Negretti et al., 2021 (B). (C–H) Heatmap and violin plots of collected SCMF/myofibroblast marker gene’s expression within mesenchymal subcell clusters as calculated on data from Zepp et al., 2021 (C–E) and Negretti et al., 2021 (F–H).
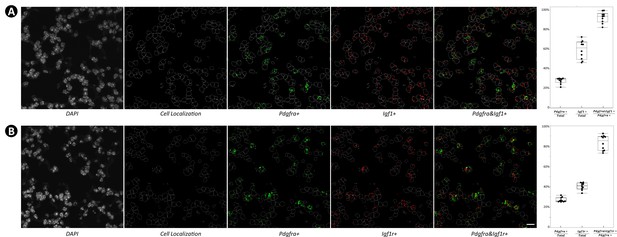
Majority of Pdgfra+ cells are Igf1+ (A) and Igf1r+ (B).
Monochrome 4′,6-diamidino-2-phenylindole (DAPI) staining was imported into ImageJ where the outline of each cell was traced. Under this outline, cellular localization of Pdgfra, Igf1, and Igf1r was analyzed and quantified. A total of 10 images from each group were used, and data was presented as box plots. Scale bar: 20 um for all images.
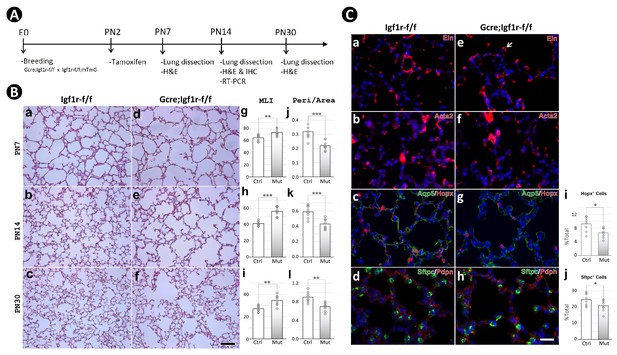
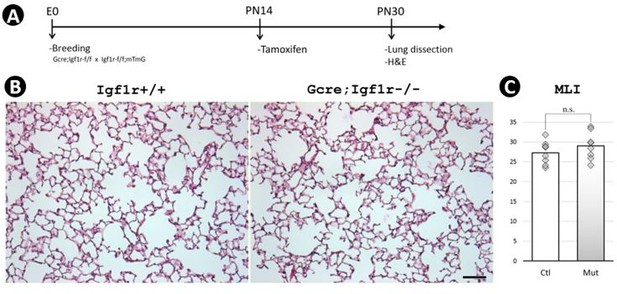
Postnatal inactivation of Igf1r from lung secondary crest myofibroblast cells.
(A) Schematic of the experimental protocol. (B) Hematoxylin and eosin (H&E) staining of lung sections from control (a–c) and Gli1CreERT2 mutant (d-f) mice and their morphometric measurements by mean linear intercept (MLI) (g–i) and peri/area ratio (j–l) at postnatal day 7 (PN7), PN14, and PN30. See Figure 2—figure supplement 1 for the definition and calculation of these indices. (C) Immunostaining of lung sections from control (a–d) and mutant (e–h) mice for elastin (a,e), ACTA2 (b,f), AQP5/HOPX (c,g), and SFTPC/PDPN (d,h), and the comparison of the number of AT1 (i) and AT2 (j) cells between the control and mutant. Quantitative data was presented as mean values +/-SD with data for each experiemntal group collected from eight lobes from four different lungs. p-Value: * stands for 0.05–0.01, ** for 0.01–0.001, *** for <0.001, n.s. for not significant. The same designation is used throughout the paper. Scale bar: 100 um in B and 25 um in C.
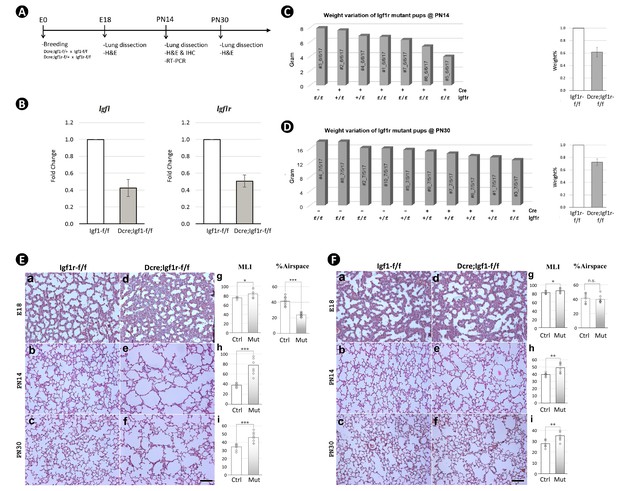
Mesoderm constitutive deletion of Igf1 and Igf1r from mouse lung.
(A) Schematic of the experimental protocol. (B) Impact of inactivation of Igf1 (left) and Igf1r (right) by Twist2cre on their respective transcripts in postnatal day 14 (PN14) lungs. Data is represented as mean ± SD of three lungs from each experimental group. (C) Body weight variation of pups at PN14 of one single litter from the breeding of Twist2cre;Igf1rflox/- and Igf1rflox/flox (left graph). Body weight comparison of pups between the control and the homozygous mutant (right graph). Data is represented as mean ± SD of four mice from each experimental group. (D) Same experiment was done in C for pups at PN30. (E) H&E staining of lung sections from Igf1r control (a–c) and Twist2cre mutant (d-f) mice and their morphometric measurements (mean linear intercept [MLI] and %Airspace – see Figure 2—figure supplement 2E,F for more information) (g–i) at embryonic day 18 (E18), PN14, and PN30. (F) H&E staining of lung sections from Igf1 control (a–c) and Twist2cre mutant (d-f) mice and their morphometric measurements (g–i) at E18, PN14, and PN30. Quantitative data was presented as mean values +/-SD with each experiemntal group in B represented by three lungs, in C, D by four pups, and in E, F by four lungs. p-Value: * stands for 0.05–0.001, ** for 0.01–0.001, *** for <0.001, n.s. for not significant. The same designation is used throughout the paper. Scale bar: 100 um for all images.
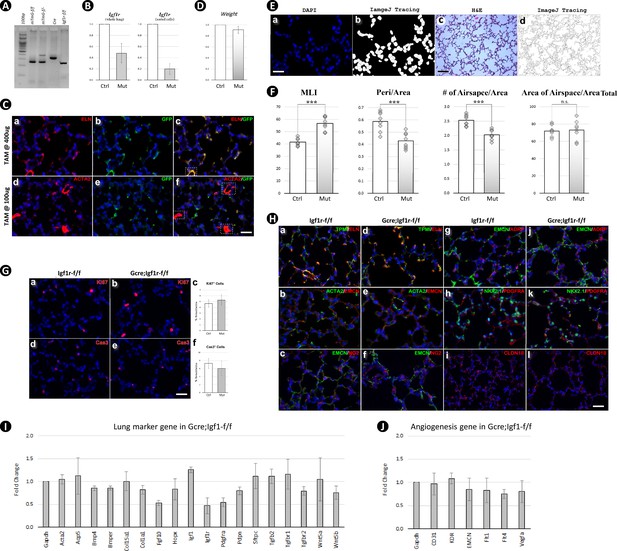
Postnatal inactivation of Igf1r in secondary crest myofibroblasts.
(A) Genotyping of pups from the breeding between Gli1CreERT2;Igf1rflox/flox and Igf1rflox/flox;Rosa26mTmG mice. (B) Igf1r expression in control and mutant lungs at postnatal day 14 (PN14) using whole lung (left) and fluorescence-activated cell sorting GFP+ cells (right). Data is represented as mean ± SD of three lungs from each experimental group. (C) Immunostaining for ELN, ACTA2, and green fluorescent protein (GFP) on PN14 lung sections from Gli1CreERT2;Rosa26mTmG mice, which were tamoxifen-treated at PN2 with 400 ug (a–c) and 100 ug (d-f) per pup. Smooth muscle fibers (outlined in c&f) were labeled by GFP at higher but not at lower tamoxifen (TAM) dosage. Scale bar: 25 um for all images. (D) Weight comparison between Ctrl and Mut at PN14. Data is represented as mean ± SD of three mice from each experimental group. (E) A sample of how to use ImageJ on cell counting (a,b) and morphometric measurements (c,d) where number of separated airspaces (N), perimeter (Peri), and area (Area) of each airspace can be calculated. Scale bar: 25 um in (a) and 100 um in c. (F) Morphometric comparison between Ctrl and Mut at PN14 using different indexes. Mean linear intercept (MLI) is defined as in Crowley et al., 2019, and the others are quantified as below: , , %Airspace=(area of airspaces/area total) /(area total). Data is represented as mean ± SD of four lungs from each experimental group. (G) Immunostaining for Ki67 (a,b) and cleaved Caspase-3 (d,e) on control and mutant lungs at PN14. The percentage of cells identified from above staining was compared between the control and mutant (c,f). Data is represented as mean ± SD of three lungs from each experimental group. Scale bar: 25 um for all images. (H) Immunostaining for mesenchymal marker ELN (a,d), TPM1 (a,d), ACTA2 (b,e), PDGFRA (h,k), epithelial marker NKX2.1 (h,k), CLDN18 (i,l), endothelial marker EMCN (b,c,e,f,g,j), lipofibroblast marker ADRP (g,j), and pericyte marker NG2 (c,f) using control and mutant lungs at PN14. Scale bar: 50 um for i, l and 25 um for all the other images. (I) Comparative expression of selected lung marker genes between control and mutant lungs at PN14. Data is represented as mean ± SD of three lungs from each experimental group. (J) Comparative expression of angiogenesis-related genes between control and mutant lungs at PN14. Data is represented as mean ± SD of three lungs from each experimental group. DAPI: 4′,6-diamidino-2-phenylindole.
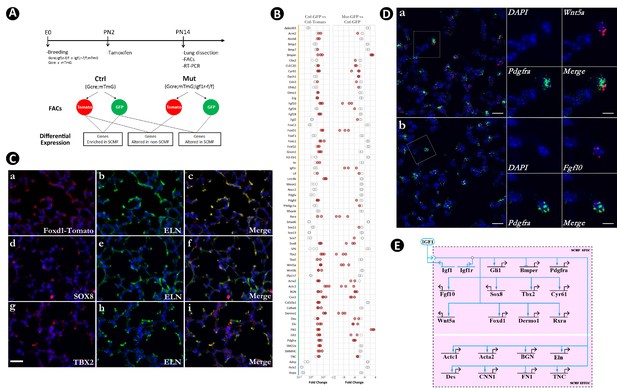
Identification of secondary crest myofibroblasts (SCMF) genes altered in Gli1CreERT2;Igf1rflox/flox mutant lungs.
(A) Schematic of the experimental protocol. (B) RT-PCR data from the selected genes displaying their enrichment in SCMF and alterations in the mutant. Genes marked with the orange line: regulatory genes selected from LungMap database; genes with the green line: common SCMF markers; genes with the blue line: non-SCMF genes. Data was presnted as dot plots with the measurements from three lungs. Red circles: data points meeting the cutoff criteria described in the text; empty circles: data points failing the cutoff criteria. This designation is used throughout the manuscript. (C) Spatial expression of Foxd1-Tomato/ELN (a–c), SOX8/ELN (d-f), and TBX2/ELN (g–i) in the alveolar compartment of postnatal day 14 (PN14) lungs as detected by immunostaining. Specific antibodies were used for SOX8, TBX2, and ELN. RFP antibody was used for Foxd1-Tomato on lungs dissected from Foxd1GFPCreERT2;CAGTomato mice. Scale bar: 25 um for all images. (D) Spatial expression of Wnt5a/Pdgfra (a) and Fgf10/Pdgfra (b) in the alveolar compartment of PN14 lungs as detected by RNAscope. Outlined area on the left is magnified on the right. Scale bars: 20 um under the whole view and 10 um under the magnified view. (E) Biotapestry network illustration of altered SCMF genes and their connections to IGF1 signaling within SCMF. The source of IGF1 can be both autocrine and paracrine. Genes (nodes) are shown in the territories (colored boxes) in which they are expressed. Edges show regulation by the originating upstream factor and are either positive (arrow) or repressive (bar). Signalings across cell membranes are indicated as double arrow heads. DAPI: 4′,6-diamidino-2-phenylindole; FACS: fluorescence-activated cell sorting.
-
Figure 3—source data 1
Table of genes with their RT-PCR data showing their level of expression in secondary crest myofibroblasts (SCMF), enrichment in SCMF, and differential expression between control and mutant lungs, and annotation of their cellular expression in the lung based on LungMAP scRNAseq data.
Cells in red: data points which meet the cutoff criteria designated in the paper or the cell type where the gene is annotated as expressing.
- https://cdn.elifesciences.org/articles/77522/elife-77522-fig3-data1-v3.xlsx
-
Figure 3—source data 2
List of the primers used in the paper.
Primers are for Mus musculus by default and for Homo sapiens when denoted with Hs.
- https://cdn.elifesciences.org/articles/77522/elife-77522-fig3-data2-v3.xlsx
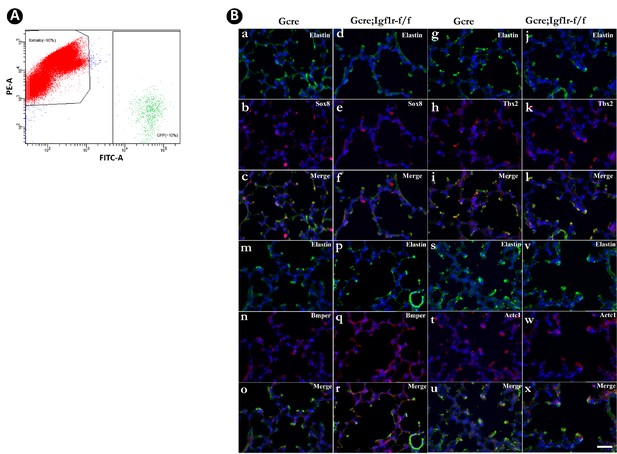
Identification of secondary crest myofibroblasts (SCMF) genes altered in Gli1CreERT2;Igf1rflox/flox mutant lung.
(A) GFP + and Tomato + cells are separated by fluorescence-activated cell sorting. (B) Spatial expression of SOX8/ELN (a–f), TBX2/ELN (g–l), BMPER/ELN (m–r), and ACTC1/ELN (s–x) in the alveolar compartment in control and mutant lungs at postnatal day 14 as detected by immunostaining. Scale bar: 25 um for all images.
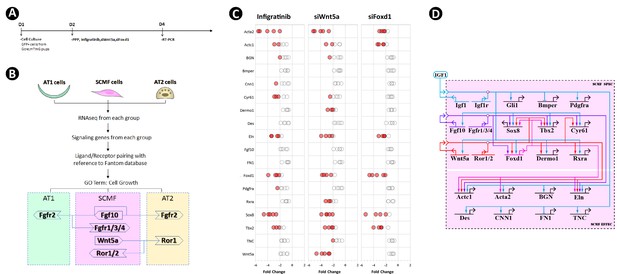
Crossregulation of the altered secondary crest myofibroblasts (SCMF) genes.
(A) Schematic of the experimental protocol. (B) Flow chart of the secretome-receptome analysis among SCMF, AT1, and AT2 cells. The ligands and receptors were identified from the following RNAseq datasets: GSE126457 for SCMF (Li et al., 2019), GSE182886 for AT2, and GSE106960 for AT1 (Wang et al., 2018b). (C) RT-PCR data from the altered SCMF genes demonstrating their response to the treatments by Infigratinib, siWnt5a, and siFoxd1. Data was presnted as dot plots with the measurements from five treatments. (D) Biotapestry network illustration of the crossregulation among the altered SCMF genes.
-
Figure 4—source data 1
List of the ligands and receptors used for secretome-receptome analysis.
- https://cdn.elifesciences.org/articles/77522/elife-77522-fig4-data1-v3.xlsx
-
Figure 4—source data 2
List of the inhibitors and their concentrations used in cell culture treatment.
- https://cdn.elifesciences.org/articles/77522/elife-77522-fig4-data2-v3.xlsx
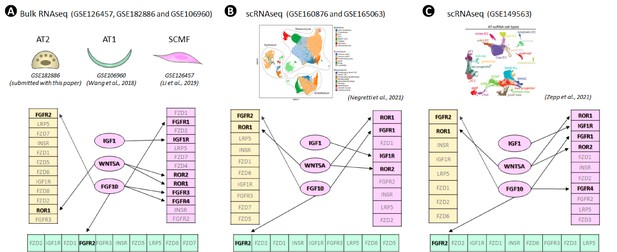
Secretome-receptome analysis of cellular communications between IGF1, WNT5A, and FGF10 ligands from secondary crest myofibroblasts (SCMF) and receptors from SCMF, AT2, and AT1 as calculated on different data sources: bulk RNAseq data.
(A), scRNAseq data from Negretti et al., 2021 (B), and scRNAseq data from Zepp et al., 2021 (C). Receptors are rendered as magenta for those from SCMF, yellow for AT2, and cyan for AT1 and are arranged based on their abundance within each cell type with higher quantities on top/left and lower quantities on bottom/right. Ligands are connected to the receptors predicted to be restrictive and decisive for the function of the respective signaling (labeled in black). Other receptors (labeled in gray) are ignored at this time due to the following: (1) their lower abundance—only the top 150 highly expressed receptors from each cell type are considered; (2) less binding affinity—IGF1R vs INSR; (3) broad expression—LRP5 and FZD proteins are ubiquitously expressed across all cell types studied here; (4) known gene spatial expression—Fgfr2 is specifically expressed in alveolar epithelium. See Figure 4—source data 1 for the full list of these signaling genes selected for each cell type and from each dataset.
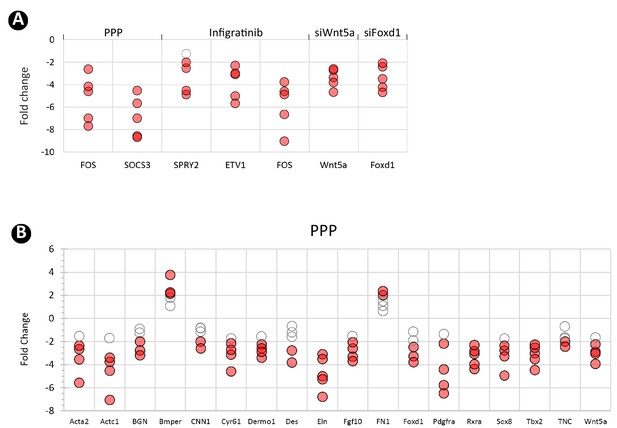
Secondary crest myofibroblasts (SCMF) cell culture treatments.
(A) RT-PCR data showing the effect of various treatments (top) on their respective targeted genes (bottom). (B) RT-PCR data from the altered SCMF genes demonstrating their response to the treatment from picropodophyllin (PPP). Data was presnted as dot plots with the measurements from five treatments.
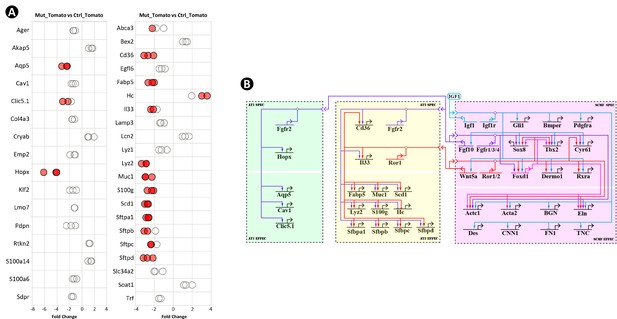
Epithelial genes affected in Gli1CreERT2;Igf1rflox/flox mutant lung and their connections to IGF1 signaling from secondary crest myofibroblasts (SCMF).
(A) RT-PCR data from selected AT1 and AT2 genes revealing their alteration in the mutant lung. Data was presnted as dot plots with the measurements from three lungs. (B) The IGF1 signaling gene regulatory networks during alveologenesis consisting of genes downstream of IGF1 signaling from three cell types (SCMF, AT2, and AT1) and the intracellular and intercellular regulatory connections among them.
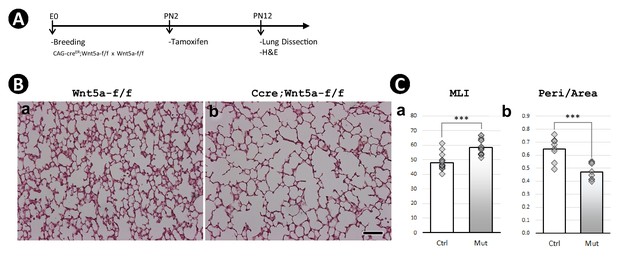
Postnatal inactivation of Wnt5a.
(A) Schematic of the experimental protocol. (B) H&E staining of lung sections obtained from control (a) and Wnt5a−/− (b) mice as generated and described in Li et al., 2020 and analyzed for this study. (C) Morphometric measurements by mean linear intercept (MLI) (c) and peri/area ratio (d). Data was presented as mean values +/-SD of four lungs. Scale bar: 100 um for all images.
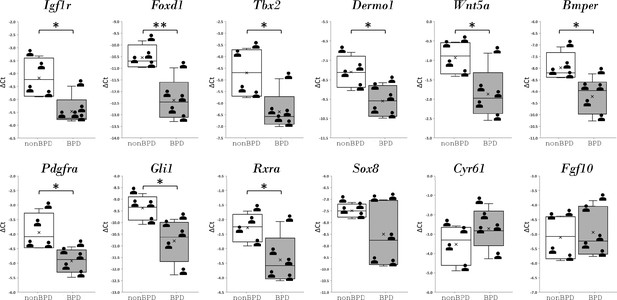
Regulatory genes from IGF1 signaling gene regulatory networks and their expression in human bronchopulmonary dysplasia (BPD) lungs.
Data was presnted as box plots with nonBPD represnted by three lungs and BPD by four lungs.
-
Figure 6—source data 1
List of clinical data for human neonatal lung samples.
- https://cdn.elifesciences.org/articles/77522/elife-77522-fig6-data1-v3.xlsx
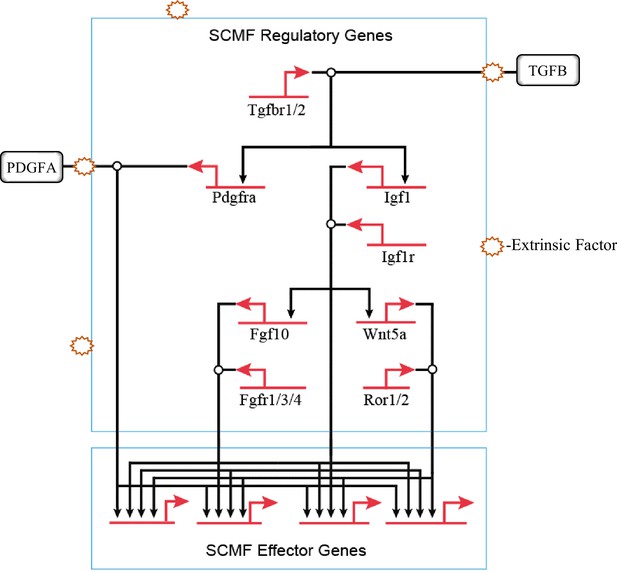
The hierarchical connections among the signaling pathways within secondary crest myofibroblasts (SCMF) during alveologenesis.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-ACTA2 (rabbit polyclonal) | Abcam | Cat#: AB5694 | IF(1:300) |
| Antibody | Anti-ACTC1 (mouse monoclonal) | Santa Cruz | Cat#: SC-58670 | IF(1:100) |
| Antibody | Anti-ADRP (rabbit monoclonal) | Abcam | Cat#: AB108323 | IF(1:200) |
| Antibody | Anti-AQP5 (rabbit polyclonal) | Alomone | Cat#: AQP-005 | IF(1:100) |
| Antibody | Anti-BMPER (mouse monoclonal) | Santa Cruz | Cat#: SC-377502 | IF(1:200) |
| Antibody | Anti-CAS3 (rabbit monoclonal) | Cell Signaling | Cat#: 9664 | IF(1:100) |
| Antibody | Anti-ELN (rabbit polyclonal) | Abcam | Cat#: AB21600 | IF(1:200) |
| Antibody | Anti-EMCN (mouse polyclonal) | R&D Systems | Cat#: AF4666 | IF(1:100) |
| Antibody | Anti-GFP (mouse monoclonal) | Santa Cruz | Cat#: SC-9996 | IF(1:100) |
| Antibody | Anti-HOPX (rabbit polyclonal) | Santa Cruz | Cat#: SC-30216 | IF(1:50) |
| Antibody | Anti-KI67 (mouse polyclonal) | R&D Systems | Cat#: AF7649 | IF(1:50) |
| Antibody | Anti-NKX2.1 (mouse monoclonal) | Seven Hills | Cat#: 8G7G3-1 | IF(1:50) |
| Antibody | Anti-NG2 (rabbit polyclonal) | Abcam | Cat#: AB5320 | IF(1:100) |
| Antibody | Anti-PDGFRa (rabbit monoclonal) | Cell Signaling | Cat#: 3174 | IF(1:50) |
| Antibody | Anti-PDPN (hamster monoclonal) | Thermo Fisher | Cat#: 14-5381-82 | IF(1:300) |
| Antibody | Anti-RFP (rabbit polyclonal) | Rockland | Cat#: 600-401-379S | IF(1:300) |
| Antibody | Anti-SFTPC (rabbit polyclonal) | Abcam | Cat#: AB3786 | IF(1:200) |
| Antibody | Anti-SOX8 (mouse monoclonal) | Santa Cruz | Cat#: SC-374446 | IF(1:50) |
| Antibody | Anti-TBX2 (mouse monoclonal) | Santa Cruz | Cat#: SC-514291 | IF(1:50) |
| Antibody | Anti-TPM1 (mouse monoclonal) | Sigma Aldrich | Cat#: T2780 | IF(1:500) |
| Chemical compound, drug | Tamoxifen | Sigma | Cat#: T5648 | 8 mg/ml |
| Chemical compound, drug | Infigratinib | Selleckchem | Cat#: S2788 | 1 uM |
| Chemical compound, drug | PPP | Selleckchem | Cat#: S7668 | 4 uM |
| Commercial assay, kit | Next Ultra DNA Library Prep Kit | New England Biolabs | Cat#: E7370 | |
| Commercial assay, kit | RNAscope Multiplex Fluorescent Reagent Kit V2 | Advanced Cell Diagnostics | Cat#: 323100 | |
| Sequence-based reagent | siFoxd1: siRNA to Foxd1(SMARTpool) | Dharmacon | Cat#: L-046204-00-0005 | 20 nM |
| Sequence-based reagent | siWnt5a: siRNA to Wnt5a(SMARTpool) | Dharmacon | Cat#: L-065584-01-0005 | 20 nM |
| Sequence-based reagent | siRNA non-targeting control | Dharmacon | Cat#: D-001810-01-05 | 20 nM |
| Sequence-based reagent | RNAscope probe: Igf1 | Advanced Cell Diagnostics | Cat#: 443901-C1 | 1:750 |
| Sequence-based reagent | RNAscope probe: Igf1r | Advanced Cell Diagnostics | Cat#: 417561-C3 | 1:500 |
| Sequence-based reagent | RNAscope probe: Pdgfra | Advanced Cell Diagnostics | Cat#: 480661-C2 | 1:750 |
| Sequence-based reagent | RNAscope probe: Fgf10 | Advanced Cell Diagnostics | Cat#: 446371-C1 | 1:750 |
| Sequence-based reagent | RNAscope probe: Wnt5a | Advanced Cell Diagnostics | Cat#: 316791-C3 | 1:500 |
| Sequence-based reagent | RNAscope 3-plex Positive Control Probe | Advanced Cell Diagnostics | Cat#: 320881 | 1:1500 |
| Sequence-based reagent | RNAscope 3-plex Negative Control Probe | Advanced Cell Diagnostics | Cat#: 320871 | 1:1500 |
| Strain, strain background (Mus musculus) | Rosa26mTmG | The Jackson Laboratory | Cat#: 007676 | |
| Strain, strain background (M. musculus) | CAGTomato | The Jackson Laboratory | Cat#: 007914 | |
| Strain, strain background (M. musculus) | Twist2Cre | The Jackson Laboratory | Cat#: 008712 | |
| Strain, strain background (M. musculus) | Igf1flox/flox | The Jackson Laboratory | Cat#: 016831 | |
| Strain, strain background (M. musculus) | Igf1rflox/flox | The Jackson Laboratory | Cat#: 012251 | |
| Strain, strain background (M. musculus) | CAGCreER | The Jackson Laboratory | Cat#: 004453 | |
| Strain, strain background (M. musculus) | Wnt5aflox/flox | Kuruvilla Laboratory | N/A | |
| Strain, strain background (M. musculus) | Foxd1GFPCreERT2 | McMahon Laboratory | N/A | |
| Strain, strain background (M. musculus) | Gli1CreERT2 | The Jackson Laboratory | Cat#: 007913 | |
| Software, algorithm | Image J | NIH | https://imagej.nih.gov/ij/ | |
| Software, algorithm | STAR 2.5 | Dobin et al., 2013 | PMCID:PMC3530905; https://github.com/alexdobin/STAR; Dobin, 2022 | |
| Software, algorithm | JMP pro 15 | Statistical Discovery | https://www.jmp.com/en_us/software/predictive-analytics-software.html | |
| Software, algorithm | Fantom5 Cell Connectome | FANTOM5 project | https://fantom.gsc.riken.jp/5/suppl/Ramilowski_et_al_2015/vis/#/hive | |
| Software, algorithm | Imaris | BitPlane | http://www.bitplane.com/imaris/imaris | |
| Software, algorithm | R 3.2 | R Project | https://www.r-project.org/ | |
| Software, algorithm | LAS X | Leica | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ | |
| Software, algorithm | BioTapestry | Institute for Systems Biology | http://www.biotapestry.org/ |